We identify two new binding partners for the OCRL1 protein that is mutated in Lowe syndrome and type 2 Dent disease, which we call IPIP27A and B. The IPIPs are required for receptor recycling in the endocytic pathway, suggesting that defects in this process lead to the aforementioned disorders.
Abstract
Mutation of the inositol polyphosphate 5-phosphatase OCRL1 results in two disorders in humans, namely Lowe syndrome (characterized by ocular, nervous system, and renal defects) and type 2 Dent disease (in which only the renal symptoms are evident). The disease mechanisms of these syndromes are poorly understood. Here we identify two novel OCRL1-binding proteins, termed inositol polyphosphate phosphatase interacting protein of 27 kDa (IPIP27)A and B (also known as Ses1 and 2), that also bind the related 5-phosphatase Inpp5b. The IPIPs bind to the C-terminal region of these phosphatases via a conserved motif similar to that found in the signaling protein APPL1. IPIP27A and B, which form homo- and heterodimers, localize to early and recycling endosomes and the trans-Golgi network (TGN). The IPIPs are required for receptor recycling from endosomes, both to the TGN and to the plasma membrane. Our results identify IPIP27A and B as key players in endocytic trafficking and strongly suggest that defects in this process are responsible for the pathology of Lowe syndrome and Dent disease.
INTRODUCTION
Phosphoinositide (PI) lipids play a key role in many cellular processes, including intracellular signaling, actin dynamics, and membrane traffic (Di Paolo and De Camilli, 2006; Balla et al., 2009). There are seven distinct species of PI, generated through the action of specific PI kinases and phosphatases that act upon the 3, 4, or 5 positions of the inositol ring of phosphatidylinositol. The different PIs are enriched at distinct cellular compartments where they serve to recruit and/or activate proteins that determine compartment function and identity (Behnia and Munro, 2005; Di Paolo and De Camilli, 2006; Balla et al., 2009). This recruitment and/or activation is usually achieved through the specific recognition of PI headgroups by modular binding domains found on proteins (Balla, 2005; Lemmon, 2008). An increasing number of diseases can be attributed to defective PI metabolism, highlighting the importance of this process (Vicinanza et al., 2008; McCrea and De Camilli, 2009).
OCRL1 is an inositol polyphosphate 5-phosphatase that preferentially hydrolyzes PtdIns(4,5)P2 and PtdIns(3,4,5)P3, PIs found predominantly at the plasma membrane (Attree et al., 1992; Schmid et al., 2004; Lowe, 2005). Mutation of OCRL1 causes two X-linked disorders in humans, oculocerebrorenal syndrome of Lowe (OCRL) and type 2 Dent disease (Attree et al., 1992; Hoopes et al., 2005). These syndromes are characterized by renal tubular dysfunction, with Lowe syndrome also resulting in neurological and ocular defects, including mental retardation, epileptic seizures, congenital cataracts, and glaucoma (Nussbaum and Suchy, 2001; Schurman and Scheinman, 2009). OCRL1 has several domains including an N-terminal PH (pleckstrin homology) domain, a central 5-phosphatase domain, and C-terminal ASH (ASPM, SPD-2, Hydin) and RhoGAP-like domains. It is localized to the trans-Golgi network (TGN), endosomes, plasma membrane ruffles, and clathrin-coated membrane-trafficking intermediates (Olivos-Glander et al., 1995; Ungewickell et al., 2004; Choudhury et al., 2005; Faucherre et al., 2005; Erdmann et al., 2007). Targeting of OCRL1 to the membrane requires interaction with Rab GTPases, which bind to the ASH domain. Rab5 and Rab6 target OCRL1 to endosomes and the TGN respectively, although binding to several other Rabs has also been reported (Hyvola et al., 2006; Fukuda et al., 2008). OCRL1 binds to clathrin heavy chain and the plasma membrane AP2 adaptor subunit α-adaptin via distinct peptide motifs located in its N- and C-terminal regions (Ungewickell et al., 2004; Choudhury et al., 2005, 2009; Mao et al., 2009). The interaction with clathrin concentrates OCRL1 into clathrin-coated buds and vesicles (Choudhury et al., 2009; Mao et al., 2009). The C-terminal RhoGAP-like domain of OCRL1 interacts with Cdc42 and Rac1, which may stabilize membrane association or recruit OCRL1 to actin-rich membrane domains (Faucherre et al., 2003, 2005; Erdmann et al., 2007). Recently, the endocytic signaling adaptor APPL1 has also been identified as an OCRL1 binding protein (Erdmann et al., 2007).
OCRL1 is related to the inositol polyphosphate 5-phosphatase Inpp5b, with which it shares a similar domain organization and substrate specificity (Matzaris et al., 1994; Shin et al., 2005; Erdmann et al., 2007; Williams et al., 2007). Inpp5b also binds to several of the proteins that interact with OCRL1 and has a similar localization, although notably it lacks binding to the clathrin machinery and is absent from clathrin-coated trafficking intermediates (Erdmann et al., 2007; Williams et al., 2007). OCRL1 and Inpp5b appear to have partially overlapping functions since deletion of OCRL1 in mice fails to give a phenotype, whereas deletion of Inpp5b gives a mild one (male sterility), yet deletion of both 5-phosphatases is embryonic lethal (Janne et al., 1998).
The combined interaction and localization data described earlier in text imply a role for OCRL1 in membrane trafficking within the endocytic pathway, at the TGN, and possibly in actin dynamics or signaling from endomembranes. Functional studies suggest that OCRL1 participates in trafficking between endosomes and the TGN (Choudhury et al., 2005; Cui et al., 2010) and possibly in receptor endocytosis (Choudhury et al., 2009; Mao et al., 2009). Analysis of Lowe syndrome fibroblasts indicate that OCRL1 is important for cell migration, although whether this is a consequence of altered membrane traffic or changes in actin dynamics is currently unclear (Suchy and Nussbaum, 2002; Coon et al., 2009). It has also been suggested that OCRL1 can regulate signaling through altered intracellular calcium fluxes (Suchy et al., 2009). Despite these studies, however, the cellular role of OCRL1 remains poorly defined; consequently, the disease mechanisms of Lowe syndrome and type 2 Dent disease are not known.
Here, we report the identification and characterization of inositol polyphosphate phosphatase interacting protein of 27 kDa (IPIP27)A and B, two novel PH domain proteins that interact with OCRL1. These proteins were recently described by De Camilli and colleagues, who used the names Ses1 and 2 (for sesquipedalian, meaning an overly long name for a simple thing) (Swan et al., 2010). We show here that the IPIPs are key regulators of endosomal trafficking, linking OCRL1 to the recycling of receptors at sorting and recycling endosomes. Our studies strongly support the hypothesis that defects in endocytic membrane traffic give rise to the symptoms of Lowe syndrome and Dent disease.
RESULTS
Identification of IPIP27A and B as binding partners for OCRL1 and Inpp5b
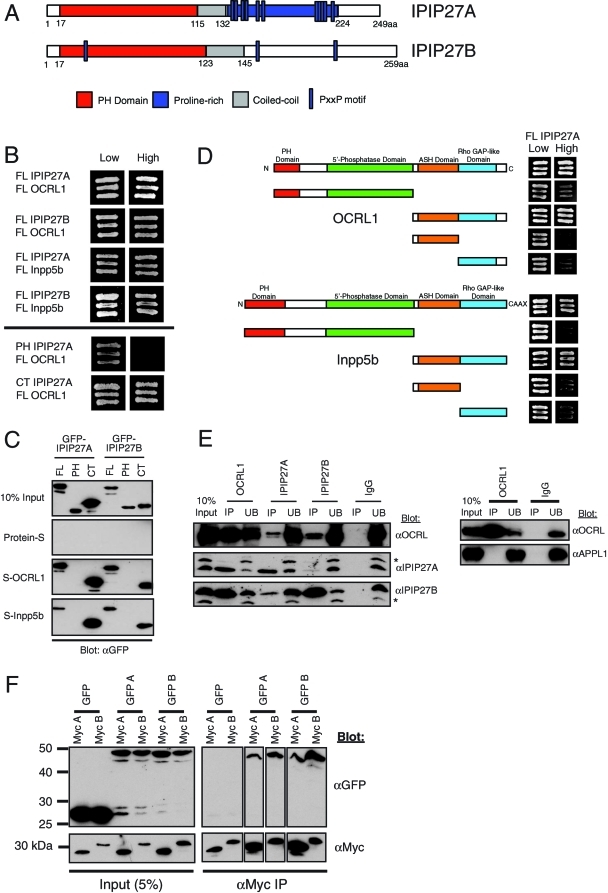
To determine the cellular role of OCRL1, we searched for new interaction partners. In a genome-wide yeast two-hybrid screen using Drosophila melanogaster proteins, it was reported that the single orthologue of OCRL1 and Inpp5b interacts with a small predicted PH-domain protein, CG12393 (Giot et al., 2003). Whereas insect species have only a single orthologue of CG12393, vertebrates have two versions of this protein, which we name IPIP27A and B (Figure 1A and Supplemental Figure 1). These proteins are the same as Ses1 and 2 recently reported by the De Camilli group (Swan et al., 2010). Both IPIP27A and B interact with OCRL1 and Inpp5b in a directed yeast two-hybrid assay, and binding is mediated by the C-terminal regions of the IPIPs (Figure 1B). These interactions were confirmed using pull-down experiments, with recombinant OCRL1 and Inpp5b efficiently binding to full-length and C-terminal regions of GFP-tagged IPIP27A and B, but not to the PH domains (Figure 1C). The regions of OCRL1 and Inpp5b responsible for binding to the IPIPs were mapped using yeast two-hybrid and pull-down experiments to the C terminus, with both the ASH and RhoGAP-like domains required for efficient binding (Figure 1D). These findings are in agreement with those reported by Swan et al. (2010).
FIGURE 1:
Interaction of OCRL1 and Inpp5b with IPIP27A and B. (A) Schematic view of human IPIP27A and B showing the predicted PH and coiled-coil domains, and the PxxP motifs found in both proteins. (B) Full-length PH domain and C terminus constructs of IPIP27A and B were tested for interaction with full-length OCRL1 and Inpp5b in the yeast two-hybrid system. Interaction results in growth on high selection medium. (C) GFP-tagged full-length PH domain or C terminus of IPIP27A and B were expressed in HeLa cells and tested for interaction with insect cell expressed in full-length, S-tagged OCRL1 and Inpp5b coupled to beads. Bound proteins were detected by Western blotting with anti-GFP antibodies. (D) Fragments of OCRL1 and Inpp5b were tested for interaction with full-length IPIP27A in the yeast two-hybrid system. (E) Endogenous OCRL1, IPIP27A, and IPIP27B were immunoprecipitated (IP) under native conditions from a HeLa cell extract, and bound proteins were detected by Western blotting with the indicated antibodies. UB, 10% of the unbound fraction. Asterisks indicate background bands that cross-react with the IPIP antibodies. (F) Dimerization of IPIP27 was assessed using extracts from HeLa cells coexpressing GFP- or Myc-tagged IPIP27A and/or IPIP27B followed by native immunoprecipitation with anti-GFP or anti-Myc antibodies and Western blotting with antibodies to these tags.
To confirm that endogenous OCRL1 and IPIPs interact in vivo, we performed native coimmunoprecipitation experiments. As shown in Figure 1E, antibodies to OCRL1 efficiently coimmunoprecipitated both IPIP27A and B, whereas antibodies to either IPIP27A or B coimmunoprecipitated OCRL1. Note that the majority of either IPIP27 was brought down with OCRL1 antibodies, suggesting that a large proportion of cellular IPIPs are found in a complex with OCRL1. In contrast, we could not detect coimmunoprecipitation of OCRL1 and APPL1 under the same conditions, suggesting that the interaction between these proteins is weak or transient (Figure 1E). Comparatively lower levels of OCRL1 were found in the IPIP27 immunoprecipitates, suggesting that a minority of cellular OCRL1 is bound to the IPIPs at any one time. Interestingly, IPIP27A coimmunoprecipitated with IPIP27B and vice versa, suggesting that they associate with one another in vivo (Figure 1E). This suggestion was confirmed by coexpressing green fluorescent protein (GFP)- or myc-tagged IPIP27A and B and performing coimmunoprecipitation. As shown in Figure 1F, both IPIPs can homodimerize, with IPIP27B dimerizing more efficiently than IPIP27A, most likely due to its longer coiled-coiled region. The IPIPs were also able to form heterodimers (Figure 1F). Interestingly, IPIP27 heterodimer formation was more efficient than IPIP27A homodimerization, although not as efficient as IPIP27B homodimerization, consistent with IPIP27B having a higher propensity than IPIP27A to dimerize.
IPIP27A and B share a conserved motif for OCRL1 binding
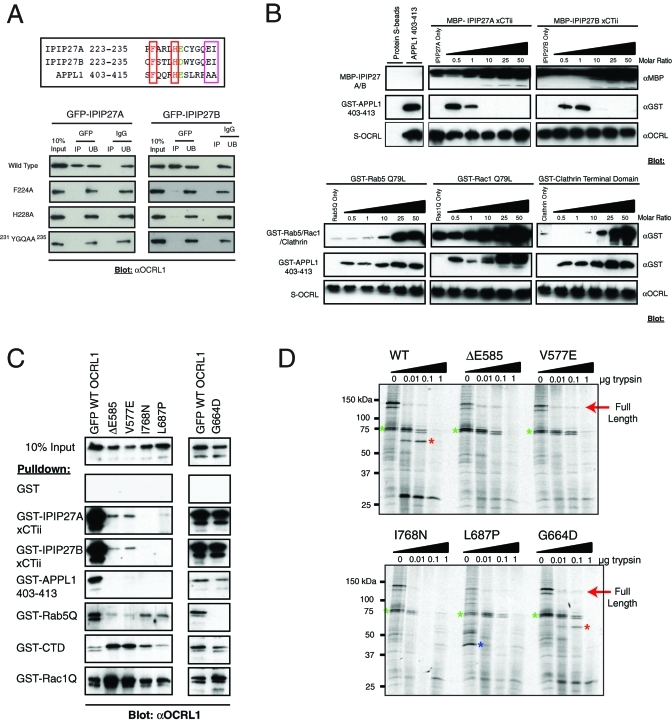
Swan et al. (2010) identified a conserved C-terminal F&H motif as the OCRL1 binding site in IPIP27/Ses, a motif that is also required for APPL1 binding to OCRL1 (see Supplemental Figure 1 and Figure 2A). As expected, pull-down experiments using truncated versions of IPIP27A and B indicated that binding to OCRL1 and Inpp5b was observed only in the presence of the C-terminal region containing the F&H motif (Supplemental Figure 2). Mutation of either the F or H residues to alanine in IPIP27A or B significantly decreased binding to OCRL1, in agreement with the findings of Swan et al. (2010) (Figure 2A). Mutation of the conserved glutamate–isoleucine (EI) pair of residues also strongly reduced binding to OCRL1 (Figure 2A), consistent with an important role for these residues in conferring high-affinity binding to OCRL1 (Swan et al., 2010). Peptide binding experiments previously suggested that the IPIPs and APPL1 bind to a common binding site in the C-terminal region of OCRL1 (Swan et al., 2010). To confirm this suggestion, we performed competition binding experiments with recombinant proteins. Purified recombinant OCRL1 bound to purified IPIP27A and B, confirming that these proteins interact directly (Figure 2B). Purified OCRL1 also bound to purified APPL1, as observed previously (Erdmann et al., 2007). Addition of a molar excess of IPIP27A or B, however, efficiently competed for this interaction, with binding completely abolished at a 10-fold excess of IPIP27A or B (Figure 2B). In contrast, binding was retained with up to a 50-fold molar excess of Rab5, Rac1, or clathrin heavy-chain terminal domain, that all bind to the ASH-RhoGAP regions of OCRL1, indicating that the competition of APPL1 by IPIPs is specific. These results, together with those of Swan et al. (2010), strongly suggest the IPIPs and APPL1 share a common mode of binding to OCRL1.
FIGURE 2:
Mutational analysis of the IPIP27-OCRL/Inpp5b interaction. (A, top) Sequence alignment of the C terminus of IPIP27A, B and amino acids 403–415 of APPL1. The conserved F and H residues are boxed in red, and the EI residues found only in the IPIPs are boxed in pink. (A, bottom) GFP-tagged IPIP point mutants were expressed in HeLa cells and immunoprecipitated under native conditions using anti-GFP antibodies, and bound OCRL1 was detected by Western blotting. (B) Competition binding assays were performed using the indicated recombinant proteins. A constant amount of S-tagged OCRL1 was coupled to beads, and an equal molar ratio of the minimal OCRL1 binding domain of APPL1 (aa 403–413) was added alone or in the presence of an increasing amount of IPIP27A or B C-terminal constructs (top) or recombinant Rab5 Q79L, Rac1 Q79L, or clathrin terminal domain. Bound proteins were detected by Western blotting with the indicated antibodies. (C) The indicated GFP-tagged Lowe syndrome missense mutants were expressed in HeLa cells, and binding to the indicated GST-tagged bait proteins was monitored by Western blotting. (D) The indicated GFP-tagged OCRL1 missense mutants were expressed in 35S-labeled HeLa cells, immunoprecipitated, and subjected to digestion with increasing amounts of trypsin. Digested proteins were analyzed by SDS–PAGE and autoradiography. G664D is an engineered mutant defective in Rab binding, whereas the others are found in Lowe syndrome patients. The colored asterisks indicate prominent digestion products.
Lowe syndrome mutations abolish interaction with IPIP27
A number of missense mutations have been identified in the ASH-RhoGAP domains of OCRL1 (http://research.nhgri.nih.gov/lowe/). Several of these mutations have been shown to abolish interaction with APPL1, suggesting that loss of APPL1 binding may contribute to Lowe syndrome (Erdmann et al., 2007; McCrea and De Camilli, 2009). It has recently been demonstrated, however, that the same missense mutations also abolish IPIP27/Ses binding (Swan et al., 2010). We obtained a similar result using V577E and ΔE585 mutations that reside in the ASH domain, and L687P and I768N mutations in the RhoGAP-like domain. All mutations abolished both IPIP27 and APPL1 binding (Figure 2C). Structural analysis suggests that these mutations may impair folding of the OCRL1 C terminus (Erdmann et al., 2007). We therefore performed limited proteolysis on the GFP-tagged wild-type OCRL1 and the Lowe syndrome mutants. Each of the missense mutants exhibited a distinct tryptic digestion pattern to that of the wild-type protein, suggesting that folding of the protein is indeed impaired (Figure 2D). This effect likely explains why binding to Rab5, clathrin, and Rac1 is also affected by these mutations (Figure 2C). In contrast, the Rab binding mutant G664D did not affect binding to IPIP27 or the other interaction partners, and gave a similar digestion pattern to wild-type OCRL1 as previously reported (Hyvola et al., 2006).
Localization of IPIP27A and B
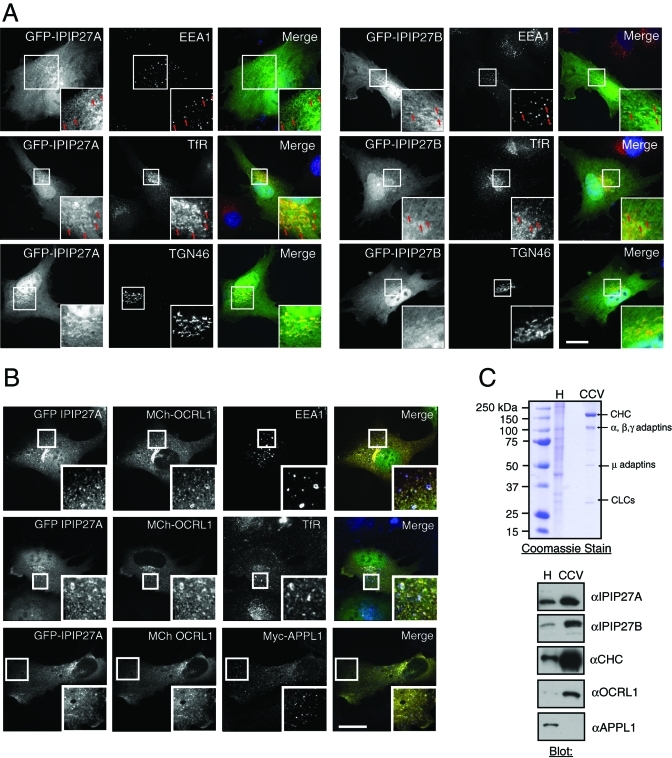
Unfortunately we were unable to localize endogenous IPIP27 as our antibodies failed to work in immunofluorescence microscopy (unpublished data). We therefore transiently expressed GFP-tagged proteins at low levels in hTERT-RPE1 and HeLa cells. Western blotting indicated that the GFP-tagged IPIPs were expressed at comparable levels to the endogenous proteins, and we observed a similar localization regardless of the type of tag used (Supplemental Figure 3). Both IPIP27A and B exhibited the same localization. In addition to cytosolic and nuclear pools, there was localization to early endosomes, indicated by overlap with EEA1 (Figure 3A). This finding supports those of Swan et al. (2010), who reported colocalization of IPIP27/Ses with EEA1 and WDFY2, which mark Rab5-positive early endosomes. We also observed a high degree of overlap with the transferrin receptor (TfR), both in punctate early endosomes and in the perinuclear recycling compartment. Consistent with this observation, the IPIPs overlapped with Rab11, a marker of perinuclear recycling endosomes (Supplemental Figure 4A). We also observed partial overlap with TGN46 and Golgin-97 in the perinuclear region, suggesting localization to the TGN (Figure 3A and Supplemental Figure 3C). We failed to detect localization of either IPIP to late endosomes or lysosomes (Supplemental Figure 4A). To better resolve the perinuclear staining, we treated cells with nocodazole to depolymerize microtubules and disperse Golgi and recycling endosomes. Both IPIP27A and B were present on dispersed TGN and recycling endosome elements, indicating localization to both compartments (Supplemental Figure 4B). Thus, we conclude that IPIP27A and B are localized to early and recycling endosomes and to the TGN.
FIGURE 3:
IPIP27A and B are localized to early and recycling endosomes and the TGN and present in clathrin-coated vesicles. (A) Immunofluorescence microscopy of hTERT-RPE1 cells expressing low levels of GFP-tagged IPIP27A or B (green) and labeled with antibodies to EEA1, TfR, or TGN46 (red). Arrows indicate colocalization of IPIP27 with endosomal markers. Scale bar = 10 μm. (B) hTERT-RPE1 cells coexpressing GFP-IPIP27A and mCherry-OCRL1 were labeled with antibodies to EEA1, TfR, or coexpressed Myc-tagged APPL1 (red). Scale bar = 10 μm. (C) Coomassie blue staining and Western blotting of human placenta homogenate (H) and purified clathrin-coated vesicles (CCV). The positions of clathrin heavy chain (CHC), clathrin light chains (CLCs), and adaptin subunits are indicated.
To further examine IPIP distribution, they were coexpressed at low levels with OCRL1. IPIP27A and B colocalize extensively with OCRL1 on endosomes containing EEA1 and TfR (Figure 3B and unpublished data). There is little overlap between coexpressed IPIP27A or B and APPL1, in line with the findings of Swan et al. (2010), indicating localization to different endosomal subpopulations. OCRL1 overlaps more extensively with coexpressed IPIP27A and B compared with APPL1, consistent with the IPIPs being the predominant OCRL1-binding partners in vivo (Figure 3B). Previous work has shown that OCRL1 is enriched in clathrin-coated vesicles (Ungewickell et al., 2004; Choudhury et al., 2005, 2009). Both IPIP27A and B are also enriched in purified clathrin-coated vesicles, with a similar degree of enrichment to that seen with OCRL1 (Figure 3C). In contrast, we could not detect APPL1 in the vesicle fraction. Together, our binding and localization results are consistent with a role for the IPIPs in clathrin- and possibly nonclathrin-mediated membrane trafficking, most likely at early and recycling endosomes and at the TGN.
Membrane targeting of IPIP27
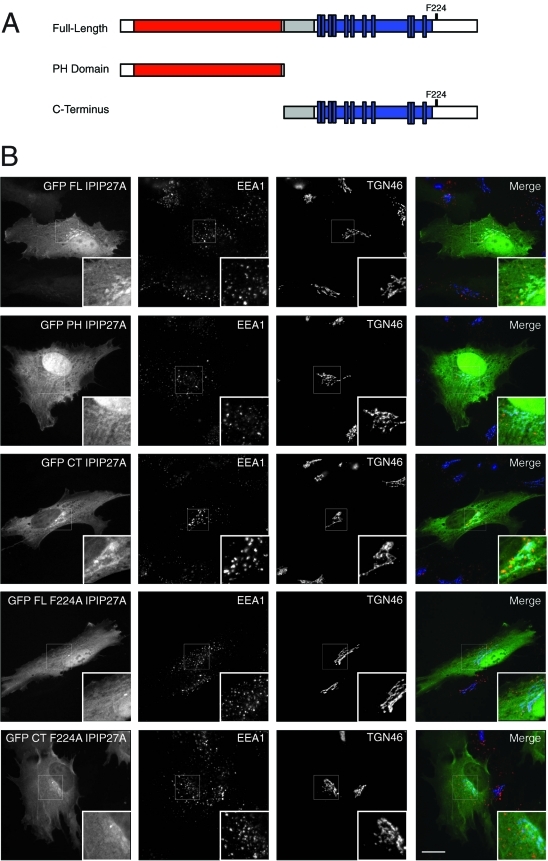
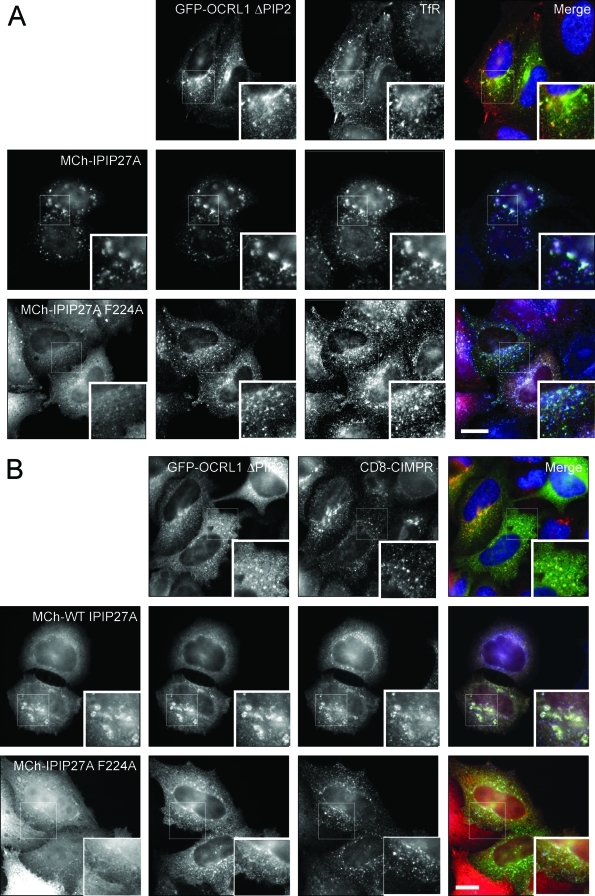
To determine which domains of IPIP27A and B are required for targeting the proteins to endosomes and the TGN, GFP-tagged versions comprising the PH or C-terminal domains were expressed in cells and their localization analyzed (Figure 4, A and B). The PH domains of IPIP27A and B were weakly targeted to these compartments, whereas the C-terminal domains of both proteins gave a stronger localization, similar to that seen with full-length IPIP27 (Figure 4B and unpublished data). To determine whether this localization is due to interaction with membrane-associated OCRL1 and Inpp5b, versions of IPIP27 containing the F224A mutation that abolishes OCRL1/Inpp5b binding were analyzed. As shown in Figure 4B, full-length and C-terminal IPIP27 F224A exhibited dramatically reduced membrane targeting, suggesting that OCRL1/Inpp5b interaction is a major determinant in IPIP27 localization. In support of this finding, we observe a significant enhancement of IPIP27 recruitment onto endosomal and TGN membranes when OCRL1 is coexpressed with wild-type IPIP27 but not the F224A mutant (Supplemental Figure 5A). The OCRL1/Inpp5b interaction is therefore important for IPIP27 targeting in vivo. Additional interactions are involved, however, most likely with the PH domain and other binding sites in the C-terminal region of the IPIPs. Conversely, binding to the IPIPs is not required for OCRL1 recruitment to the membrane, since depletion of the IPIPs alone or together does not diminish OCRL1 targeting (Supplemental Figure 5B). This finding is consistent with previous work showing that binding to Rab GTPases targets OCRL1 to the TGN and endosomes (Hyvola et al., 2006).
FIGURE 4:
Membrane targeting of IPIP27. (A) Schematic view of IPIP27A domains used for analysis of membrane targeting. (B) Immunofluorescence microscopy of hTERT-RPE1 cells expressing low levels of GFP-tagged, full-length IPIP27A or truncated versions comprising the PH domain or downstream C-terminal region with or without the F224A mutation and labeled with antibodies to EEA1 (red) and TGN46 (blue). Scale bar = 10 μm.
Depletion of IPIP27A causes enlargement of early endosomes
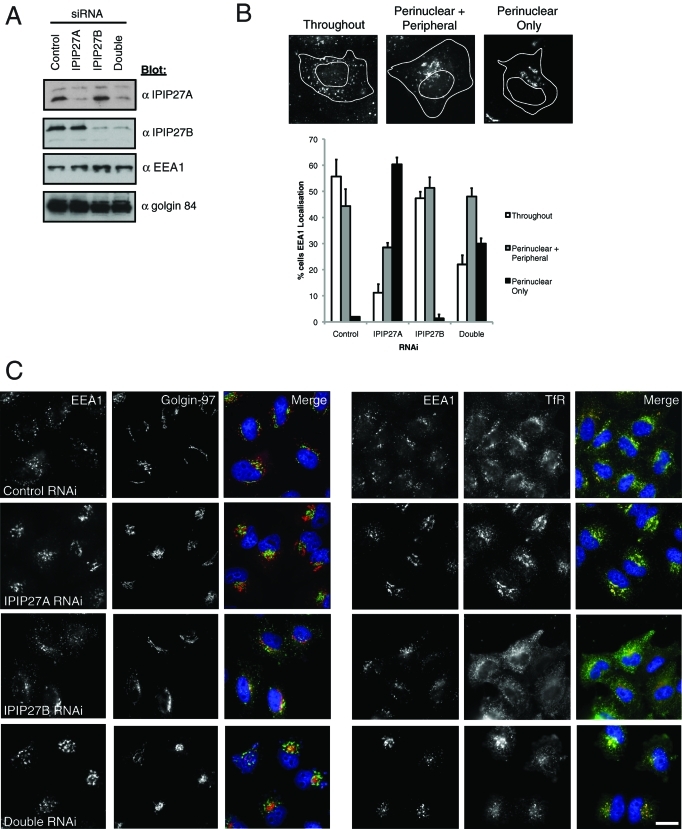
To determine the functional role of IPIP27A and B, they were depleted separately or in combination using RNA interference. Depletion of both IPIPs was specific and efficient (Figure 5A). Importantly we obtained the same results using two independent oligos for each protein (details of small interfering RNA [siRNA] oligos are in Materials and Methods). Depletion of IPIP27A altered endosome morphology, resulting in enlarged EEA1-positive early endosomes that clustered in the perinuclear region (Figure 5, B and C). The Golgi also appeared more compact in the IPIP27A-depleted cells (Figure 5C). The clustered EEA1-positive endosomes contained TfR, which was redistributed to these structures (Figure 5C). Depletion of IPIP27B did not induce endosomal clustering, whereas the depletion of both IPIPs together resulted in a slightly weaker phenotype (Figure 5, A and B). Depletion of IPIP27B, however, did affect the steady-state distribution of TfR, which accumulated in endosomes located in the cell periphery (Figure 5C). In the double-depleted cells, TfR gave a phenotype resembling a combination of that observed with the single knock-downs (Figure 5C). These observations indicate a role for IPIP27A in endosomal organization and suggest that the IPIPs participate in TfR trafficking.
FIGURE 5:
Depletion of IPIP27A alters endosomal morphology. (A) IPIP27A and B were depleted separately or together from HeLa cells for 72 h, and depletion was monitored by Western blotting with the indicated antibodies. (B) Examples of early endosomal morphology in RNAi-treated cells as assessed by EEA1 staining and quantification of the phenotype. Results are from three experiments with 100 cells counted per experiment, and are shown as the mean + standard deviation. (C) Immunofluorescence microscopy of the RNAi-treated cells after labeling with antibodies to EEA1 and Golgin-97 or TfR. Scale bar = 10 μm.
IPIP27A and B are required for TfR recycling
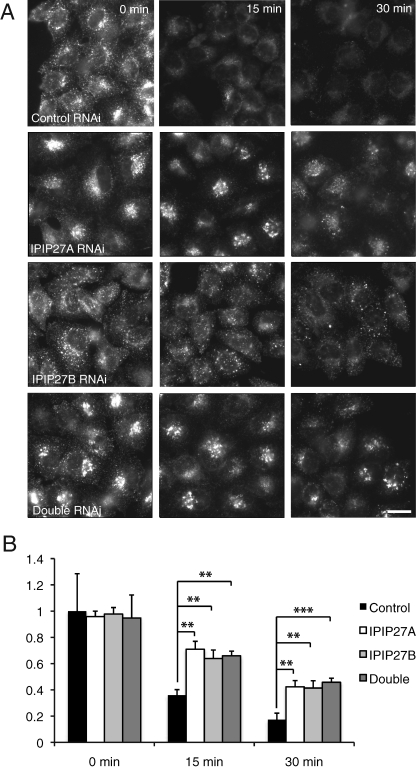
The simplest explanation for the redistribution of TfR into clustered perinuclear or peripheral endosomes upon IPIP27A or B depletion is that the IPIPs are required for trafficking of the receptor out of these compartments. To test this hypothesis, we directly analyzed recycling of fluorescently labeled transferrin that had been internalized into IPIP-depleted cells. Depletion of IPIP27A or B alone or together had little effect on the amount of transferrin internalized into cells over a 30-min time period, indicating that the IPIPs are not essential for transferrin endocytosis (Figure 6A). The recycling of internalized transferrin was clearly impaired, however, upon depletion of either IPIP alone or when they were codepleted (Figure 6, A and B). Transferrin was retained in clustered perinuclear endosomes upon depletion of IPIP27A and in more peripheral endosomes when IPIP27B was depleted, consistent with the redistribution of TfR observed under these conditions (Figures 5C and 6A).
FIGURE 6:
Depletion of IPIP27A impairs transferrin recycling. (A) Control, IPIP27A and/or B RNAi-treated HeLa cells were allowed to internalize Alexa 488-conjugated transferrin for 30 min at 37ºC, washed, and incubated for an additional 0, 15, or 30 min at 37ºC prior to fixation and analysis by fluorescence microscopy. Scale bar = 10 μm. (B) Quantitation of cell-associated Alexa 488–conjugated transferrin at various times of incubation. Results are from three experiments with 20 cells counted per experiment, and are shown in arbitrary units (AU) as the mean + standard deviation. **p < 0.001, ***p < 0.0005.
IPIP27 depletion impairs trafficking from endosomes to the TGN
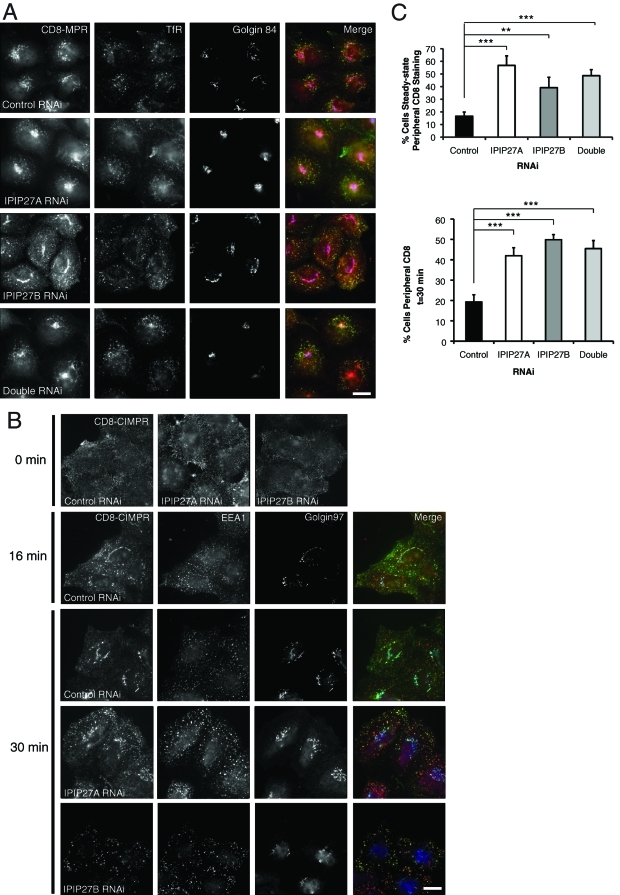
We next wanted to test whether the IPIPs are required for the endosomal trafficking of other receptors. Previous work has suggested a role for OCRL1 in trafficking between endosomes and the TGN (Ungewickell et al., 2004; Choudhury et al., 2005; Cui et al., 2010), leading us to test whether the IPIPs are also involved in this trafficking step. A major receptor that follows this route is the cation-independent mannose 6-phosphate receptor (CIMPR), which transports newly synthesized lysosomal hydrolases from the TGN to endosomes. The CIMPR undergoes continual cycling between the TGN and endosomes, as well as a lower amount of trafficking via the plasma membrane. To study CIMPR trafficking we exploited a cell line stably expressing a chimeric CD8-CIMPR reporter (Seaman, 2004). Depletion of IPIP27A or B in these cells resulted in partial redistribution of CD8-CIMPR to endosomes, suggesting that endosome to TGN trafficking may be impaired (Figure 7, A and C). This finding was confirmed by incubating cells with an antibody that binds to the CD8-CIMPR ectodomain at the cell surface and by monitoring uptake and delivery from endosomes to the TGN by fluorescence microscopy. In control cells the CD8-CIMPR was efficiently internalized into endosomes at early times and was delivered to the TGN after a 30-min uptake (Figure 7, B and C). In contrast, there remained a significant amount of CIMPR retained within endosomes at this time in IPIP27-depleted cells, indicating reduced delivery to the TGN. Interestingly, depletion of IPIP27B also reduced the amount of CD8 antibody bound to the cell surface prior to uptake. This result indicates a lower amount of receptor at the cell surface, suggesting an additional role for IPIP27B in recycling of the CIMPR to the plasma membrane (Figure 7B).
FIGURE 7:
CD8-CIMPR trafficking in IPIP27-depleted cells. (A) CD8-CIMPR expressing HeLaM cells were depleted of IPIP27A and/or B and labeled with antibodies to CD8 (CD8-CIMPR, green), TfR (red), and Golgin-84 (blue). (B) HeLaM CD8-CIMPR RNAi-treated cells were incubated on ice with anti-CD8 antibody and either fixed immediately (0 min) or warmed to 37ºC for 16 or 30 min prior to fixation and labeling with antibodies to EEA1 (red) or Golgin-97 (blue). CD8 was detected with an appropriate secondary antibody (green). Bars = 10 μm. (C) Quantitation of CD8-CIMPR steady-state distribution and transport to TGN, expressed as the percentage of cells with endosomal CD8-CIMPR. Results are from three experiments with 100 cells counted per experiment and are shown as the mean + standard deviation. **p < 0.001; ***p < 0.0005.
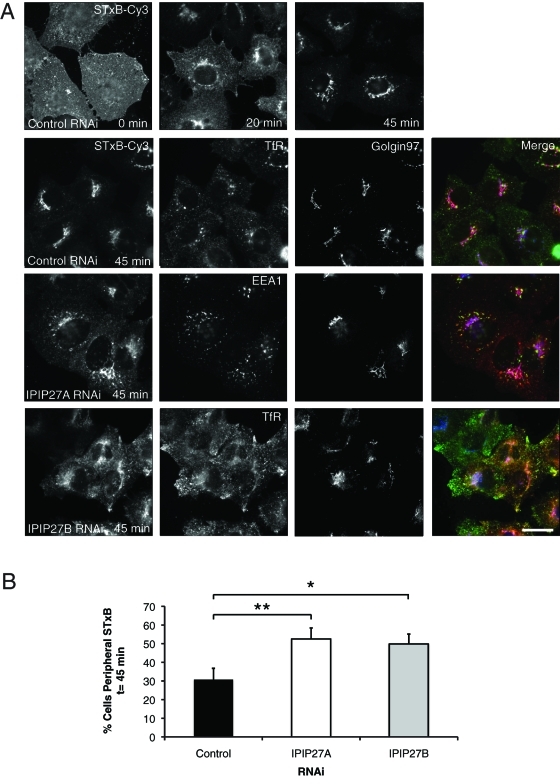
Another way to measure endosome-to-TGN trafficking is to use the Shiga toxin B-subunit (STxB). STxB is internalized via non-clathrin-mediated endocytosis and delivered to early endosomes, from where it is transported to the TGN in a clathrin-dependent manner (Saint-Pol et al., 2004). We exploited this assay to determine whether the IPIPs are required for trafficking of other cargoes from endosomes to the TGN. In control cells, STxB was efficiently transported to the Golgi apparatus after a 45-min internalization (Figure 8, A and B). In contrast, in cells depleted of IPIP27A or B, a significant amount of STxB was retained within cytoplasmic puncta corresponding to endosomes at 45-min internalization (Figure 8, A and B). IPIP27 depletion therefore impairs endosome-to-TGN trafficking of STxB. Thus, IPIP27A and B are required for efficient endosome-to-TGN trafficking of both CIMPR and STxB.
FIGURE 8:
Shiga toxin trafficking in IPIP27-depleted cells. (A) RNAi-treated HeLa cells were incubated on ice to allow surface binding of STxB-Cy3 (0 min) prior to warming to 37ºC for 20 or 45 min. Cells were labeled with antibodies to TfR or EEA1 (green) and Golgin-97 (blue) prior to analysis by immunofluorescence microscopy. Bar = 10 μm. (B) Quantitation of STxB-Cy3 transport to the TGN, expressed as the percentage of cells with endosomal STxB-Cy3 at t = 45 min. Results are from three experiments with 50 cells counted per experiment and are shown as the mean + standard deviation. *p < 0.01; **p < 0.001.
IPIP27 depletion causes missorting of lysosomal hydrolases
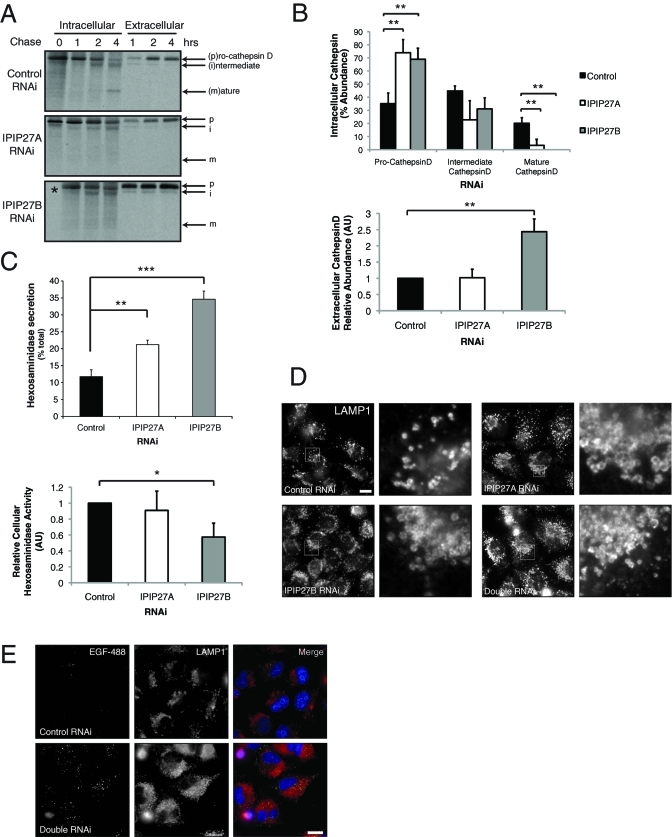
Our results indicate that CIMPR trafficking from endosomes to the TGN is impaired by IPIP27 depletion. We should therefore expect reduced levels of CIMPR in the TGN, with consequent missorting of newly synthesized lysosomal hydrolases. To test this expectation, we monitored processing of the lysosomal hydrolase cathepsin-D, which is synthesized as a 53-kDa precursor that undergoes cleavage to 47 kDa intermediate and 31 kDa mature forms upon delivery to endosomes and then lysosomes, respectively (Davidson, 1995). Depletion of IPIP27A or B resulted in a dramatic reduction in the amount of cathepsin D processing, indicating reduced delivery to endosomes and lysosomes (Figure 9, A and B). There was also increased secretion of the cathepsin-D precursor, consistent with having reduced levels of CIMPR in the TGN. Interestingly, this was most apparent for IPIP27B depletion, suggesting a stronger defect in CIMPR recycling. We observed a similar result when analyzing the sorting of another lysosomal hydrolase hexosaminidase. Depletion of either IPIP caused increased secretion of hexosaminidase compared with controls, with a concomitant decrease in the amount of cell-associated hexosaminidase activity, and again the effect was more pronounced upon IPIP27B depletion (Figure 9C).
FIGURE 9:
IPIP27 depletion causes lysosomal hydrolase missorting. (A) Processing of newly synthesized cathepsin-D in RNAi-treated HeLa cells. Following pulse-labeling with [35S]methionine/cysteine and incubation at 37ºC for the indicated times, cathepsin-D was immunoprecipitated from the cells (intracellular) or medium (extracellular) and detected by autoradiography. (B) Quantitation of cathepsin-D processing and secretion. Results are from three experiments and expressed in arbitrary units (AU) as the mean + standard deviation. **p < 0.001. (C) Hexosaminidase activity in IPIP27 RNAi-treated HeLa cell extracts or the medium following incubation at 37ºC for 6 h was assayed as indicated in Materials and Methods. Results are expressed as the mean + standard error of the mean from four experiments. *p < 0.01; **p < 0.001; ***p < 0.0005. (D) Immunofluorescence microscopy of IPIP27-depleted HeLa cells labeled with antibodies to LAMP1. Bar = 10 μm. (E) Immunofluorescence microscopy of control or IPIP27-depleted cells that have internalized Alexa 488-conjugated EGF (green) for 4 h at 37ºC prior to fixation and labeling with an antibody to LAMP1. Bar = 10 μm.
Our results indicate a reduced level of mature lysosomal hydrolases in IPIP-depleted cells. Impaired degradation of lysosomal substrates, as seen in lysosomal storage disorders, leads to an increase in the number and size of lysosomes in affected cells (Karageorgos et al., 1997; Parkinson-Lawrence et al., 2010). To determine whether the loss of cellular hydrolases observed in IPIP27-depleted cells gives a similar effect, labeling for the lysosomal marker LAMP1 (lysosome-associated membrane protein 1) was performed. As shown in Figure 9D, the IPIP27-depleted cells have increased numbers of LAMP1-positive lysosomes, and the lysosomes appear larger than in the control. These results are consistent with an impairment of lysosomal substrate degradation upon depletion of the IPIPs. In line with this hypothesis, we observe a reduction in the ability of IPIP27-depleted cells to degrade internalized epidermal growth factor (EGF) (Figure 9E). The undegraded EGF present in IPIP27-depleted cells only partially overlaps with lysosomes, suggesting delivery to lysosomes may be impaired, as recently reported for lysosomal storage disorders (Fraldi et al., 2010).
Interaction between IPIP27 and OCRL1 is important for endosome morphology
To address the functional importance of the interaction between IPIP27 and OCRL1/Inpp5b, we adopted an overexpression strategy. Wild-type and mutant forms of IPIP27 were coexpressed in cells with OCRL1 lacking the entire 5-phosphatase domain (ΔPIP2) or containing a point mutation in the 5-phosphatase domain that renders it catalytically inactive (D499A) (Jefferson and Majerus, 1996), and effects on endosome morphology and receptor distribution were analyzed. The OCRL1 ΔPIP2 mutant acts as a dominant negative in its own right, causing endosome enlargement and redistribution of the TfR and CIMPR to the aberrant endosomes, as described in earlier studies (Choudhury et al., 2005; Hyvola et al., 2006) (Figure 10, A and B). The effects on both endosomal morphology and receptor protein distribution, however, are greatly exacerbated by coexpression with IPIP27A. To confirm that IPIP27 binding to OCRL1 is required to induce these effects, IPIP27A F224A was coexpressed with OCRL1 ΔPIP2. As shown in Figure 10, A and B, the effect of coexpressing IPIP27 with OCRL1 ΔPIP2 on endosome morphology and receptor distribution is abolished by the F224A mutation. Similar results were observed with IPIP27B (unpublished data). In contrast to the ΔPIP2 mutant, OCRL1 D499A alone had little effect on endosome morphology and receptor distribution (Supplemental Figure 6). Coexpression with IPIP27A again caused endosome enlargement, albeit to a lesser degree than with the OCRL1 ΔPIP2 mutant, and there was redistribution of both TfR and CIMPR to the aberrant endosomal compartments, consistent with a defect in their trafficking (Supplemental Figure 6). Again, these effects were abolished by the F224A mutation in IPIP27A, and similar results were seen with IPIP27B (Supplemental Figure 6 and unpublished data). Together, these results strongly suggest that the interaction between IPIP27 and OCRL1 is important for endosomal morphology and function.
FIGURE 10:
Co-overexpression of IPIP27 with OCRL1 ΔPIP2 alters endosomal morphology and receptor distribution. CD8-CIMPR HeLaM cells expressing GFP-OCRL1 ΔPIP2 (green) without or with coexpression of mCherry wild-type (WT) IPIP27A or the F224A mutant (red) were labeled with antibodies to TfR (blue) (A) or CD8 (CD8-CIMPR, blue) (B). Bars = 10 μm.
DISCUSSION
In this article we describe the identification of IPIP27A and B as new interaction partners for the inositol polyphosphate 5-phosphatases OCRL1 and Inpp5b. We identify the OCRL1/Inpp5b binding site in the IPIPs and show that they are localized to endosomes and the TGN. Our IPIP27 binding and localization results are in agreement with those of De Camilli and colleagues, who refer to the IPIPs as Ses1 and 2 (Swan et al., 2010). In this study we now show that these proteins are key regulators of endocytic trafficking.
Early and recycling endosomes represent hubs for receptor sorting and trafficking within the endocytic pathway (Bonifacino and Rojas, 2006; Grant and Donaldson, 2009; Hsu and Prekeris, 2010). Nutrient receptors such as TfR are recycled back to the plasma membrane, whereas others are targeted to the lysosome for degradation or sorted into carriers destined for the TGN, as in the case of CIMPR (Bonifacino and Rojas, 2006; Johannes and Popoff, 2008). The IPIPs are localized to both sorting and recycling endosomes, and our studies indicate a role in multiple trafficking routes out of these compartments. We observe a strong effect of IPIP27 depletion upon CIMPR trafficking, with consequent defects in lysosomal hydrolase sorting. It has previously been reported that depletion of OCRL1 results in CIMPR missorting, although the effects are more subtle than we observe with IPIP27 depletion (Choudhury et al., 2005; Cui et al., 2010). Importantly, Lowe syndrome patients have elevated levels of lysosomal hydrolases in their plasma, indicating that missorting occurs in the disease state (Ungewickell and Majerus, 1999). We propose that some Lowe syndrome symptoms, such as those of the CNS, may arise through lysosomal dysfunction. It is interesting to note that neurological problems are commonly observed in lysosomal storage disorders (Parkinson-Lawrence et al., 2010).
We also observe defects in recycling of plasma membrane receptors upon IPIP27 depletion. Defective endocytic recycling to the plasma membrane could explain the renal symptoms of Lowe syndrome, since loss of the multiligand endocytic receptor megalin or its impaired recycling from endosomes gives a similar renal dysfunction to that observed in Lowe and Dent patients (Christensen and Birn, 2002; Norden et al., 2002; Christensen et al., 2003; Lowe, 2005). A recent study failed to observe defects in trafficking of ectopically expressed megalin in OCRL1-depleted MDCK cells, although recycling was not specifically analyzed (Cui et al., 2010). Whether this is also true for endogenous megalin in the kidney proximal tubule remains to be investigated. Lowe syndrome patients appear to have less megalin at the cell surface, consistent with a recycling defect, although altered intramembrane proteolysis could also be responsible (Norden et al., 2002; Cui et al., 2010). Further experiments using an appropriate model will be required to more thoroughly test the involvement of OCRL1 and IPIP27 in megalin trafficking.
OCRL1 and Inpp5b appear able to partially or, in the case of the mouse, fully compensate for the loss of each other (Janne et al., 1998). How this compensation occurs is unclear, since OCRL1 is associated with the clathrin trafficking machinery whereas Inpp5b is not (Ungewickell et al., 2004; Choudhury et al., 2005; Erdmann et al., 2007; Williams et al., 2007; Choudhury et al., 2009; Mao et al., 2009). Our results with the IPIPs provide a potential explanation for this apparent conundrum. The recycling of receptors within the endosomal system is complex, with more than one trafficking route possible for the same receptor (Bonifacino and Rojas, 2006; Johannes and Popoff, 2008; Grant and Donaldson, 2009; Hsu and Prekeris, 2010). For example, TfR can be recycled to the plasma membrane from both early and recycling endosomes, whereas the CIMPR can be trafficked from different endosomes to the TGN. Deficiency of OCRL1 or Inpp5b likely affects distinct trafficking routes out of the endosome, with OCRL1 participating in clathrin-dependent and Inpp5b participating in clathrin-independent trafficking. The loss of either enzyme could therefore be compensated by the ability of cargo to use an alternative endosomal recycling pathway. The extent of defect observed would depend on the relative levels of the two phosphatases and the nature of the receptors and trafficking routes they follow, which is largely dependent on cell type, explaining why Lowe syndrome affects only certain tissues. Because the IPIPs bind to both OCRL1 and Inpp5b, their depletion would be expected to inhibit pathways used by both phosphatases, which may explain the strong trafficking defects we observe when these proteins are depleted.
The mechanism by which the IPIPs function in receptor recycling at endosomes is currently unclear. Their presence in clathrin-coated vesicles is consistent with a role at a late stage of carrier formation, or possibly in events that occur after carrier fission. The presence of numerous PxxP motifs in the C-terminal regions of the IPIPs suggests that they act as adaptors, linking OCRL1 and Inpp5b to the machinery required for the formation or maturation of endosome-derived carriers. Endocytic recycling requires several proteins that bind to PtdIns(4)P and PtdIns(4,5)P2 (Mills et al., 2003; Zimmermann et al., 2005; Jovic et al., 2009), and both lipids have been localized to endosomal recycling intermediates (Brown et al., 2001; Jovic et al., 2009). OCRL1 and Inpp5b, via interaction with the IPIPs, likely participate in controlling the levels of these lipids in a highly temporally and spatially controlled manner to ensure efficient formation or maturation of carriers. The majority of Rab binding partners for OCRL1 and Inpp5b are involved in endocytic recycling (Rab6, Rab8, Rab10, Rab14, Rab22A, Rab35), supporting a role for the phosphatases in this process (Hyvola et al., 2006; Fukuda et al., 2008; Grant and Donaldson, 2009; Lowe and Barr, unpublished data).
It has previously been reported that all known missense mutations in the C-terminal region of OCRL1 abolish binding to APPL1, suggesting that loss of this interaction may be responsible for the defects in Lowe syndrome (Erdmann et al., 2007; McCrea and De Camilli, 2009). Our results indicate that binding to the IPIPs is also abolished by these mutations, and further analysis indicates altered interactions with other known binding proteins. We could show that OCRL1 folding is impaired by the mutations, suggesting a more global disruption in protein function rather than a specific effect on protein interactions. This hypothesis would fit with the observations that many of the disease-causing missense mutations in OCRL1 lead to loss of protein expression (http://research.nhgri.nih.gov/lowe/).
APPL1 is associated with signaling endosomes that appear transiently during growth factor receptor endocytosis (Miaczynska et al., 2004). APPL1-positive signaling endosomes lie upstream of sorting endosomes, and their maturation is controlled by a PI switch (Zoncu et al., 2009). Loss of APPL1 and termination of signaling are mediated by the production of PtdIns(3)P, which is abundant at the sorting endosome (Zoncu et al., 2009). Our results suggest that the IPIPs contribute to this process, since they localize to sorting endosomes and share a common binding site on OCRL1 and Inpp5b with APPL1. Moreover, they bind more strongly to OCRL1 and Inpp5b than does APPL1. The IPIPs are therefore ideally placed to displace APPL1 from the membrane, as would occur upon fusion of a signaling endosome with the sorting endosome, to help ensure the strict demarcation that exists between these types of endosomes.
MATERIALS AND METHODS
Materials and antibodies
All reagents were obtained from Sigma (St. Louis, MO) or Merck (Whitehouse Station, NJ) unless stated otherwise. Protease inhibitors (cocktail set III) were obtained from Merck (Whitehouse Station, NJ) and used at 1:250. Cy3-STxB was provided by Ludger Johannes (CNRS, Paris, France). Polyclonal antibodies to IPIP27A and B were raised in sheep using glutathione S-transferase (GST)-tagged C-terminus IPIP27A (CT; residues 124–249) or IPIP27B (CT, residues 133–259) and were affinity purified using maltose binding protein (MBP)-tagged, full-length IPIP27A or CT IPIP27B protein covalently coupled to Affigel 15 (Bio-Rad Laboratories, Hercules, CA). Other polyclonal antibodies were sheep anti-OCRL1 (Choudhury et al., 2005, 2009) and rabbit anti–Golgin-97 (Nobuhiro Nakamura, Kyoto Sangyo University, Japan); rabbit anti-APPL1 (Joseph Testa, Fox Chase Cancer Center, Philadelphia, PA); rabbit anti-GM130, sheep anti–Golgin-84, sheep anti-GFP (Diao et al., 2003), sheep anti-GST (Hyvola et al., 2006), and sheep anti-TGN46 (Vas Ponnambalam, University of Leeds, UK); goat anti-EEA1 (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-CIMPR (Paul Luzio, University of Cambridge, UK); and rabbit anti-Rab11 (Sigma). Monoclonal antibodies were mouse anti-CD63, mouse anti-TfR, and mouse anti-lysosome–associated membrane protein-1 (LAMP1; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), mouse anti-CD8 (clone UCHT-4, Sigma), mouse anti-EEA1 (BD Biosciences, Sparks, MD), mouse anti–clathrin X22 (Liz Smythe, University of Sheffield, UK), and mouse anti-MBP (New England Biolabs, Ipswich, MA). Fluorophore (Alexa 594, Alexa 488, Cy3, and Cy5) and horseradish peroxidase–conjugated secondary antibodies were purchased from Molecular Probes (Eugene, OR), Tago Immunologicals (Camarillo, CA), or Invitrogen (Carlsbad, CA).
Molecular biology and yeast two-hybrid analysis
All constructs were made using standard molecular biology techniques. Full-length human IPIP27A and B complementary DNA (cDNA) was obtained from the MRC Geneservice as IMAGE clones 4943538 (FAM109A Acc. No: Q8N4B1) and 5205871 (FAM109B Acc. No: Q6ICB4). Full-length, PH domain (aa 1–120 IPIP27A, 1–135 IPIP27B), C terminus (aa 121–249 IPIP27A, 129–259 IPIP27B) and C-terminal truncations were cloned into pGAD-T7 (BD Biosciences) for yeast two-hybrid analysis or pGEX4T-2 or pMAL-C2 for bacterial expression of GST or MBP-tagged proteins, respectively. IPIP27A (aa 192–249, xCTii) and B (193–259, xCTii) fragments encoding the extreme C terminus were cloned into pGEX4T-2 or pMAL-C2 for production in bacteria. Full-length, PH domain (aa 1–114 IPIP27A, 1–123 IPIP27B), and C-terminus (aa 114–259 IPIP27A, 122–259) IPIP27 constructs were cloned into pEGFP-C2 (BD Biosciences) and modified pcDNA3.1 vectors (Stratagene, La Jolla, CA) containing an N-terminal MCherry or Myc tag for expression in mammalian cells. Mammalian and yeast two-hybrid expression constructs of OCRL1 and Inpp5b have been described previously (Choudhury et al., 2005,2009; Williams et al., 2007). The plasmid encoding MApple-Rab11 was a gift from Neftali Rodriguez and Philip Woodman (University of Manchester, UK). Point mutations were introduced by PCR using site-directed mutagenesis Quikchange method (Stratagene). Primer sequences for all manipulations are available upon request. All constructs were verified by DNA sequencing using the ABI PRISM Big Dye Terminator kit (version 2; PE Applied Biosystems (Waltham, MA). Yeast two-hybrid analysis was performed as previously described (Choudhury et al., 2005).
Cell culture and transfection
HeLa and hTERT-RPE1 cells were grown at 37°C and 5% CO2 in Dulbecco’s modified Eagle Medium (DMEM) or 1:1 Hams F12:DMEM respectively, supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1 mM l-glutamine. HeLa M cells stably expressing CD8-MPR were grown in the presence of G418 at 0.5 mg/ml. For nocodazole treatment, cells were incubated at 37ºC for 2 h in the presence of nocodazole at 5 μg/ml. Transient transfections were performed using FUGENE 6 (Roche Biochemicals, Indianapolis, IN) according to the manufacturer’s instructions. For low-level expression, 5–25 ng of cDNA was cotransfected with 0.5–1.0 μg of carrier DNA (pBluescript II SK).
RNA interference
RNA interference was performed on HeLa cells using Interferin (Polyplus Transfection, New York, NY) and siRNA oligos (Thermo Fisher Scientific, Dharmacon Products, Lafayette, CO) Lamin A (target sequence AACUGGACUUCCAGAAGAACA) and luciferase (GL2; Eurogentec, Freemont, CA) were used as negative controls. IPIP27A and B were targeted with 20 nM Dharmacon SMARTpool oligos or single oligos derived from the SMARTpool: IPIP27A oligo 1 (target sequence GUGCUGACCUAGUGGCGGA) or oligo 4 (GGUGACAGACUCAGCCCAA), or IPIP27B oligo 4 (UGGCCGAAGAUGCUGGUUU). IPIP27B was also targeted with oligo 5 (CGGCAGUGUUGGCAGAAGA). Similar results were obtained with the two independent oligos targeting each protein. Unless indicated otherwise, the results shown were obtained with oligo 4. Cells were analyzed 72 h after transfection.
Hexosaminidase assay
HeLa cells were washed 2× with cold phosphate-buffered saline (PBS) before incubation in DMEM (without phenol red) containing bovine serum albumin (BSA) at 2 mg/ml and 2.5 mM mannose 6-phosphate for 6 h at 37ºC, 5% CO2. Medium was collected, and the cells were washed 2× with cold PBS and extracted in 250 μl of cell extraction buffer (10 mM sodium phosphate, pH 6.0, 0.5% (wt/vol) Triton X-100, 0.15 M NaCl) for 10 min on ice. Lysates and media were cleared by centrifugation in a microfuge for 5 min at 4°C at 13,500 rpm. Medium (0.5 ml) was added to 120 μl of 5× assay buffer (0.5 M sodium acetate, pH 4.4, 0.5% (wt/vol) Triton X-100, 5 mM 4-nitrophenyl-N-acetyl-beta-d-glucosaminide). Cell extract (50 μl) was added to 570 μl of 1× assay buffer (diluted from 5× assay buffer in water). Solutions were incubated overnight at 37ºC. Reactions were stopped by adding 600 μl of stop buffer (0.5 M glycine, 0.5 M Na2CO3 pH 10.0), and absorbance was measured at 405 nm using a spectrophotometer. Data were analyzed using GraphPad Instat software (GraphPad Software, La Jolla, CA), and statistics were performed using the one-way analysis of variance (ANOVA) test.
Cathepsin-D trafficking
HeLa cells were incubated for 37ºC in labeling medium (DMEM without Met or Cys, containing 10% (vol/vol) dialyzed FBS, 1 mM glutamine, 0.2 U/ml penicillin, 100 µg/ml streptomycin for 1 h followed by a 20-min incubation in labeling medium containing 50 μCi/ml 35S Met/Cys. Cells were washed and incubated in chase medium (DMEM, 10 mM HEPES, pH 7.4, BSA at 1 mg/ml, 1 mM cold Met, 1 mM cold Cys, 2.5 mM mannose 6-phosphate) for between 0 and 4 h at 37ºC. The medium was removed, and protease inhibitors were added and placed on ice. Cells were extracted in lysis buffer (20 mM HEPES, pH 7.4, 0.15 M NaCl, 0.5% (wt/vol) Triton X-100, 2 mM EDTA, protease inhibitors) for 15 min on ice. Medium and cell lysate samples were centrifuged at 13,500 rpm for 10 min at 4ºC to clarify and were incubated with 2.5 μl of anti–cathepsin D, and 20 μl of protein A-sepharose were added to supernatants and incubated overnight at 4ºC. Beads were washed 3× with cold lysis buffer and eluted by boiling in 30 μl of 2× SDS nonreducing sample buffer. Proteins were analyzed by SDS–PAGE and autoradiography, and bands were quantitated using ImageJ software. Statistics were performed using the one-way ANOVA test with GraphPad Instant software.
Shiga Toxin trafficking
HeLa cells grown on coverslips were placed cell-side up on parafilm and incubated with 40 μl of carbonate-free DMEM (10% (vol/vol) FBS, 1 mM glutamine, 0.2 U/ml penicillin, 100 µg/ml streptomycin) containing Cy3-STxB at 0.4 μg/ml for 30 min on ice. Cells were washed 2× with cold PBS and either fixed directly or incubated for various times at 37ºC prior to fixation.
CD8-MPR trafficking
HeLaM stably expressing CD8-MPR were incubated in carbonate-free DMEM (10% [vol/vol] FBS, 1mM glutamine, 0.2 U/ml penicillin, 100 µg/ml streptomycin) for 15 min on ice. Coverslips were placed cell-side up on parafilm and incubated with 40 μl of carbonate-free DMEM containing 0.8 μg of mouse anti-CD8 antibody for 30 min on ice. Cells were washed 2× in cold PBS and either fixed directly or incubated at 37ºC for various times prior to fixation.
Transferrin trafficking
HeLa cells were preincubated for 30 min at 37ºC in serum-free DMEM (1 mM glutamine, BSA at 2 mg/ml) and then incubated for a further 30 min in the same medium containing Alexa 488-Tf at 5 μg/ml. Cells were washed 3× in PBS and incubated in DMEM containing unlabelled holo-transferrin at 100 μg/ml for 15 or 30 min at 37ºC before fixation.
EGF degradation
HeLa cells were grown overnight at 37ºC in serum-free DMEM (1 mM glutamine, BSA at 2 mg/ml) and then incubated for 4 min at 37ºC in the same medium containing Alexa 488-EGF at 5 ng/ml. Cells were washed 3× in PBS and incubated in DMEM for a further 4 h at 37ºC before fixation.
Immunofluorescence microscopy
For immunofluorescence microscopy, cells were grown on coverslips and fixed in 3% (wt/vol in PBS) paraformaldehyde at room temperature for 20 min. Coverslips were incubated for 20 min at room temperature with primary antibodies diluted in PBS containing BSA at 0.5 mg/ml. Cells were washed 3× with PBS and incubated for a further 20 min at room temperature with fluorophore-conjugated secondary antibodies diluted in PBS/BSA. In some experiments, the DNA dye Hoechst 33342 (200 ng/ml) was included in the second incubation. Coverslips were mounted in Mowiol 4-88 (Merck, Whitehouse Station, NJ), allowed to dry, and analyzed using an Olympus BX60 upright microscope equipped with a MicroMax cooled, slow-scan CCD camera (Roper Scientific, Sarasota, FL) driven by Metaview software (University Imaging Corporation, Sunnyvale, CA). Images were processed using Adobe Photoshop CS2 and quantified using ImageJ software. Statistics were performed using the one-way ANOVA test with GraphPad Instant software.
Immunoprecipitation
Extracts were prepared from HeLa cells by washing the cells in PBS, then extracting them in 1 ml of HMNT (20 mM HEPES, pH 7.4, 5 mM MgCl2, 0.1 M NaCl2, 0.5% (wt/vol) Triton X-100, and Protease Inhibitor Cocktail III) for 15 min on ice. Cell extracts were clarified by centrifugation at 13,500 rpm for 15 min at 4ºC in a microfuge. The appropriate purified immunoglobulin G (IgG) (2 μg) and 20 μl of protein G or A-Sepharose (50% slurry) were added, and, after incubation for 1–4 h at 4ºC, beads were washed 3× with HMNT prior to elution in SDS sample buffer.
Protein pull-downs
Binding was performed by incubating 250 μl of HMNT HeLa cell extract with 10 μg of bait protein bound to protein S-agarose, GSH-Sepharose, or amylose resin for 1 h at 4°C. Competition binding with recombinant proteins was performed by coupling 1 μg of His/S-tagged OCRL1 to protein S-agarose in 100 μl of HMNT containing BSA at 100 μg/ml. GST APPL1 (aa 403–413; 0.3 μg) was added along with increasing molar ratios of the relevant MBP or GST-tagged protein (between 0.3 and 30 μg) and was incubated at 4°C for 2 h. After washing 3× times with HMNT, bound proteins were eluted by boiling in SDS sample buffer and analyzed by Western blotting with appropriate antibodies.
Supplementary Material
Acknowledgments
We thank our colleagues for providing reagents as noted in the text. We thank Philip Woodman for critically reading the manuscript. This work was supported by a BBSRC (Biotechnology and Biological Sciences Research Council)-funded PhD studentship awarded to CJN and a Wellcome Trust project grant (082923) awarded to ML.
Abbreviations used:
- APPL1, adaptor protein, phosphotyrosine binding, PH domain, and leucine zipper containing 1
ASH, ASPM, SPD-2, Hydin
- BSA
bovine serum albumin
- cDNA
complementary DNA
- CIMPR
cation-independent mannose 6-phosphate receptor
- EGF
epidermal growth factor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- IPIP27
inositol polyphosphate phosphatase interacting protein of 27 kDa
- LAMP1
lysosome-associated membrane protein 1
- MBP
maltose binding protein
- OCRL
oculocerebrorenal syndrome of Lowe
- PBS
phosphate-buffered serum
- PH
pleckstrin homology
- PI
phosphoinositide
- STxB
Shiga toxin B-subunit
- siRNA
small interfering RNA
- TfR
transferrin receptor
- TGN
trans-Golgi network
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0730) on January 13, 2011.
REFERENCES
- Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL, Lewis RA, McInnes RR, Nussbaum RL. The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- Balla T, Szentpetery Z, Kim YJ. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R et al. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell. 2005;16:3467–3479. doi: 10.1091/mbc.E05-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M. Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem. 2009;284:9965–9973. doi: 10.1074/jbc.M807442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen EI, Birn H. Megalin and cubilin: multifunctional endocytic receptors. Nat Rev Mol Cell Biol. 2002;3:256–266. doi: 10.1038/nrm778. [DOI] [PubMed] [Google Scholar]
- Christensen EI, Devuyst O, Dom G, Nielsen R, Van Der Smissen P, Verroust P, Leruth M, Guggino WB, Courtoy PJ. Loss of chloride channel ClC-5 impairs endocytosis by defective trafficking of megalin and cubilin in kidney proximal tubules. Proc Natl Acad Sci USA. 2003;100:8472–8477. doi: 10.1073/pnas.1432873100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon BG, Mukherjee D, Hanna CB, Riese DJ 2nd, Lowe M, Aguilar RC. Lowe syndrome patient fibroblasts display Ocrl1-specific cell migration defects that cannot be rescued by the homologous Inpp5b phosphatase. Hum Mol Genet. 2009;18:4478–4491. doi: 10.1093/hmg/ddp407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Guerriero CJ, Szalinski CM, Kinlough CL, Hughey RP, Weisz OA. OCRL1 function in renal epithelial membrane traffic. Am J Physiol Renal Physiol. 2010;298:F335–F345. doi: 10.1152/ajprenal.00453.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson HW. Wortmannin causes mistargeting of procathepsin D. Evidence for the involvement of a phosphatidylinositol 3-kinase in vesicular transport to lysosomes. J Cell Biol. 1995;130:797–805. doi: 10.1083/jcb.130.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Diao A, Rahman D, Pappin DJ, Lucocq J, Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell. 2007;13:377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucherre A, Desbois P, Nagano F, Satre V, Lunardi J, Gacon G, Dorseuil O. Lowe syndrome protein Ocrl1 is translocated to membrane ruffles upon Rac GTPase activation: a new perspective on Lowe syndrome pathophysiology. Hum Mol Genet. 2005;14:1441–1448. doi: 10.1093/hmg/ddi153. [DOI] [PubMed] [Google Scholar]
- Faucherre A, Desbois P, Satre V, Lunardi J, Dorseuil O, Gacon G. Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum Mol Genet. 2003;12:2449–2456. doi: 10.1093/hmg/ddg250. [DOI] [PubMed] [Google Scholar]
- Fraldi A et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–3620. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- Giot L et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes RR Jr et al. Dent Disease with mutations in OCRL1. Am J Hum Genet. 2005;76:260–267. doi: 10.1086/427887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol. 2010;22:528–534. doi: 10.1016/j.ceb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J. 2006;25:3750–3761. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Suchy SF, Bernard D, MacDonald M, Crawley J, Grinberg A, Wynshaw-Boris A, Westphal H, Nussbaum RL. Functional overlap between murine Inpp5b and Ocrl1 may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J Clin Invest. 1998;101:2042–2053. doi: 10.1172/JCI2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson AB, Majerus PW. Mutation of the conserved domains of two inositol polyphosphate 5-phosphatases. Biochemistry. 1996;35:7890–7894. doi: 10.1021/bi9602627. [DOI] [PubMed] [Google Scholar]
- Johannes L, Popoff V. Tracing the retrograde route in protein trafficking. Cell. 2008;135:1175–1187. doi: 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Jovic M, Kieken F, Naslavsky N, Sorgen PL, Caplan S. Eps15 homo-logy domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol Biol Cell. 2009;20:2731–2743. doi: 10.1091/mbc.E08-11-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgos LE, Isaac EL, Brooks DA, Ravenscroft EM, Davey R, Hopwood JJ, Meikle PJ. Lysosomal biogenesis in lysosomal storage disorders. Exp Cell Res. 1997;234:85–97. doi: 10.1006/excr.1997.3581. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lowe M. Structure and function of the Lowe syndrome protein OCRL1. Traffic. 2005;6:711–719. doi: 10.1111/j.1600-0854.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- Mao Y, Balkin DM, Zoncu R, Erdmann KS, Tomasini L, Hu F, Jin MM, Hodsdon ME, De Camilli P. A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J. 2009;28:1831–1842. doi: 10.1038/emboj.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzaris M, Jackson SP, Laxminarayan KM, Speed CJ, Mitchell CA. Identification and characterization of the phosphatidylinositol-(4, 5)-bisphosphate 5-phosphatase in human platelets. J Biol Chem. 1994;269:3397–3402. [PubMed] [Google Scholar]
- McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- Mills IG, Praefcke GJ, Vallis Y, Peter BJ, Olesen LE, Gallop JL, Butler PJ, Evans PR, McMahon HT. EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J Cell Biol. 2003;160:213–222. doi: 10.1083/jcb.200208023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden AG et al. Urinary megalin deficiency implicates abnormal tubular endocytic function in Fanconi syndrome. J Am Soc Nephrol. 2002;13:125–133. doi: 10.1681/ASN.V131125. [DOI] [PubMed] [Google Scholar]
- Nussbaum R, Suchy SF. In: Lowe syndrome. In: Metabolic and molecular basis of inherited diseases. Scriver CR, Beauder AL, Sly WS, Valle D, editors. IV, ed. New York: McGraw-Hill; 2001. pp. 6257–6266. vol. [Google Scholar]
- Olivos-Glander IM, Janne PA, Nussbaum RL. The oculocerebrorenal syndrome gene product is a 105-kD protein localized to the Golgi complex. Am J Hum Genet. 1995;57:817–823. [PMC free article] [PubMed] [Google Scholar]
- Parkinson-Lawrence EJ, Shandala T, Prodoehl M, Plew R, Borlace GN, Brooks DA. Lysosomal storage disease: revealing lysosomal function and physiology. Physiology (Bethesda) 2010;25:102–115. doi: 10.1152/physiol.00041.2009. [DOI] [PubMed] [Google Scholar]
- Saint-Pol A et al. Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev Cell. 2004;6:525–538. doi: 10.1016/s1534-5807(04)00100-5. [DOI] [PubMed] [Google Scholar]
- Schmid AC, Wise HM, Mitchell CA, Nussbaum R, Woscholski R. Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett. 2004;576:9–13. doi: 10.1016/j.febslet.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Schurman SJ, Scheinman SJ. Inherited cerebrorenal syndromes. Nat Rev Nephrol. 2009;5:529–538. doi: 10.1038/nrneph.2009.124. [DOI] [PubMed] [Google Scholar]
- Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HW et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchy SF, Cronin JC, Nussbaum RL. Abnormal bradykinin signaling in fibroblasts deficient in the PIP(2) 5-phosphatase, ocrl1. J Inherit Metab Dis. 2009;32:280–288. doi: 10.1007/s10545-009-1058-3. [DOI] [PubMed] [Google Scholar]
- Suchy SF, Nussbaum RL. The deficiency of PIP2 5-phosphatase in Lowe syndrome affects actin polymerization. Am J Hum Genet. 2002;71:1420–1427. doi: 10.1086/344517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan LE, Tomasini L, Pirruccello M, Lunardi J, De Camilli P. Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1. Proc Natl Acad Sci USA. 2010;107:3511–3516. doi: 10.1073/pnas.0914658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell A, Ward ME, Ungewickell E, Majerus PW. The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc Natl Acad Sci USA. 2004;101:13501–13506. doi: 10.1073/pnas.0405664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell AJ, Majerus PW. Increased levels of plasma lysosomal enzymes in patients with Lowe syndrome. Proc Natl Acad Sci USA. 1999;96:13342–13344. doi: 10.1073/pnas.96.23.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA. Phosphoinositides as regulators of membrane trafficking in health and disease. Cell Mol Life Sci. 2008;65:2833–2841. doi: 10.1007/s00018-008-8353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Choudhury R, McKenzie E, Lowe M. Targeting of the type II inositol polyphosphate 5-phosphatase INPP5B to the early secretory pathway. J Cell Sci. 2007;120:3941–3951. doi: 10.1242/jcs.014423. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Zhang Z, Degeest G, Mortier E, Leenaerts I, Coomans C, Schulz J, N’Kuli F, Courtoy PJ, David G. Syndecan recycling [corrected] is controlled by syntenin-PIP2 interaction and Arf6. Dev Cell. 2005;9:377–388. doi: 10.1016/j.devcel.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Perera RM, Balkin DM, Pirruccello M, Toomre D, De Camilli P. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 2009;136:1110–1121. doi: 10.1016/j.cell.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.