Through proteomic identification of sumoylated proteins, combined with detailed molecular and cellular analyses of conditional and null smt3/smt3 mutants, the small ubiquitin-like modifier, SUMO, is shown to play key roles in cellular growth and stress adaptation of the major fungal pathogen Candida albicans.
Abstract
Posttranslational modifications of proteins play critical roles in the control of cellular differentiation, development, and environmental adaptation. In particular, the covalent attachment of the small ubiquitin-like modifier, SUMO, to target proteins (sumoylation) regulates cell cycle progression, transcription, nucleocytoplasmic transport, and stress responses. Here we combine proteomic, molecular, and cellular approaches to examine the roles of sumoylation in the major fungal pathogen of humans, Candida albicans. Using an N-terminally FLAG-tagged SUMO, 31 sumoylated proteins were identified in C. albicans with roles in stress responses (e.g., Hsp60, Hsp70 family members, Hsp104), the cytoskeleton and polarized growth (e.g., Tub1, Cct7, Mlc1), secretion, and endocytosis (e.g., Lsp1, Sec24, Sec7). The output from this proteomic screen was entirely consistent with the phenotypes of C. albicans mutants in which the single SUMO-encoding locus (SMT3) was inactivated or down-regulated. C. albicans smt3/smt3 cells displayed defects in growth, morphology, cell separation, nuclear segregation, and chitin deposition, suggesting important roles for sumoylation in cell cycle control. Smt3/smt3 cells also displayed sensitivity to thermal, oxidative, and cell wall stresses as well as to the antifungal drug caspofungin. Mutation of consensus sumoylation sites in Hsp60 and Hsp104 affected the resistance of C. albicans to thermal stress. Furthermore, signaling via the cell integrity pathway was defective in C. albicans smt3/smt3 cells. These observations provide mechanistic explanations for many of the observed phenotypic effects of Smt3 inactivation upon C. albicans growth and environmental adaptation. Clearly sumoylation plays key roles in fundamental cellular processes that underpin the pathogenicity of this medically important fungus.
INTRODUCTION
All organisms must respond effectively to environmental change if they are to survive. In particular, fungal pathogens have developed robust stress responses that help them to counteract the antimicrobial defenses of their human host and promote the colonization of specific niches. For example, the major opportunistic pathogen of humans, Candida albicans, is a relatively harmless commensal of the oral cavity, gastrointestinal tract, and genitalia of humans (Odds, 1988; Calderone, 2002). However, defects in host immunity, physical injury, or medical intervention can allow C. albicans to access different internal organs and tissues, causing potentially fatal infections. C. albicans encounters potentially damaging reactive oxidative species in some niches through the action of host defenses (Enjalbert et al., 2007). However, C. albicans has evolved effective oxidative stress responses that promote survival in the host (Alonso-Monge et al., 1999; Hwang et al., 2002; Chauhan et al., 2006; Brown et al., 2009). Furthermore, evolutionarily conserved thermal adaptation mechanisms help C. albicans tune the levels of essential chaperones to the temperature of host niches (Nicholls et al., 2009). These and other types of stress response combine to promote the virulence of this major pathogen.
Posttranslational modifications of proteins play critical roles in the cellular adaptation of all organisms as well as their growth, division, differentiation, and development. Such modifications include phosphorylation, ubiquitination, sumoylation, glycosylation, and methylation. These modifications provide essential mechanisms by which the functions, activities, and stabilities of preexisting proteins can be rapidly and specifically modulated, thereby controlling dynamic cellular processes. Phosphorylation is known to be essential for the oxidative and thermal stress adaptation of C. albicans (Smith et al., 2004; Nicholls et al., 2009). However, the impact of sumoylation in growth and environmental adaptation has not been investigated in this major pathogen.
SUMO is a small ubiquitin-like modifier that is covalently attached to proteins to modulate their activity. SUMO belongs to a family of structurally related proteins of which ubiquitin is the most prominent member (Schwartz and Hochstrasser, 2003; Kerscher et al., 2006). The tagging of proteins with polyubiquitin often targets them for degradation via the 26S proteasome, whereas the addition of a single ubiquitin tag modulates the activity of other proteins, thereby affecting vesicular trafficking or protein kinase activation, for example (Pickart, 2001). SUMO tags have only 8–15% sequence identity with ubiquitin, but they fold into a similar globular structure in which the carboxy-terminal tail is exposed, revealing the carboxy-terminal glycine residue necessary for ligation (Bayer et al., 1998).
The SUMO and ubiquitin conjugation pathways are distinct, but they share some similarities. First, SUMO is activated by a heterodimeric activating enzyme, E1, consisting of the Aos1 and Uba2 proteins in S. cerevisiae. Both of these proteins share sequence similarity to the corresponding ubiquitin-activating enzymes (Johnson et al., 1997; Okuma et al., 1999). Second, Ubc9, which shares strong sequence similarity to ubiquitin-conjugating E2 enzymes, catalyzes the formation of an isopeptide bond between the carboxy terminus of SUMO and an ε-lysine residue on the target protein (Gong et al., 1997; Johnson and Blobel, 1997; Schwarz et al., 1998). Third, specific cysteine proteases, one of which is encoded by the ULP1 gene, cleave SUMO-substrate bonds to release SUMO from its target protein (Li and Hochstrasser, 1999). The genes encoding Aos1, Uba2, Ubc9, and Ulp1 are all required for cell growth and division in yeast (Johnson, 2004).
Unlike mammalian cells, Saccharomyces cerevisiae encodes only one form of SUMO, Smt3. SMT3 is an essential gene in S. cerevisiae, as is the protein SUMO-1 in mammalian cells (Johnson and Hochstrasser, 1997). Smt3 is conjugated to target proteins via an isopeptide bond to lysine residues that lie within the consensus sequence Ψ-K-X-E, where Ψ is a large hydrophobic amino acid, K is the lysine to which SUMO is conjugated, X is any amino acid, and E is glutamic acid (Melchior, 2000; Yeh et al., 2000; Muller et al., 2001). Sumoylation of target proteins in this manner can regulate their function through a number of different mechanisms. These include the regulation of their subcellular localization, the modulation of their protein–protein interactions, and the inhibition of ubiquitin attachment or of other lysine-targeting modifications (Melchior, 2000; Hay, 2001; Kim et al., 2002). In mammalian cells, the abundance of SUMO conjugates increases in response to heat and oxidative stress (Mao et al., 2000; Saitoh and Hinchey, 2000). Furthermore, data in both budding and fission yeast point to broad roles for sumoylation and desumoylation in the regulation of many biological processes, including cell cycle progression, intracellular transport, stress responses, signal transduction, transcription, and the DNA damage response (Schwienhorst et al., 2000; Seeler and Dejean, 2003; Wohlschlegel et al., 2004; Zhou et al., 2004).
The C. albicans Smt3 protein has 62% sequence identity with S. cerevisiae Smt3 and 48% identity with human SUMO-1. Presumably SUMO plays vital roles in growth and stress adaptation in this pathogen, as it does in its benign cousin S. cerevisiae. Therefore our aim in this study was to examine the roles of SMT3 in this pathogen by screening for sumoylation targets in C. albicans using a proteomic approach. Proteins with roles in growth and stress adaptation were identified. Consistent with this, we have shown C. albicans cells lacking Smt3 grow with elongated buds, show defects in cell separation and nuclear segregation, and display aberrant signaling and a range of stress sensitivities. Furthermore we have demonstrated that mutations of the consensus sumoylation sites in two of our identified sumoylation targets, Hsp104 and Hsp60, essentially replicate the temperature-sensitive and morphological phenotypes of Smt3-depleted cells. Our data indicate that sumoylation plays important roles in key cellular processes required for the pathogenicity of this clinically important fungus.
RESULTS
Numerous proteins are sumoylated in C. albicans
To determine the roles that sumoylation might be playing in C. albicans, we set out to identify sumoylation targets in this fungal pathogen. First we epitope tagged Smt3 in C. albicans to facilitate identification of sumoylated proteins in yeast (MLC01; Table 1). The carboxy-terminal region of S. cerevisiae Smt3 is processed and covalently attached to the lysine residues of substrate proteins. Therefore we FLAG tagged C. albicans Smt3 at its amino terminus, expressing the epitope-tagged construct from the ACT1 promoter on pACT1-FLAG-SMT3 (Materials and Methods). This epitope tagging did not affect the growth of MLC01 cells compared with their parental control (THE1 cells).
Table 1:
C. albicans strains
| Strain | Genotype | Source |
|---|---|---|
| BWP17 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG | Wilson et al. (1999) |
| THE1 | ade2::hisG/ade2::hisG, ura3::λ imm434/ura3::λ imm434, ENO1/eno1::ENO1-tetR-ScHAP4AD-3XHA-ADE2 | Nakayama et al. (2000) |
| MLC01 | ade2::hisG/ade2::hisG, ura3::λ imm434/ura3::λ imm434, ENO1/eno1::ENO1-tetR-ScHAP4AD-3XHA-ADE2 RPS1::ACT1p-FLAG-SMT3 | This study |
| MLC02 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-SMT3/SMT3 | This study |
| MLC04 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-SMT3/smt3::loxP-ARG4-loxP | This study |
| MLC10 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-SMT3/SMT3 | This study |
| MLC13 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-SMT3/smt3::loxP-ARG4-loxP | This study |
| MLC33 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, SMT3/smt3::loxP-URA3-loxP | This study |
| MLC34 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, SMT3/smt3::loxP-URA3-loxP | This study |
| MLC37 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, smt3::loxP-URA3-loxP/smt3::loxP-ARG4-loxP | This study |
| MLC38 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, smt3::loxP-URA3-loxP/smt3::loxP-ARG4-loxP | This study |
| MLC39 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, smt3::loxP-URA3-loxP/smt3::loxP-ARG4-loxP | This study |
| MLC43 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG, RPS1::pACT1p-FLAG-SMT3 (URA3) | This study |
| MLC46 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG, HSP60/hsp60::loxP-ARG-loxP | This study |
| MLC49 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG, HSP104/ hsp104::loxP-ARG-loxP | This study |
| MLC52 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG, HSP60-HIS1/ hsp60::loxP-ARG-loxP | This study |
| MLC55 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG, HSP60K324R-HIS1/ hsp60::loxP-ARG-loxP | This study |
| MLC56 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG, HSP104 -HIS1/ hsp104::loxP-ARG-loxP | This study |
| MLC59 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG, HSP104K356R -HIS1/ hsp104::loxP-ARG-loxP | This study |
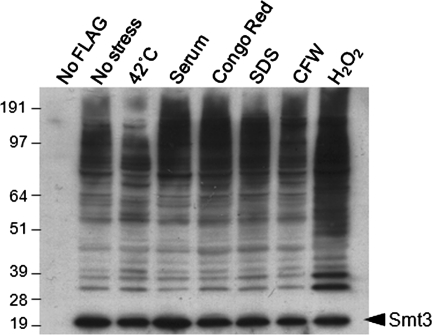
Before embarking on our proteomic screen for SUMO targets, we examined sumoylation in C. albicans under a range of experimental conditions. These were heat shock (30–42°C for 1 h), oxidative stress (50 mM H2O2 for 1 h), cell wall stress (SDS, Congo red, and calcofluor white for 1 h), morphogenesis (10% fetal calf serum [FCS] for 1 h), and untreated control. (A concentration of 50 mM H2O2 was chosen [Bossis and Melchior, 2006] because SUMO conjugation increases at high doses of oxidative stress, whereas sumoylation is decreased in response to low doses of H2O2 [≤1 mM].) Protein extracts were then prepared from these cells and subjected to Western blotting with an anti-FLAG antibody.
Minimal background was observed in control cells lacking the FLAG tag (THE1; Table 1) (Figure 1). A large number of bands were observed in untreated MLC01 cells, indicating that numerous proteins are sumoylated in unstressed C. albicans cells. Interestingly, significant differences were observed in the sumoylation banding patterns of untreated, heat-shocked, and peroxide-treated cells (Figure 1). For example, following peroxide stress, a number of new bands between 50 and 100 kDa were observed. These differences could result from cell cycle–regulated changes in the sumoylation of target proteins or from changes in the synthesis, modification, or degradation of target proteins. The observed changes in sumoylation patterns following heat shock and peroxide treatment were consistent with our working hypothesis that sumoylation probably plays significant roles during stress adaptation in C. albicans. Therefore we pursued sumoylation targets in untreated, heat-shocked, and peroxide-treated cells.
FIGURE 1:
Many C. albicans proteins are sumoylated. Cells expressing FLAG-Smt3 (MLC01) were grown for 5 h and then stressed for 1 h with the appropriate stress at the concentrations indicated in Materials and Methods. The parental C. albicans strain (THE1) was used for the no-FLAG control. Protein extracts were prepared and analyzed by Western blotting with an α-FLAG antibody. The band presumed to be FLAG-Smt3 is highlighted.
Identification of sumoylation targets
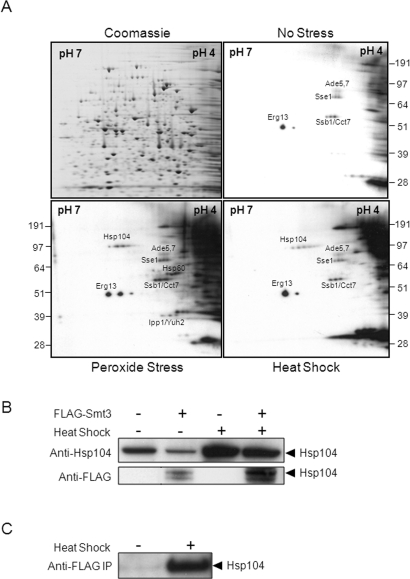
Proteins were extracted from untreated MLC01 (FLAG-Smt3) cells and from equivalent cells that were exposed to a 30–42°C heat shock or to 50 mM H2O2 for 1 h. These protein extracts were subjected to two-dimensional (2-D) gel electrophoresis, and replicate gels were stained with Coomassie blue or subjected to Western blotting using an anti-FLAG antibody. Sumoylated proteins were identified by aligning the Western blots and Coomassie-stained gels (Figure 2A). Spots that displayed reproducible effects in two independent replicate experiments were then cut from the Coomassie-stained gels, and the corresponding proteins were identified by tryptic digestion followed by tandem mass spectrometry (Materials and Methods).
FIGURE 2:
Identification of sumoylated proteins in C. albicans using a proteomic screen. (A) Cells expressing FLAG-Smt3 (MLC01) were grown for 5 h and then exposed to stress for 1 h. Protein extracts were prepared, run on replicate 2-D gels, and either stained with Coomassie blue or subjected to Western blotting with an α-FLAG antibody: Western blots of no-stress control, peroxide-treated cells (50 mM H2O2), and heat-shocked cells (30°C to 42°C). Autoradiographs were aligned with the Coomassie-stained gels, spots chosen for analysis, and the corresponding proteins identified by tryptic digestion and LC-MS/MS. The identities of some sumoylation targets are shown. (B) THE1 cells (FLAG-Smt3 −) and MLC01 cells (FLAG-Smt3 +) were heat shocked for 1 h, analyzed by Western blotting with an anti-Hsp104 antibody, and compared with untreated cells. Membranes were then reprobed with an anti-FLAG antibody. The band corresponding to the molecular mass of Hsp104 is highlighted. (C) Untreated and heat-shocked MLC01 cells were immunoprecipitated with an anti-FLAG antibody, and lysates were analyzed by Western blotting with the anti-Hsp104 antibody. The highlighted band corresponds to the molecular mass of Hsp104.
Table 2 presents those protein identifications that met several strict data quality criteria: a Mascot score of >35 (p < 0.05) and an excess of limit-digested peptides (EDLP) score of ≥1 (Stead et al., 2006). For those C. albicans proteins that were identified using our proteomic approach, we also looked for experimental evidence for sumoylation of their S. cerevisiae orthologue. In addition, we screened for potential sumoylation sites in the C. albicans protein sequence using the program SUMOsp version 2.0 (Ren et al., 2009) (Table 2).
Table 2:
Identification of sumoylation targets in C. albicans
| Sample ref. | Accession number | Protein name | Mol. wt. (Da) | pI | Function | Sumoylated in S. cerevisiae? | Predicted sumoylation site | Sumoylation detected in absence of stress? |
|---|---|---|---|---|---|---|---|---|
| Constitutively sumoylated | ||||||||
| 1 (1) | CA5549 | ERG13 | 49,952 | 5.67 | Ergosterol biosynthesis | Yes | Type I: Ψ-K-X-E position 311 | Yes |
| 8 (10) | CA0585 | ADE5,7 | 86,120 | 5.14 | Enzyme of adenine biosynthesis | ND | Type I: Ψ-K-X-E position 107 | Yes |
| 9 (11) | CA4672 | NSP1 | 74,656 | 5.15 | Essential component of the nuclear pore complex | ND | Type II: nonconsensus position 421, 672, and 715 (medium) | Yes |
| 9 (11) | CA1911 | SSE1 (MSI3) | 78,818 | 5.2 | Protein of the HSP70 family | Yes | Type I: Ψ-K-X-E position 94, 393, 610, and 630 Type II: nonconsensus position 681 | Yes |
| 30 (28) | CA3534 | SSB1 | 66,580 | 5.25 | Putative HSP70 family heat shock protein | Yes | Type I: Ψ-K-X-E position 245 | Yes |
| 30 (28) | CA2480 | CCT7 | 61,011 | 5.15 | Cytosolic chaperonin Cct ring complex, required for the assembly of actin and tubulins | ND | Type I: Ψ-K-X-E position 254 and 478 | Yes |
| 34 (8) | CA5920 | RPS12 | 15,973 | 4.63 | Putative ribosomal protein | ND | Type II: nonconsensus position 54 | Yes |
| 34 (8) | CA6058 | ATP16 | 17,571 | 5.15 | Subunit of the mitochondrial F1F0 ATP synthase | ND | Type II: nonconsensus position 115 (low) | Yes |
| 34 (8) | CA2454 | RPL23A | 13,199 | 10.1 | Putative ribosomal protein | ND | Type I: Ψ-K-X-E position 245 | Yes |
| 34 (8) | CA3045 | MLC1 | 15,899 | 4.64 | Protein with microtubule-dependent localization to the Spitzenkörper | ND | No sites predicted | Yes |
| Increased sumoylation in response to heat | ||||||||
| 22 (3) | CA3730 | SEC72 | 23,411 | 5.01 | Component of endoplasmic reticulum protein translocation complex | ND | Type I: Ψ-K-X-E position 31 and 103 | Yes |
| Increased sumoylation in response to H2O2 | ||||||||
| 6 (2) | CA0685 | ADO1 | 38,345 | 5.04 | Adenosine kinase | ND | Type I: Ψ-K-X-E position 19 | No |
| 6 (2) | CA0622 | LSP1 | 35,569 | 4.89 | Component of eisosomes, associated with endocytosis | ND | Type I: Ψ-K-X-E position 63 | No |
| 6 (2) | CA1024 | CAR1 | 34,649 | 4.88 | Arginine degradation | ND | Type II: nonconsensus position 13, 56, and 178 (low) | No |
| 6 (2) | CA5773 | DOT5 | 29,177 | 4.69 | Nuclear thiol peroxidase | ND | Type I: Ψ-K-X-E position 208 Type II: nonconsensus position 204 | No |
| 6 (2) | CA2582 | TAL1 | 35,661 | 4.63 | Transaldolase | Yes | Type I: Ψ-K-X-E position 271 | No |
| 7 (5) | CA0870 | IPP1 | 32,336 | 5.15 | Putative inorganic pyrophosphatase | Yes | Type I: Ψ-K-X-E position 58 and 233 | No |
| 7 (5) | CA0784 | YUH2 | 33,859 | 5.28 | Ubiquitin C-terminal hydrolase | ND | Type I: Ψ-K-X-E position 261Type II: nonconsensus position 311 | No |
| 10 (12) | CA1239 | HSP60 | 60,374 | 5.22 | Mitochondrial heat shock protein | Yes | Type I: Ψ-K-X-E position 324 | No |
| 12 (14) | CA4139 | THR4 | 57,723 | 5.18 | Threonine synthase | ND | Type I: Ψ-K-X-E position 505 | No |
| Increased sumoylation in response to heat and H2O2 | ||||||||
| 11 (13) | CA2474 | PDC11 | 62,744 | 5.39 | Pyruvate decarboxylase | Yes | Type II: nonconsensus position 177, 271, and 275 (medium) | No |
| 11 (13) | CA4875 | LAT1 | 50,145 | 5.73 | Component of pyruvate dehydrogenase complex | ND | Type I: Ψ-K-X-E position 153Type II: nonconsensus position 154 | No |
| 33 (7) | CA4474 | SSC1 | 69,876 | 5.48 | Heat shock protein | Yes | Type I: Ψ-K-X-E position 269 and 294 | No |
| 33 (7) | CA3534 | SSB1 | 66,580 | 5.25 | Putative HSP70 family heat shock protein | Yes | Type I: Ψ-K-X-E position 245 | No |
| 33 (7) | CA0838 | TFP1 | 63,060 | 5.24 | Subunit of vacuolar H+ −ATPase | ND | Type I: Ψ-K-X-E position 117 | No |
| 35 (9) | CA2239 | IPF10029 | 48,029 | 5.05 | Putative Cys-Gly dipeptidase involved in glutathione degradation | ND | Type I: Ψ-K-X-E position 71 and 336 | No |
| 35 (9) | CA4456 | ATP1 | 52,881 | 8.49 | Protein similar to ATP synthase α subunit | ND | TypeII: nonconsensus position 170, 419, and 529 (medium) | No |
| 35 (9) | CA5546 | TUB1 | 50,162 | 4.98 | α-Tubulin | Yes | Type II: nonconsensus position 97, 305, 337, and 340 (low) | No |
| 35 (9) | CA4331 | SGT1 | 47,368 | 4.92 | Cochaperone protein involved in kinetochore assembly | ND | Type I: Ψ-K-X-E position 119 and 245 | No |
| 23 (4) | CA5135 | HSP104 | 100,171 | 5.42 | Functional homologue of S. cerevisiae Hsp104p | ND | Type I: Ψ-K-X-E position 356 | No |
| 23 (4) | CA0958 | SEC24 | 102,683 | 5.26 | ER to Golgi transport | ND | Type I: Ψ-K-X-E position 214 | No |
This was not an exhaustive proteomic screen for sumoylated target proteins in C. albicans. For example, some sumoylated proteins in C. albicans might lie outside the isoelectric point (pI) range of our 2-D gels, or they might not be expressed under the conditions examined. Furthermore, only a small fraction of a particular target protein might be sumoylated in vivo (Hannich et al., 2005). Alternatively, some sumoylation targets might be low-abundance proteins that lie below the sensitivity of our analyses. Nevertheless, our aim in these experiments was to identify some proteins that are constitutively sumoylated in stressed and control cells and some proteins whose sumoylation increases either in response to heat shock or peroxide stress. Some of the gel spots we analyzed contained more than one protein, and it is conceivable that one or more of the proteins in these spots represent the sumoylation target. Nevertheless, a number of interesting sumoylation targets were identified in our proteomic screen.
Our proteomic approach revealed C. albicans proteins whose orthologues have been shown to be sumoylated in S. cerevisiae (Table 2). These included heat shock proteins (Hsp60, Ssb1, Ssc1, and Sse1), a key microtubule protein (Tub1), and metabolic enzymes (Erg13 and Pdc11). These proteins lend weight to the validity of our list of sumoylation targets in C. albicans. In addition, this list includes many proteins whose orthologues are not known to be sumoylated in S. cerevisiae but whose sequences contain potential sumoylation sites (Table 2). These include additional heat shock and stress proteins (Dot5, Hsp104, and Tfp1), cell cycle– and microtubule-related proteins (Cct7 and Sgt1), proteins associated with secretion and endocytosis (Lsp1, Sec24, and Sec72), an essential component of the nuclear pore complex (Nsp1), and a ubiquitin hydrolase (Yuh1) as well as additional metabolic enzymes (Ade5/7, Atp1, Atp16, Car1, Lat1, and Thr4).
Some of the sumoylation targets we identified appeared to be constitutively sumoylated in C. albicans based on the intensity of their signals on Western blots, whereas the sumoylation of other targets appeared to increase in response to heat shock and/or oxidative stress (Figure 2, Table 2). In many cases the functions of these sumoylation targets appeared to correlate with these sumoylation patterns. For example, the sumoylation of several heat shock proteins (Hsp60, Ssb1, Ssc1, and Hsp104) increased in response to stress.
To test the validity of our proteomic analyses, we further examined the link between Smt3 and Hsp104, which to our knowledge has not been shown to be sumoylated in S. cerevisiae. First, protein extracts from heat-shocked and control C. albicans cells expressing FLAG-Smt3 (MLC01; Table 1) and from control cells lacking the FLAG tag (THE1) were examined by Western blotting. Hsp104 was analyzed using an antibody kindly provided by Mick Tuite (Zenthon et al., 2006), revealing that Hsp104 levels increased in C. albicans upon heat shock, as expected (Figure 2B). A heat shock–inducible band of an equivalent size was observed in cells expressing FLAG-Smt3, but not in untagged control cells, when the same membranes were reprobed with the anti-FLAG antibody (Figure 2B). This observation was consistent with the idea that Hsp104 is sumoylated following heat shock.
The sumoylation of Hsp104 was tested further by coimmunoprecipitation. MLC01 lysates were immunoprecipitated with an anti-FLAG antibody, and these FLAG-Smt3 immunoprecipitates were then examined by Western blotting. The anti-Hsp104 antibody detected a band of ∼110 kDa in the heat-shocked lysates but not in the nonstressed controls (Figure 2C), confirming that C. albicans Hsp104 is sumoylated in response to heat shock.
SMT3 is not essential for viability in C. albicans
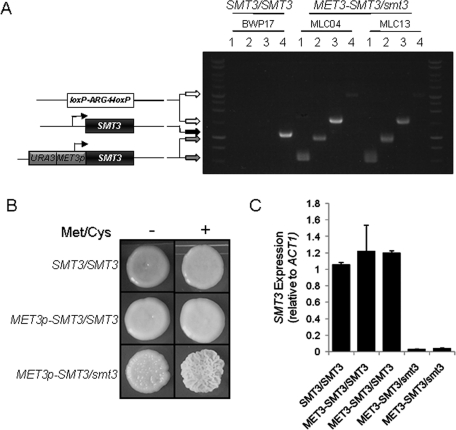
Our proteomic data supported our working hypothesis that sumoylation plays important roles in growth and environmental adaptation in C. albicans. To test this further, we wanted to examine the phenotypes of C. albicans smt3/smt3 mutants. However, SUMO conjugation is essential for viability in S. cerevisiae, as SMT3, UBA2, AOS1, UBC9, and ULP1 are all essential genes in this yeast (Dohmen et al., 1995; Seufert et al., 1995; Johnson et al., 1997; Li and Hochstrasser, 1999). We reasoned, therefore, that the single SUMO-encoding gene in C. albicans, SMT3, might also be essential for viability. Thus we first generated a conditional smt3 mutation in C. albicans using the methionine/cysteine-repressible MET3 promoter (Care et al., 1999). As C. albicans is constitutively diploid, the first SMT3 allele was placed under the control of the MET3 promoter, and the second SMT3 allele was deleted to create the independent conditional C. albicans MET3p-SMT3/smt3 mutants MLC04 and MLC13 (Figure 3A).
FIGURE 3:
Conditional C. albicans MET3p-SMT3/smt3 mutants can grow in the presence of methionine and cysteine. (A) Construction of the methionine-conditional C. albicans mutants MLC4 and MLC13 from the parental strain BWP17 (Table 1). One SMT3 allele was disrupted by insertional inactivation using the loxP-ARG4-loxP cassette, and the other allele was placed under the control of the MET3 promoter. Cartoons represent the structure of these alleles, and the arrows indicate the lengths of the diagnostic PCR products on the agarose gel: PCR reactions 1, primers MET3p-F and SMT3d-R (Supplemental Material); PCR reactions 2, primers MET3p-F and SMT3d2-R; PCR reactions 3, primers SMT3d2-F and LALd-R; PCR reactions 4, primers SMT3d2-F and SMT3d2-R. (B) Growth of the conditional C. albicans MET3p-SMT3/smt3 mutants in the presence of methionine and cysteine on plates. YPD plates contained (+) or lacked (−) 2.5 mM methionine (Met) and cysteine (Cys): SMT3/SMT3 (BWP17), MET3p-SMT3/SMT3 (MLC02), and MET3p-SMT3/smt3 (MLC04) (Table 1). (C) qRT-PCR quantification of SMT3 mRNA levels relative to the internal ACT1 mRNA control in cells grown with 2.5 mM Met/Cys for 4 h. Independent MET3p-SMT3/SMT3 (MLC02, MLC10) and MET3p-SMT3/smt3 strains (MLC04, MLC13) were analyzed.
To test the impact of down-regulating SMT3 in C. albicans, the wild-type strain (SMT3/SMT3), the heterozygous strain (MET3p-SMT3/SMT3), and the conditional mutants (MET3p-SMT3/smt3) were plated on yeast peptone dextrose (YPD) medium containing or lacking 2.5 mM methionine and cysteine. As expected, all strains exhibited normal growth in the absence of methionine and cysteine (Figure 3B). Also, the control strains grew normally in the presence of methionine and cysteine. Unexpectedly, given the essentiality of sumoylation in S. cerevisiae (Dohmen et al., 1995; Seufert et al., 1995; Johnson et al., 1997; Li and Hochstrasser, 1999), the C. albicans MET3p-SMT3/smt3 cells also grew in the presence of methionine and cysteine, albeit displaying a wrinkly colonial phenotype (Figure 3B). This suggested that SMT3 is not essential for the growth of C. albicans. The strains were also examined in liquid culture, where they continued to grow in the presence of methionine and cysteine even after overnight incubation.
It was possible that the continued growth of MET3p-SMT3/smt3 cells in the presence of methionine and cysteine was due to ineffective repression of the MET3p-SMT3 allele under these conditions. Therefore we tested this by quantitative real-time PCR (qRT-PCR), measuring SMT3 mRNA levels relative to the internal ACT1 mRNA control (Figure 3C). qRT-PCR was performed on RNA extracted from wild-type cells (BWP17: SMT3/SMT3) (Table 1), the conditional heterozygotes (MLC02 and MLC10: MET3p-SMT3/SMT3), and the conditional mutants (MLC04 and MLC13: MET3p-SMT3/smt3) grown for 4 h in either the absence or the presence of 2.5 mM methionine and cysteine. As expected, following repression by methionine and cysteine, SMT3 mRNA levels remained high in control cells but were reduced to negligible levels in the conditional MET3p-SMT3/smt3 mutants (Figure 3C).
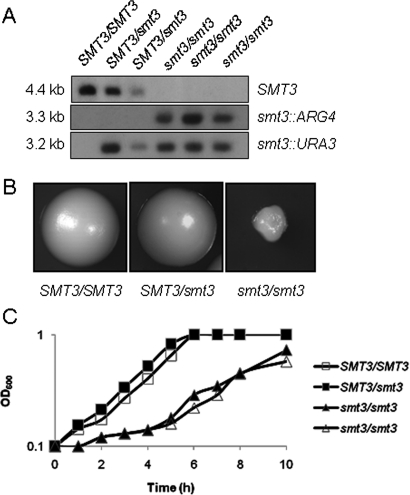
Although the MET3p-SMT3 allele was effectively repressed by methionine and cysteine in the conditional mutant, some residual SMT3 expression remained (Figure 3C). Hence it remained formally possible that this residual SMT3 expression was sufficient to support the growth of C. albicans (Figure 3B). Consequently, we created independent C. albicans smt3/smt3 null mutants (smt3::URA3/smt3::ARG4: MLC37, MLC38, MLC39; Table 1), confirming their genotypes by Southern blotting (Figure 4A) and diagnostic PCR. These smt3/smt3 cells grew slowly on plates and in liquid media (Figure 4, B and C), suggesting that SMT3 is required for normal growth of C. albicans. Nevertheless, these mutants were viable, indicating that SMT3 is not an essential gene in C. albicans.
FIGURE 4:
SMT3 is not an essential gene in C. albicans. (A) Confirmation of C. albicans smt3/smt3 mutants by Southern blotting of HindIII-digested C. albicans genomic DNA with PCR-amplified probes against the SMT3, ARG4, and URA3 open reading frames: SMT3/SMT3 (BWP17), SMT3/smt3 (MLC33 and MLC34), and smt3/smt3 (MLC37, MLC38, MLC39). (B) Colonial growth of C. albicans smt3 mutants: SMT3/SMT3 (BWP17), SMT3/smt3 (MLC33), and smt3/smt3 (MLC37). Images were taken at the same magnification. (C) Growth of C. albicans smt3 mutants in liquid YPD at 30°C: SMT3/SMT3, open boxes (BWP17); SMT3/smt3, closed boxes (MLC33); smt3/smt3, open and closed triangles (MLC37, MLC38).
Depletion of Smt3 causes cell cycle–related phenotypes
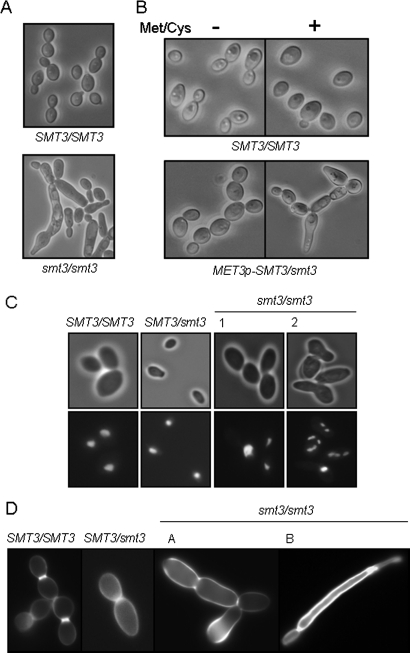
SUMO is required for the normal execution of the cell cycle in S. cerevisiae. Mutants deficient in SUMO conjugation accumulate at G2/M in the cell cycle with short spindles, unseparated sister chromatids, and undivided nuclei (Johnson et al., 1997; Li and Hochstrasser, 1999; Biggins et al., 2001). Furthermore, we had identified sumoylation targets with important roles in growth and cell division (e.g., Tub1 and Mlc1; Table 2). We reasoned, therefore, that Smt3-deficient C. albicans cells might also display cell cycle–related defects. Notably, smt3/smt3 colonies were significantly smaller and displayed rough, asymmetrical phenotypes compared with the larger, smooth, waxy appearance of control colonies (Figure 4B). Smt3/smt3 cells also showed aberrant morphological phenotypes in liquid culture. They displayed significant size variation, often forming enlarged, elongated buds and often remaining attached end-to-end in a pseudohyphal-like growth pattern (Figure 5A). Hence smt3/smt3 cells displayed heterogeneous behavior with regard to cell separation. Repression of the MET3p-SMT3 allele in the conditional MET3p-SMT/smt3 cells phenocopied this behavior of the smt3/smt3 null mutant (Figure 5B). Therefore Smt3 depletion has a significant impact upon cell growth and separation in C. albicans.
FIGURE 5:
C. albicans smt3 mutants display cell cycle–related defects. (A) Light microscopy of C. albicans smt3/smt3 cells grown in YPD at 30°C reveals morphological abnormalities and cell separation defects: SMT3/SMT3 (BWP17) and smt3/smt3 (MLC37). (B) C. albicans MET3p-SMT3/smt3 cells grown with 2.5 mM methionine and cysteine (Met/Cys) phenocopy smt3 null mutants: SMT3/SMT3 (BWP17) and MET3p-SMT3/smt3 (MLC04). (C) DAPI staining of C. albicans smt3/smt3 cells grown in YPD at 30°C reveals aberrant nuclear segregation in some cells: SMT3/SMT3 (BWP17), SMT3/smt3 (MLC33), and smt3/smt3 (MLC37). (D) Calcofluor white staining of C. albicans smt3/smt3 cells reveals aberrant chitin deposition in some cells: SMT3/SMT3 (BWP17), SMT3/smt3 (MLC33), and smt3/smt3 (MLC37). Images were taken at the same magnification and with the same exposures.
To examine the effects of Smt3 depletion upon nuclear segregation, smt3/smt3 cells were stained with DAPI. Interestingly, smt3/smt3 cells displayed significant heterogeneity, with some enlarged cells being multinucleate and others containing a single nucleus (Figure 5C). Therefore Smt3 depletion caused population heterogeneity with respect to nuclear segregation. Once again, this behavior was phenocopied by the conditional MET3p-SMT3/smt3 mutant during growth with methionine and cysteine. The observed defects in growth and nuclear segregation were consistent with our identification of tubulin (Tub1), a Spitzenkörper protein (Mlc1), and a nuclear pore complex component (Nsp1) as sumoylation targets in C. albicans (Table 2).
In S. cerevisiae, the septins Cdc3 and Cdc11 are modified by Smt3 late in the budding cell cycle at the G2/M transition (Johnson and Blobel, 1999; Takahashi et al., 1999). In C. albicans, CDC11 inactivation generates multinucleate cells (Johnson and Blobel, 1999; Takahashi et al., 1999). Furthermore, septin-interacting proteins are known to be modified by SUMO (Warenda and Konopka, 2002; Martin and Konopka, 2004). These observations suggest that Smt3 depletion in C. albicans might lead to the formation of a defective septum. To test this, C. albicans smt3/smt3 cells were stained with calcofluor white to examine the chitin content of the cell wall and septal plate. As expected, wild-type cells displayed normal distributions of chitin in their cell walls and at septal junctions (Figure 5D). In contrast, smt3/smt3 cells displayed heterogeneous patterns of chitin deposition. Some smt3/smt3 cells displayed no obvious septum between cell compartments while others displayed chitin mislocalization or abnormally high amounts of chitin in their cell walls (Figure 5D). Again, similar phenotypes were observed in the conditional MET3p-SMT3/smt3 mutant after growth in the presence of 2.5 mM methionine and cysteine. These phenomena are consistent with the idea that Smt3 depletion causes defective localization of the chitin synthetic machinery. This was reinforced by our identification of proteins involved in secretion and endocytosis as sumoylation targets (Lsp1, Sec24, Sec7, and Tub1; Table 2).
Taken together, our molecular, cellular, and proteomic data indicate that sumoylation plays important roles in the regulation of cell division in C. albicans.
SMT3 is essential for morphogenesis in C. albicans
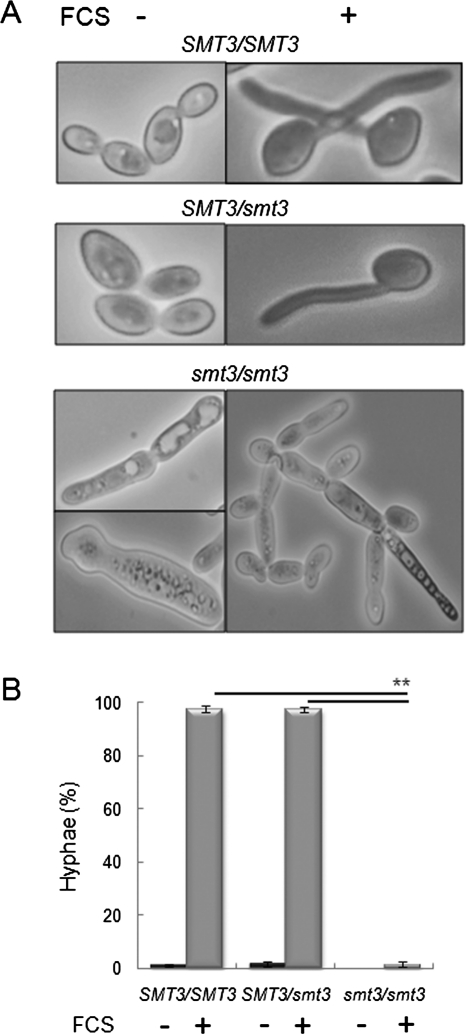
Yeast–hypha morphogenesis is a major virulence attribute of C. albicans. Smt3/smt3 cells displayed aberrant morphologies under conditions that promote the growth of budding cells (Figure 5A). Therefore we asked whether smt3/smt3 cells could form hyphae following serum stimulation. In contrast to the control strains (SMT3/SMT3 and smt3/SMT3), the smt3/smt3 mutant was unable to form true hyphae, with these cells becoming only slightly elongated in the presence of serum (Figure 6).
FIGURE 6:
C. albicans smt3 mutants are unable to form hyphae. (A) Light microscopy of C. albicans smt3/smt3 cells grown with 10% serum at 37°C reveals their inability to form true hyphae: SMT3/SMT3 (BWP17), SMT3/smt3 (MLC33), and smt3/smt3 (MLC37). (B) Percentage of true hyphal cells in YPD at 30°C (FCS −) or in YPD plus 10% serum at 37°C (FCS +): SMT3/SMT3 (BWP17), SMT3/smt3 (MLC33), and smt3/smt3 (MLC37). **p < 0.01 (Student’s t test).
SMT3 is required for the adaptation of C. albicans to different stresses
The SUMO conjugation/deconjugation pathway is regulated by different stresses, and various treatments, including heat shock and ethanol and oxidative (H2O2) stress, enhance global sumoylation patterns in S. cerevisiae (Zhou et al., 2004). C. albicans displays robust stress responses that help to protect it against environmental insults and that contribute to its pathogenicity (Alonso-Monge et al., 1999; Hwang et al., 2002; Smith et al., 2004; Hromatka et al., 2005; Enjalbert et al., 2006). Furthermore, we identified several proteins involved in stress adaptation in our proteomics screen (Cct7, Dot5, Hsp60, Hsp104, Ssb1, Ssc1, Sse1, and Tfp1; Table 2). Therefore we examined the impact of Smt3 inactivation upon the resistance of C. albicans to temperature, oxidative, osmotic, cell wall, and unfolded protein stresses as well as to the antifungal drug caspofungin.
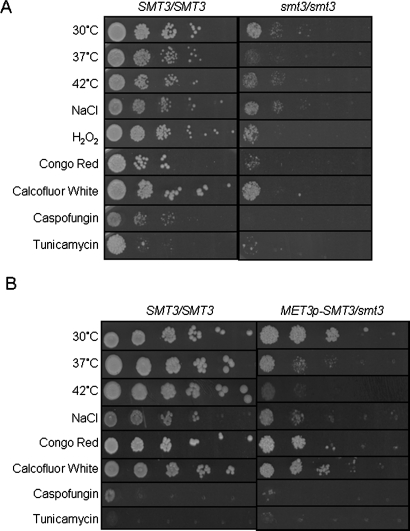
Exponentially growing wild-type (BWP17) and smt3/smt3 cells (MLC37) were spotted onto YPD plates and grown under the appropriate stress conditions. As described above (Figure 4B), smt3/smt3 cells grow more slowly than wild-type cells, and this phenotype was replicated in these stress experiments (Figure 7A). Comparing smt3/smt3 cells in the presence and absence of stress reveals that the inactivation of Smt3 consistently reduced the ability of C. albicans to grow at 37°C and 42°C. This was consistent with the observation that various heat shock proteins are sumoylated in C. albicans (Hsp60, Hsp104, Ssb1, Ssc1, and Sse1; Table 2) as well as in S. cerevisiae (Panse et al., 2004; Wohlschlegel et al., 2004; Zhou et al., 2004; Hannich et al., 2005). Although there was no obvious effect upon osmotic stress resistance, Smt3 inactivation did render C. albicans more sensitive to peroxide. This observation might have physiological significance given the importance of reactive oxygen species in the antimicrobial activity of macrophages, for example (Miller and Britigan, 1997). Furthermore, smt3/smt3 cells consistently displayed cell wall stress phenotypes. They grew more slowly in the presence of Congo red and the antifungal drug caspofungin, which both target glucan biosynthesis. They also showed slight sensitivity to a low concentration of calcofluor white, which affects chitin synthesis (Figure 7A).
FIGURE 7:
C. albicans smt3 mutants are sensitive to a range of stresses. (A) Sensitivity of an smt3/smt3 null mutant to stresses. Serial dilutions of exponentially growing cells were spotted onto YPD plates containing the appropriate stress at the concentrations indicated in Materials and Methods: SMT3/SMT3 (BWP17) and smt3/smt3 (MLC37). (B) Sensitivity of a MET3p-SMT3/smt3 conditional mutant to stresses. Serial dilutions were spotted onto SC plates containing the appropriate stress and containing 2.5 mM methionine and cysteine: SMT3/SMT3 (BWP17) and MET3p-SMT3/smt3 (MLC04). These strains grew at similar rates on control plates that lacked methionine and cysteine (Supplemental Material).
These phenotypes were also examined by comparing the behavior of wild-type and MET3p-SMT3/smt3 cells in the presence and absence of methionine and cysteine. The impact of peroxide stress upon the conditional MET3p-SMT3/smt3 mutant was not tested because of the confounding effects of the reducing agents methionine and cysteine. As expected, wild-type (BWP17) and MET3p-SMT3/smt3 cells (MLC04) displayed no significant differences under nonrepressing conditions, that is, in the absence of methionine and cysteine (Supplemental Material). Wild-type cells were especially sensitive to tunicamycin in the presence of methionine and cysteine because both tunicamycin and reducing agents trigger the unfolded protein response in C. albicans (Wimalasena et al., 2008). Nevertheless, the conditional mutant displayed temperature and cell wall stress sensitivities under repressing conditions (Figure 7B), thereby mirroring the smt3/smt3 null mutant with respect to these phenotypes. Taken together, these data confirm that sumoylation plays significant roles in heat adaptation, the oxidative stress response, and cell wall remodeling.
SMT3 inactivation perturbs Mkc1 activation
As described above, C. albicans smt3/smt3 cells display variable chitin deposition (Figure 5D) and susceptibility to cell wall stresses (Figure 7). How might sumoylation impact these phenotypes? In C. albicans the cell integrity (Pkc1) pathway is known to stimulate chitin synthase activity and elevate cell wall chitin in response to cell wall stresses (Munro et al., 2007). Both caspofungin and calcofluor white activate the Pkc1 pathway in C. albicans (Liu et al., 2005; Bruno et al., 2006; Munro et al., 2007). We reasoned therefore that sumoylation might affect Pkc1 signaling. To test this, we examined the effects of disrupting SMT3 upon the activation of the mitogen-activated protein kinase (MAPK) on this pathway, Mkc1.
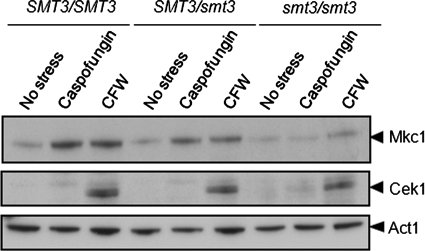
Wild-type (BWP17), SMT3/smt3 (MLC33), and smt3/smt3 (MLC37) cells were either untreated (controls), exposed to caspofungin (0.032 μg/ml for 5 min), or incubated with calcofluor white (100 μg/ml for 2 h). These concentrations were chosen because they are known to activate Pkc1 signaling in C. albicans (Munro et al., 2007). Interestingly, the inactivation of Smt3 led to an attenuation of Mkc1 phosphorylation in response to both caspofungin and calcofluor white (Figure 8). Indeed, no significant Mkc1 activation was observed in response to the antifungal drug caspofungin.
FIGURE 8:
Mkc1 activation is compromised in C. albicans following inactivation of Smt3. Western blotting of the phosphorylated forms of Mkc1 and Cek1 in C. albicans following treatment with caspofungin for 5 min or calcofluor white (CFW) for 2 h: SMT3/SMT3 (BWP17), SMT3/smt3 (MLC33), and smt3/smt3 (MLC37). The Cek1 blot is a longer exposure of the same blot used to detect Mkc1.
The p42/44 antibody also detects the phosphorylated form of Cek1, which is the MAPK on a major morphogenetic signaling pathway in C. albicans (Csank et al., 1998). The disruption of SMT3 did not affect Cek1 activation in response to calcofluor white, indicating that the impact of Smt3 upon Mkc1 activation is specific. We conclude that caspofungin- and calcofluor white–mediated activation of Pkc1 signaling is dependent upon sumoylation.
Mutating sumoylation sites in target proteins replicates some smt3/smt3 phenotypes
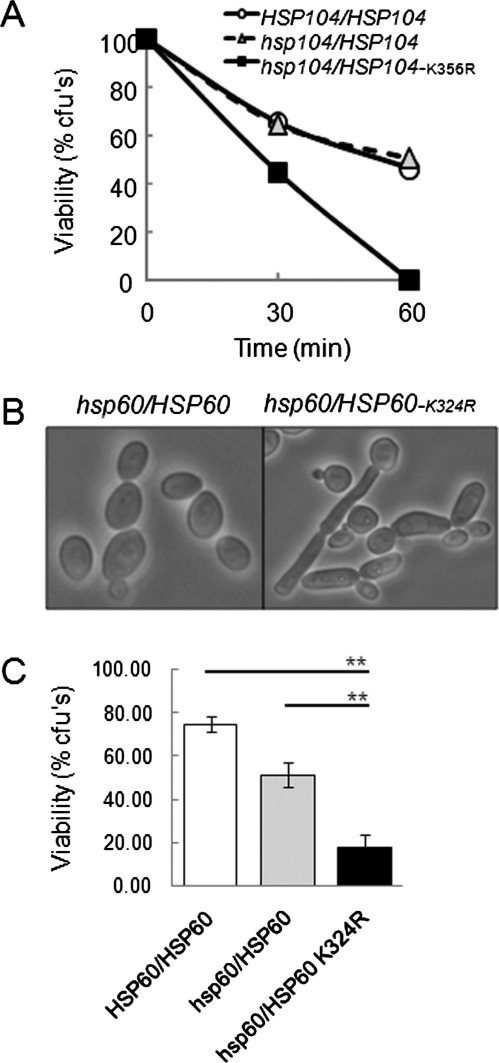
Our proteomics screen identified a number of sumoylated proteins (Table 2), some of which were sumoylated following heat or peroxide stress. We also showed that sumoylation is important for the cells’ ability to respond to stress (Figure 7A) and that the heat shock protein Hsp104 is sumoylated in response to heat shock (Figure 2, B and C). Therefore we next asked whether heat shock resistance in C. albicans is dependent upon the consensus sumoylation site in Hsp104 (Ψ-K-X-E at position 356). To achieve this we deleted the first HSP104 allele in C. albicans (MLC49; Table 1) and then mutated the second allele to remove this sumoylation site (MLC59: hsp104::LAL/HSP104K356R-HIS1). An equivalent control strain was generated that contains a wild HSP104 allele (MLC56: hsp104::LAL/HSP104-HIS1). When we compared the heat shock sensitivity of these strains, the hsp104/HSP104K356R mutant was reproducibly more sensitive to this stress than the control strains (Figure 9A). Thus Hsp104 sumoylation is important for resistance to heat shock in C. albicans, and the lack of Hsp104 sumoylation in smt3/smt3 cells probably contributes to the thermal sensitivity of this mutant (Figure 7).
FIGURE 9:
Mutation of sumoylation sites in Hsp104 and Hsp60 mimics phenotypes of the C. albicans smt3 null mutant. (A) Sensitivity of an Hsp104 sumoylation mutant to a 30–42ºC heat shock. Cell viability was assayed 30 and 60 min after the heat shock: HSP104/HSP104 (MLC43, open circles), hsp104/HSP104 (MLC56, gray triangles), and hsp104/HSP104-K356R (MLC59, black squares). (B) Light microscopy of a C. albicans Hsp60 sumoylation mutant grown at 30ºC reveals their abnormal morphology: hsp60/HSP60 (MLC52) and hsp60/HSP60-K324R (MLC55). (C) Sensitivity of the Hsp60 sumoylation mutant to a 30–42ºC heat shock in the presence of a respiratory inhibitor. Cells were grown in the presence of 1 μg/ml antimycin A and then subjected to a 42ºC heat shock for 30 min: HSP60/HSP60 (MLC43), hsp60/HSP60 (MLC52), and hsp60/HSP60-K324R (MLC55). **p < 0.01 (Student’s t test). These strains showed no difference in heat shock sensitivity in the absence of antimycin A (unpublished data).
Hsp60, another heat shock protein on our list of sumoylated C. albicans proteins, also contains a consensus sumoylation site (Ψ-K-X-E at position 324). We tested the effects of mutating this Hsp60 sumoylation site upon C. albicans. Once again we deleted the first allele (MLC46; Table 1), mutated the sumoylation site in the second allele (MLC55: hsp60::LAL/HSP60-K324R-HIS1), and generated the corresponding control strain (MLC52: hsp60::LAL/HSP60-HIS1). Unlike the Hsp104 sumoylation mutant, the hsp60/HSP60-K324R cells withstood a 30–42°C heat shock as well as the control strains (unpublished data). However, the Hsp60 sumoylation mutant did display an aberrant morphology. Approximately 23% of these hsp60/HSP60-K324R cells grew in an elongated, pseudohyphal-like manner in YPD at 30°C (Figure 9B), a similar phenotype to the smt3/smt3 mutant (Figure 5B). Only 3% of the control wild-type cells displayed this aberrant morphology under equivalent conditions (Figure 9B). Thus a lack of Hsp60 sumoylation probably contributes to the aberrant growth morphology of smt3/smt3 cells.
Hsp60 is a mitochondrial chaperone. Therefore we tested whether growth in the presence of the respiration inhibitor antimycin A affected the ability of the Hsp60 sumoylation mutant to resist heat shock. Interestingly, antimycin A–treated hsp60/HSP60-K324R cells were three- to fourfold more sensitive to heat shock than the corresponding control strains (Figure 9C). We conclude that Hsp60 sumoylation contributes to mitochondrial function and is important for cell survival under certain stress conditions.
DISCUSSION
Sumoylation is one of a number of protein modifications that play key roles in the regulation of growth, environmental adaptation, differentiation, and development of eukaryotic cells. We reasoned, therefore, that sumoylation probably underpins cellular processes that are critical for the pathogenicity of C. albicans. In this article we confirm this by showing that sumoylation is required for normal growth, cell division, morphogenesis, and stress adaptation in this clinically important fungus. This was achieved by combining a proteomic approach to identify sumoylation targets in C. albicans, with a molecular approach to explore the phenotypes of smt3/smt3 mutants. This was facilitated by our observation that C. albicans smt3/smt3 mutants are viable, indicating that SMT3 is not an essential gene in this pathogen. This is consistent with Schizosaccharomyces pombe, in which the Smt3 homologue, Pmt3, is not essential for viability (Tanaka et al., 1999), but contrasts with S. cerevisiae, in which sumoylation is essential (Johnson and Hochstrasser, 1997).
Several observations suggest that sumoylation plays an important role in the regulation of cell growth and division in C. albicans. First, the inactivation or depletion of Smt3 caused severe growth defects, with the cells displaying heterogeneous morphologies with many large, elongated buds (Figure 5, A and B). Many daughter cells remained joined to their mothers, suggesting the existence of cytokinesis or cell separation defects. Second, many smt3/smt3 cells were multinucleate, indicating defects in nuclear segregation (Figure 5C). Third, SMT3 inactivation caused aberrant chitin deposition (Figure 5D). Fourth, smt3/smt3 cells were unable to make hyphae (Figure 6).
How might sumoylation affect these growth and cell cycle–related events? Martin and Konopka (2004) suggested that septins may be sumoylated in C. albicans because the Cdc3, Cdc11, and Shs1/Sep7 septins are sumoylated in S. cerevisiae (Johnson and Blobel, 1999). The observation that C. albicans cdc11/cdc11 cells are enlarged, are multinucleate, and have elongated buds and aberrant chitin localization (Warenda and Konopka, 2002) is consistent with this view. However, they were unable to detect septin sumoylation in C. albicans (Martin and Konopka, 2004), and we did not identify a septin in our screen for sumoylation targets in this pathogen (Table 2). Nevertheless, they did demonstrate that a C. albicans orthologue of a septin that is sumoylated and localized to the bud neck in S. cerevisiae (Johnson and Blobel, 1999) is also located at the bud neck and septation sites in C. albicans (Martin and Konopka, 2004). This suggests that there might be a conserved function for SUMO at the bud neck in these two species, although the sumoylation of S. cerevisiae septins is not required for their essential function in cytokinesis (Johnson and Blobel, 1999; Johnson and Gupta, 2001). Alternatively, sumoylation might be critical for the regulation of cytoskeletal organization in C. albicans. Our identification of Tub1, Cct7, and Mlc1 among the sumoylation targets would be consistent with this view (Table 2). In addition, Cct3, Cct5, Cct8, Tub2, and Tub1, which are all required for the assembly of actin and tubulin, have all been identified as sumoylation targets in S. cerevisiae (Panse et al., 2004). These suggestions, which are not mutually exclusive, might account for the observed effects of SMT3 inactivation upon nuclear segregation, chitin deposition, and cell separation.
Our analyses of Hsp60 sumoylation suggest that additional processes might contribute to cell growth and morphology, albeit indirectly. Mutating the sumoylation site in this mitochondrial chaperone caused elongated, pseudohyphal-like growth (Figure 9), as did SMT3 inactivation (Figure 5). Interestingly inactivation of the stress-activated protein kinase Hog1 causes filamentous growth and interferes with mitochondrial functions (Alonso-Monge et al., 2009). Therefore the phenotypes of both smt3 and hog1 mutants suggest links between mitochondrial activity and filamentous growth in C. albicans.
C. albicans smt3 mutants also showed susceptibility to a wide range of stresses (Figure 7). These defects were specific because while C. albicans smt3/smt3 cells were less able to withstand some conditions (elevated temperatures, cell wall and oxidative stresses), they were as resistant as wild-type cells to others (osmotic stress). Similarly in mammalian cells osmotic stress has little effect on sumoylation compared with heat shock and oxidative stress (Saitoh and Hinchey, 2000). Sumoylation of the heat shock transcription factor HSF1 in mammalian cells is strongly induced by heat shock (Hong et al., 2001). In contrast, sumoylation of HsfA2 by SUMO1 represses its transcriptional activity in Arabidopsis, demonstrating that sumoylation can negatively regulate genes in some cases (Cohen-Peer et al., 2010). Interestingly, a recent study proteomic screen of sumoylated proteins in Arabidopsis identified stress proteins that were especially enriched in proteins that respond to temperature (Elrouby and Coupland, 2010). Moreover, Zhou et al. (2004) showed that several heat shock proteins are targets of sumoylation in S. cerevisiae, including Hsp26, Hsp82, Hsc82, Ssa1, Ssb1, Sse1, and Sse2. This is entirely consistent with our identification of Hsp60, Hsp104, Ssb1, Ssc1, and Sse1 as sumoylation targets in C. albicans (Table 2). Clearly, the inability to sumoylate these proteins might account for the temperature sensitivity of C. albicans smt3/smt3 cells (Figure 6). This was confirmed by mutating the sumoylation site in Hsp104, which caused cells to become more heat shock sensitive (Figure 9). This does not exclude the possibility that sumoylation of other heat shock proteins contributes to thermotolerance.
C. albicans smt3/smt3 cells also displayed reproducible sensitivity to H2O2 (Figure 6A), and sumoylation was increased in response to high doses of H2O2 (Figure 1). This is consistent with the observed effects of oxidative stress upon sumoylation in mammalian cells, plants, and S. cerevisiae (Saitoh and Hinchey, 2000; Kurepa et al., 2003; Zhou et al., 2004). However, the molecular basis for the peroxide sensitivity of C. albicans smt3/smt3 cells was not clear from the list of sumoylation targets, although this list did include Dot5, a putative nuclear thiol peroxidase (Table 2).
C. albicans smt3/smt3 cells displayed sensitivities to a range of cell wall stresses, including the antifungal drug caspofungin (Figure 6). The cell wall is the first line of defense for C. albicans against environmental insults. It has a dynamic structure whose composition is regulated during the cell cycle and in response to environmental change, and the up-regulation of chitin synthesis is a primary response of C. albicans to cell wall damage (Richard et al., 2002; Walker et al., 2008), in part through activation of the cell integrity (Pkc1) pathway (Munro et al., 2007). Interestingly C. albicans smt3/smt3 cells displayed an increase in chitin levels and inaccurate chitin deposition (Figure 5D). This was consistent with the slightly elevated susceptibility to calcofluor white (Figure 6). This aberrant chitin regulation is probably mediated partly through the defective Pkc1 signaling in smt3/smt3 cells (Figure 8) and partly through possible defects in secretion and endocytosis suggested by the identification of sumoylation targets involved in these processes (Lsp1, Sec24, Sec7, and Tub1; Table 2). Such defects can also account for the sensitivity to other stresses that impact upon the cell wall and the unfolded protein response (caspofungin, Congo red, and tunicamycin: Figure 6). Therefore our analyses of Pkc1 signaling and sumoylation targets have provided mechanistic explanations for the cellular phenotypes observed upon Smt3 inactivation.
Collectively, our data indicate that protein sumoylation is highly dynamic in C. albicans, contributing to the regulation of a wide range of cellular processes. The dynamics of sumoylation could be regulated by controlling the rates of sumoylation and desumoylation (Bossis and Melchior, 2006). While sumoylation of Hsp104 is important for resistance to heat shock in C. albicans, it remains possible that sumoylation targets pathways rather than individual proteins, as is typical of other protein modifications. In this way sumoylation could coordinate the activity of multiple components on a single pathway. Further studies of specific target proteins are required to uncover the roles of protein sumoylation in these cellular processes and in the virulence of this major human pathogen.
MATERIALS AND METHODS
Strains and growth conditions
C. albicans strains (Table 1) were grown in YPD (Sherman, 1991) or synthetic complete (SC) medium lacking methionine and cysteine (Kaiser et al., 1994). The growth medium was supplemented with 2.5 mM methionine and cysteine for MET3 promoter shutoff assays. The conditions used to examine stress phenotypes are described below.
One-dimensional (1-D) Western analysis
Total soluble protein was extracted and subjected to Western blotting using published protocols (Smith et al., 2004). Briefly, cells were resuspended in 250 μl lysis buffer (0.1 M Tris-HCl, pH 8, 10% glycerol, 1 mM dithiothreitol [DTT], pepstatin A, protease inhibitor cocktail) and sheared with glass beads in a mini-bead beater (6 × 30 s with 1-min intervals on ice). Lysates were centrifuged at 13,000 rpm for 10 min at 4°C. Proteins (15 μg) were separated by SDS–PAGE using the XCell SureLock Mini-Cell system (Invitrogen, Paisley, UK) with NuPAGE Novex Bis-Tris 4–12% precast gels (Invitrogen) in NuPAGE MOPS-SDS Running Buffer (Invitrogen) containing NuPAGE Antioxidant (Invitrogen) as per the manufacturer’s instructions. Proteins were transferred to Invitrolon PVDF Membranes (Invitrogen) in NuPAGE Transfer Buffer containing methanol using the XCell II Blot Module (Invitrogen) as per the manufacturer’s instructions. Following transfer, the membranes were rinsed in phosphate-buffered saline (PBS) and blocked in PBS-T + 5% milk (PBS 0.1%, Tween-20, 5% [wt/vol] milk) for 1 h at room temperature. The membranes were then incubated for 1 h at room temperature in PBS-T + 5% milk containing antibody. To detect FLAG-Smt3, a 1:10,000 dilution of anti-FLAG horseradish peroxidase (HRP)–conjugated antibody was used (Sigma A8592, Gillingham, UK). Hsp104 was detected using an anti-Hsp104 antibody kindly supplied by Mick Tuite (Zenthon et al., 2006) at a 1:5000 dilution in PBS-T + 5% milk. An anti-rabbit HRP-conjugated antibody was used at a 1:2000 dilution in PBS-T + 5% milk before developing. To detect Mkc1 and Cek1 phosphorylation, a 1:2000 dilution of phospho-p44/42 MAPK (Erk1/2) (Thr-202/Tyr-204) rabbit monoclonal antibody (New England Biolabs, Hitchin, Hertfordshire, UK) was used in Tris-buffered saline (TBS)–T + 5% bovine serum albumin (BSA) (TBS 0.1%, Tween-20, 5% [wt/vol] BSA). Membranes were incubated overnight at 4°C. For detection, an anti-rabbit HRP-conjugated antibody was used at a 1:2000 dilution in TBS-T + 5% BSA for 1 h at room temperature. To detect Act1, an anti-Act1 antibody (Abcam, Cambridge, UK) was used at a 1:5000 dilution in PBS-T + 5% milk. An anti-mouse HRP-conjugated antibody was used at a 1:2000 dilution in PBS-T + 5% milk before developing. Membranes were washed in PBS-T or TBS-T, and signals were detected using an enhanced chemiluminescence (ECL) Western blotting kit (Amersham, Little Chalfont, Buckinghamshire, UK) as per the manufacturer’s instructions.
2-D gel analysis
C. albicans protein extracts were prepared as described previously (Yin et al., 2004). Briefly, cells were disrupted with glass beads in 160 μl lysis solution (7.5 M urea, 2.5 M thio-urea, 1.25 mM EDTA, 1.75 μg/ ml pepstatin A, 62.5 mM DTT, 25 mM Tris-Cl, pH 10.8, and 3.7 × protease inhibitor cocktail tablets; Roche, Lewes, UK) using a bead beater. After cell disruption, proteins were further solubilized by adding 40 μl of lysis 2 (20% wt/vol 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, 50% vol/vol glycerol, 10% vol/vol carrier ampholyte, pH 4–7), and incubated on ice for 1 h. After repeated bead beating, cell debris were removed by centrifugation (20 min, 13,000 rpm, 4°C), and protein extracts were stored at −20°C.
Two-dimensional gel electrophoresis was performed as described previously (Cash and Argo, 2009). After rehydration, immobilized pH gradient (IPG) strips (pH 4–7 linear) were used to separate proteins on a Multiphor II (75–100 μg) in the first dimension according to the following protocol: 200 V for 1 min, 3500 V for 90 min, 3500 V for 65 min. After isoelectric focusing, IPG strips were equilibrated in two steps: 1) 50 mM Tris-Cl (pH 6.8), 6 M urea, 30% vol/wt glycerol, 2% wt/wt SDS, 62.5 mM DTT for 25 min, and 2) 50 mM Tris-Cl (pH 6.8), 6 M urea, 30% vol/wt glycerol, 2% wt/wt SDS, 2.5% wt/vol iodoacetamide for 25 min. Proteins were then separated in the second dimension on precast 4–12% SDS polyacrylamide gels (Invitrogen) using 3-(N-morpholino)propanesulfonic acid buffer at first with 100 V per gel for 30 min, followed by 200 V per gel for a further 90 min. Gels were fixed (2% H3PO4, 50% ethanol) overnight, rinsed three times with water, and equilibrated (1.3 M (NH4)2SO4, 2% wt/wt H3PO4, 34% wt/vol methanol) for 60 min, before staining with colloidal Coomassie blue G250 (0.67 g/l). Equivalent 2-D gels were subjected to Western blotting for FLAG-Smt3 sequences, as described above for the 1-D gels. Independent biological replicates were performed for all experiments, and only reproducible observations are reported.
Mass spectrometry
Sumoylated proteins were selected by aligning the Western blots with the corresponding Coomassie-stained gels, and these proteins were identified by liquid chromatography–tandem mass spectrometry (LC-MS/MS). The corresponding spots (1.2-mm diameter) were cut from gels and transferred to 96-well microtiter plates using an Investigator ProPic robotic workstation (Genomic Solutions, Huntingdon, UK). Proteins in gel plugs were digested with trypsin (Promega, Southampton, UK) using an Investigator ProGest robotic workstation (Genomic Solutions). Peptides were extracted, dried in a Savant SpeedVac SC110A (Thermo Fisher Scientific, Loughborough, UK), and dissolved in 0.1% formic acid for LC-MS/MS. The LC-MS system comprised an UltiMate 3000 LC system (Dionex, Camberley, UK) coupled to an HCTultra ion trap mass spectrometer with an electrospray ion source fitted with a low-flow nebulizer (Bruker Daltronics, Coventry, UK). Peptides were separated on a PepSwift monolithic capillary column (Dionex) at 2 μl/min using a linear gradient of acetonitrile. Eluent A was 3% acetonitrile in 0.05% formic acid, and eluent B was 80% acetonitrile in 0.04% formic acid. The gradient was 3–45% eluent B > 12 min at a flow rate of 2 μl/min.
Tandem mass spectra were acquired in data-dependent AutoMS(2) mode using the following parameters: MS scan range = 300–1500 m/z; averages = 3, maximum number of precursors = 3; MS(2) scan range = 100–2200 m/z, averages = 2; active exclusion on (maximum spectra = 2, release after 1 min). Peptide peaks were detected and deconvoluted automatically using DataAnalysis software (Bruker). Mass lists were used as input for Mascot MS/MS Ions searches using Mascot Server version 2.2 (Matrix Science, London, UK). Our in-house protein sequence database (containing 6166 sequences) was built from the flat file CALBI_prot.txt downloaded from the CandidaDB web server ftp://ftp.pasteur.fr/pub/GenomeDB/CandidaDB/FlatFiles. Protein annotations were taken from the Candida Genome Database (candidagenome.org) and CandidaDB (http://genolist.pasteur.fr/CandidaDB). Potential sumoylation sites were predicted using the program SUMOsp version 2.0 (Ren et al., 2009).
Our proteomics data set can be accessed via the PRoteomics IDEntifications database data repository at the European Bioinformatics Institute (www.ebi.ac.uk/pride) using accession number 13274 (Vizcaino et al., 2010).
Immunoprecipitation
Unstressed and heat-stressed protein extracts used for 2-D gel analysis were diluted in a 1:1 ratio in 2× HEPES immunoprecipitation (IP) buffer (40 mM HEPES, pH 7.5, 300 mM NaCl, 0.2% Triton X-100, and 20% glycerol). Lysates were incubated with 3 μg anti-FLAG antibody (Sigma F7425, Gillingham, UK) at 4°C overnight. Protein A Sepharose beads (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) in 1× HEPES IP buffer were added at 100 μl, and lysates were rotated at 4°C for 5 h. The supernatant was then removed, and the beads were washed three times in 1× HEPES IP buffer, resuspended in 2× SDS buffer, and boiled at 95°C for 5 min. Samples were analyzed by immunoblotting against anti-Hsp104.
Strain construction
The Smt3 protein was amino-terminally tagged with a triple FLAG epitope by replacing the green fluorescent protein (GFP) open reading frame (ORF) in the plasmid pACT1-FLAG-GFP with the SMT3 coding region to create pACT1-FLAG-SMT3. Initially, pACT1-FLAG-GFP was constructed by inserting a double-stranded oligonucleotide encoding three copies of the FLAG epitope plus a new BamHI site into the HindIII site of pACT1-GFP (Barelle et al., 2004). The SMT3 ORF was PCR amplified using the primers SMT3FL-F and SMT3FL-R (Supplemental Material) and then cloned between the HindIII and NheI sites in pACT1-FLAG-GFP to replace the GFP ORF and create pACT1-FLAG-SMT3. This plasmid was transformed into C. albicans THE1 to create the strain MLC01 (Table 1), in which FLAG-Smt3 is expressed from the C. albicans ACT1 promoter.
To generate conditional C. albicans SMT3 mutants, one SMT3 allele was placed under the control of the methionine/cysteine (Met/Cys)–repressible MET3 promoter (Care et al., 1999), and the other allele was deleted using the lox-ARG4-lox marker (Dennison et al., 2005). To achieve this, the URA3-MET3p cassette from plasmid pURA3-MET3 (Rodaki et al., 2006) was PCR amplified with Pfu Turbo (Promega) using the primers MetUS-F and MetUS-R (Supplemental Material) to generate a URA3-MET3p-SMT3 cassette with 80 base pairs of homology to the SMT 3 5′ upstream region and 80 base pairs of homology to the beginning of the SMT3 ORF. Similarly, the loxP-ARG4-loxP sequence from plasmid pLAL (Dennison et al., 2005) was PCR amplified using primers LAL-F and LAL-R (Supplemental Material) to generate an smt3::ARG4 disruption cassette with 90 base pairs of flanking homology to the 5′ upstream and 3′ downstream regions of the SMT3 gene. C. albicans strain BWP17 (Table 1) was then transformed with these cassettes as described previously (Gietz et al., 1995; Walther and Wendland, 2003) to generate strains MLC02 and MLC10 (MET3p-SMT3/SMT3). MLC02 was then transformed with the smt3::ARG4 cassette to generate the conditional mutants MLC04 and MLC13 (MET3p-SMT3/smt3::ARG4).
The two SMT3 alleles in C. albicans strains BWP17 (SMT3/SMT3; Table 1) were inactivated sequentially with the loxP-URA3-loxP (LUL) and loxP-ARG4-loxP (LAL) markers (Dennison et al., 2005) to create the homozygous smt3/smt3 null mutants MLC37, MLC38, and MLC39 (Table 1). The smt3::LUL and smt3::LAL disruption cassettes were designed to delete the complete SMT3 ORF and generated by PCR amplification with the primers LAL-F, LAL-R, LUL-F, and LUL-R (Supplemental Material). The smt3::LUL cassette was transformed into C. albicans, transformants were selected on the basis of their uridine prototrophy, and correct integration was confirmed by diagnostic PCR with primers SMT3d-F and LULd-R (Supplemental Material). The resultant heterozygote (MLC33: smt3::URA3/SMT3) was then transformed with the smt3::LAL cassette, and accurate disruption of the remaining SMT3 allele was confirmed by PCR using primers SMT3d-F and LALd-R (Supplemental Material). This yielded the homozygous null mutants MLC37, MLC38, and MLC39 (smt3::URA3/smt3::ARG4) (Table 1). The genotypes of all SMT3 mutants made in this study were confirmed both by PCR and Southern analysis using the ECL Direct Nucleic Acid Labelling and Detection System (GE Healthcare).
To generate mutants in which the consensus sumoylation sites in Hsp60 or Hsp104 were inactivated, the first HSP60 or HSP104 allele was deleted using loxP-ARG4-loxP (LAL) to create heterozygous hsp60/HSP60 and hsp104/HSP104 mutants. This was achieved by PCR amplification of the LAL cassette (using the primers HSP60-LAL-F plus HSP60-LAL-R and HSP104-LAL-F plus HSP104-LAL-R; Supplemental Material), transforming the hsp60::LAL and hsp104::LAL cassettes separately into MLC43, and selecting transformants on the basis of their arginine prototrophy. Correct integration was confirmed by diagnostic PCR (with primers HSP60d-F plus LALd-R and HSP104d-F plus LALd-R). This created strains MLC46 and MLC49, respectively (Table 1).
To mutate the sumoylation sites, HSP60 and HSP104 were first cloned into CIp20 (using primers HSP60-CTF plus HSP60-CTR and HSP104-CTF plus HSP104-CTR). Then the HSP60 sumoylation site was mutated by amplifying an HSP60-HIS1 cassette with a primer that changed the lysine at position 324 to arginine (AAA to CGA) (with primers HSP60-SDM-K324R-F and HSP60-SDM-R). (This cassette contains the 3′ proximal region of the HSP60 ORF together with the HIS1 marker.) As a control an analogous wild-type HSP60-HIS1 cassette was amplified (with primers HSP60-WT-F and HSP60-SDM-R). Both cassettes were transformed separately into MLC46, and transformants were selected via their histidine prototrophy. Confirmation of the wild-type and mutated alleles was achieved by diagnostic PCR with primers HSP60-WTd-F plus HSP60d-R for the wild-type control and HSP60-SDMd-F plus HSP60d-R for the allele with the mutated sumoylation site (Supplemental Material). Strains MLC52 (hsp60/HSP60) and MLC55 (hsp60/HSP60K324R) were thus created (Table 1).
The Hsp104 sumoylation site was mutated in a similar manner. An HSP104-HIS1 cassette was PCR amplified with a primer that changed lysine 356 to arginine (AAA to CGA) (primers HSP104-SDM-K356R-F plus HSP104-SDM-R). An analogous wild-type cassette was created in parallel (using HSP104-WT-F plus HSP104-SDM-R). Both cassettes were transformed independently into MLC49 and transformants selected on the basis of histidine prototrophy. Correct integration was confirmed by diagnostic PCR with primers HSP104-WTd-F plus HSP104d-R for the wild-type allele and HSP104-SDMd-F plus HSP104d-R for the allele with the mutated sumoylation site (Supplemental Material). Strains MLC56 (hsp104/HSP104) and MLC59 (hsp104/HSP104K356R) were thus created.
mRNA analyses
SMT3 mRNA levels were measured by qRT-PCR. C. albicans cells were grown overnight in YPD containing 2.5 mM Met/Cys at 30°C. Cells were then reinoculated at OD600 = 0.1 into 50 ml fresh YPD either containing or lacking Met/Cys and regrown at 30°C to midlog phase (OD600 = 0.5). Cells were harvested and frozen rapidly in liquid N2. RNA was extracted with Trizol (GibcoBRL, Grand Island, NY) as described previously (Hauser et al., 1998), and RNA integrity was assayed on an Agilent Bioanalyser 2100 (Stockport, UK). For qRT-PCR, samples were incubated at room temperature for 15 min in a 20-μl reaction mix containing 2 μg RNA, 2 μl DNase I buffer (Invitrogen), 1.5 μl DNase I, and 1.5 μl RNase OUT (Invitrogen). cDNA was prepared using Superscript II (Invitrogen) as per the manufacturer’s protocol. RT-PCR was performed in triplicate in optical multiwall plate 384 using the LightCycler 480 Probes Master (Roche Applied Science, Burgess Hill, UK) as per the manufacturer’s guidelines. Briefly, for the target transcripts SMT3 and ACT1, probes were chosen using the ProbeFinder Software version 2.45 (Roche, universalprobelibrary.com). PCR was performed in a 20-μl reaction containing 10 μl LightCycler 480 Probes Master Mix, 3 μl 1:5 diluted cDNA, 0.2 μl forward and reverse primers, 0.2 μl selective probe (Roche), and 6.4 μl water, PCR grade. Negative controls were performed using water instead of cDNA. Reactions were performed in a LightCycler 480 system (Roche Applied Science) using the following parameters: preincubation at 95°C for 5 min, 50 cycles of amplification at 95°C for 10 s and 60°C for 30 s, and a final cooling at 40°C for 1 min. Standard curves were prepared using four dilutions of the control, wild-type. SMT3 mRNA levels were normalized against the ACT1 mRNA (arbitrary units).
Stress phenotypes
To examine sumoylation levels during stress responses, cells were subjected to the following stresses for 1 h in YPD: heat stress (42°C heat shock), hyphal-inducing conditions (37°C + 10% FCS), Congo red (100 μg ml−1), SDS (10%), calcoflour white (100 μg ml−1), and H2O2 (50 mM).
The susceptibilities of mutants to H2O2 (5 mM), NaCl (1 M), calcofluor white (20 μg ml−1), Congo red (100 μg ml−1), tunicamycin (4.73 mM), caspofungin (0.064 μg ml−1), and thermal stress (37°C and 42°C) were tested using cells grown in YPD medium at 30°C to exponential phase (OD600 = 0.5). Cells were diluted in YPD, spotted onto YPD plates containing the appropriate supplements, and then examined after incubation for 48 h at 30°C (or at the temperatures specified). All experiments were done with two independent mutants and in duplicate.
To test the heat sensitivity of HSP104 mutants, cells were grown in YPD medium at 30°C to midexponential phase, and cultures were then split: one-half was immediately raised to 42°C by addition of an equal volume of YPD prewarmed at 54°C; the other half remained at 30°C by addition of YPD at 30°C. Samples were taken at 30 min and 60 min to assay cell viability (colony-forming units [CFUs]). Data represent means from three independent experiments.
To test the heat sensitivity of HSP60 mutants during mitochondrial stress, cells were grown in YPD medium with 1 μg/ml antimycin A (Axxora, Nottingham, UK) until midexponential phase. Cultures were then split and treated as described above for the HSP90 mutants. Samples were taken at 30 min to assay cell viability (CFUs). Data represent means from three independent experiments.
Microscopy
Samples of exponentially growing C. albicans cells (OD600 = 0.5) were collected, fixed in 3.7% paraformaldehyde, and examined by phase differential interference contrast (DIC) microscopy. Cells were stained with 2 μg/ml calcofluor white to visualize chitin. Nuclei were stained by overlaying samples with mounting media containing 1.5 μg/ml DAPI (Vector Laboratories, Peterborough, UK). All samples were examined by DIC and fluorescence microscopy using a Zeiss Axioplan 2 microscope. Images were recorded digitally using the Openlab system (Openlab v. 4.04; Improvision, Coventry, UK) using a Hamamatsu C4742-95 digital camera (Hamamatsu Photonics, Hamamatsu, Japan).
Supplementary Material
Acknowledgments
We are grateful to Mick Tuite for his generous provision of the anti-Hsp104 antibody. We also thank Brian Morgan, Jan Quinn, Neil Gow, Carol Munro, and Phil Cash for stimulating discussions and helpful advice. M.L. was supported by a Carnegie/Caledonian Scholarship from the Carnegie Trust. This work was also supported by a grant from the UK Biotechnology and Biological Research Council (BB/D009308/1). A.B. is supported by the BBSRC (BB/F00513X/1), the Wellcome Trust (080088), and the European Commission (PITN-GA-2008–214004, ERC-2009-AdG-249793). We thank Susan Budge and Jan Walker for excellent technical support.
Abbreviations used:
- 1-D
one-dimensional
- 2-D
two-dimensional
- BSA
bovine serum albumin
- CFU
colony-forming unit
- DIC
differential interference contrast
- DTT
dithiothreitol
- ECL
enhanced chemiluminescence
- EDLP
excess of limit-digested peptides
- FCS
fetal calf serum
- GFP
green fluorescent protein
- HRP
horseradish peroxidase
- IP
immunoprecipitation
- IPG
immobilized pH gradient
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- MAPK
mitogen-activated protein kinase
- ORF
open reading frame
- PBS
phosphate-buffered saline
- pI
isoelectric point
- qRT-PCR
quantitative real-time PCR
- SC
synthetic complete
- SUMO
small ubiquitin-like modifier
- TBS
Tris-buffered saline
- YPD
yeast peptone dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0632) on January 5, 2011.
REFERENCES
- Alonso-Monge R, Navarro-Garcia F, Molero G, Diez-Orejas R, Gustin M, Pla J, Sanchez M, Nombela C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge R, Carvailho S, Nombela C, Rail E, Pla J. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology. 2009;155:413–423. doi: 10.1099/mic.0.023309-0. [DOI] [PubMed] [Google Scholar]
- Barelle CJ, Manson CL, MacCallum DM, Odds FC, Gow NA, Brown AJ. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast. 2004;21:333–340. doi: 10.1002/yea.1099. [DOI] [PubMed] [Google Scholar]
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- Biggins S, Bhalla N, Chang A, Smith DL, Murray AW. Genes involved in sister chromatid separation and segregation in the budding yeast S. cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Haynes K, Quinn J. Nitrosative and oxidative stress responses in fungal pathogenicity. Curr Opin Microbiol. 2009;12:384–391. doi: 10.1016/j.mib.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2006;2:e21. doi: 10.1371/journal.ppat.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone RA. Candida and Candidiasis. Washington, DC: ASM Press; 2002. [Google Scholar]
- Care RS, Trevethick J, Binley KM, Sudbery PE. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- Cash P, Argo E. Analysis of bacterial proteins by 2DE. Methods Mol Biol. 2009;519:131–144. doi: 10.1007/978-1-59745-281-6_9. [DOI] [PubMed] [Google Scholar]
- Chauhan N, Latge JP, Calderone R. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat Rev Microbiol. 2006;4:435–444. doi: 10.1038/nrmicro1426. [DOI] [PubMed] [Google Scholar]
- Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A. Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Mol Biol. 2010;74:33–45. doi: 10.1007/s11103-010-9652-1. [DOI] [PubMed] [Google Scholar]
- Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas DY, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison PM, Ramsdale M, Manson CL, Brown AJ. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol. 2005;42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA. 2010;107:17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, MacCallum DM, Odds FC, Brown AJ. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun. 2007;75:2143–2151. doi: 10.1128/IAI.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gong L, Kamitani T, Fujise K, Caskey LS, Yeh ET. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J Biol Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li S-J, Heide H, Emili A, Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in S. cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- Hauser NC, Vingron M, Scheideler M, Krems B, Hellmuth K, Entian KD, Hoheisel JD. Transcriptional profiling on all open reading frames of S. cerevisiae. Yeast. 1998;14:1209–1221. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1209::AID-YEA311>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Hay RT. Protein modification by SUMO. Trends Biochem Sci. 2001;26:332–333. doi: 10.1016/s0968-0004(01)01849-7. [DOI] [PubMed] [Google Scholar]
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- Hromatka BS, Noble SM, Johnson AD. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell. 2005;16:4814–4826. doi: 10.1091/mbc.E05-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002;148:3705–3713. doi: 10.1099/00221287-148-11-3705. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Hochstrasser M. SUMO-1: Ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Ann Rev Cell Devel Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kim KI, Baek SH, Chung CH. Versatile protein tag, SUMO: its enzymology and biological function. J Cell Physiol. 2002;191:257–268. doi: 10.1002/jcp.10100. [DOI] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung D-Y, Vierstra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- Liu TT, Lee RE, Barker KS, Wei L, Homayouni R, Rogers PD. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother. 2005;49:2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- Martin SW, Konopka JB. SUMO modification of septin-interacting proteins in Candida albicans. J Biol Chem. 2004;279:40861–40867. doi: 10.1074/jbc.M406422200. [DOI] [PubMed] [Google Scholar]
- Melchior F. SUMO—nonclassical ubiquitin. Ann Rev Cell Devel Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- Miller RA, Britigan BE. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin’s mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJ, Gow NA. The PKC, HOG and Ca2+ signalling pathways coordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, Aoki Y. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect Immun. 2000;68:6712–6719. doi: 10.1128/iai.68.12.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S, Leach MD, Priest CL, Brown AJ. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol Microbiol. 2009;74:844–861. doi: 10.1111/j.1365-2958.2009.06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC (ed.), editor. Candida and Candidosis. London, UK:: Bailliere Tindall; 1988. [Google Scholar]
- Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- Richard M, Ibata-Ombetta S, Dromer F, Bordon-Pallier F, Jouault T, Gaillardin C. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol Microbiol. 2002;44:841–853. doi: 10.1046/j.1365-2958.2002.02926.x. [DOI] [PubMed] [Google Scholar]
- Rodaki A, Young T, Brown AJP. Effects of depleting the essential central metabolic enzyme, fructose-1,6-bisphosphate aldolase, upon the growth and viability of Candida albicans: implications for antifungal drug target discovery. Eukaryotic Cell. 2006;5:1371–1377. doi: 10.1128/EC.00115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28:321–328. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienhorst I, Johnson ES, Dohmen RJ. SUMO conjugation and deconjugation. Mol Gen Genet. 2000;263:771–786. doi: 10.1007/s004380000254. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead DA, Preece A, Brown AJ. Universal metrics for quality assessment of protein identifications by mass spectrometry. Mol Cell Proteomics. 2006;5:1205–1211. doi: 10.1074/mcp.M500426-MCP200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Iwase M, Konishi M, Tanaka M, Toh-e A, Kikuchi Y. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem Biophys Res Commun. 1999;259:582–587. doi: 10.1006/bbrc.1999.0821. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. Characterization of a fission yeast SUMO-1 homologue, Pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. The proteomics identifications database: 2010 update. Nucleic Acids Res. 2010;38:D736–742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NA. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4:e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A, Wendland J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet. 2003;42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol Biol Cell. 2002;13:2732–2746. doi: 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, Yin Z, Brown AJ, Archer DB. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol. 2008;45:1235–1247. doi: 10.1016/j.fgb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR, III. Global analysis of protein sumoylation in S. cerevisiae. J Biol Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yin Z, Stead D, Selway L, Walker J, Riba-Garcia I, McLnerney T, Gaskell S, Oliver SG, Cash P, Brown AJ. Proteomic response to amino acid starvation in Candida albicans and S. cerevisiae. Proteomics. 2004;4:2425–2436. doi: 10.1002/pmic.200300760. [DOI] [PubMed] [Google Scholar]
- Zenthon JF, Ness F, Cox B, Tuite MF. The [PSI+] prion of S. cerevisiae can be propagated by an Hsp104 orthologue from Candida albicans. Eukaryot Cell. 2006;5:217–225. doi: 10.1128/EC.5.2.217-225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in S. cerevisiae. J Biol Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.