When activated by high NaCl, the transcription factor TonEBP/OREBP increases transcription of osmoprotective genes. High NaCl activates CDK5 kinase, which directly phosphorylates TonEBP/OREBP on threonine 135. This contributes to rapid nuclear translocation of TonEBP/OREBP, accelerating transcription of its osmoprotective target genes.
Abstract
When activated by high NaCl, tonicity-responsive enhancer–binding protein/osmotic response element–binding protein (TonEBP/OREBP) increases transcription of osmoprotective genes. High NaCl activates TonEBP/OREBP by increasing its phosphorylation, nuclear localization, and transactivating activity. In HEK293 cells, mass spectrometry shows phosphorylation of TonEBP/OREBP-S120, -S134, -T135, and -S155. When those residues are individually mutated to alanine, nuclear localization is greater for S155A, less for S134A and T135A, and unchanged for S120A. High osmolality increases phosphorylation at T135 in HEK293 cells and in rat renal inner medullas in vivo. In HEK293 cells, high NaCl activates cyclin-dependent kinase 5 (CDK5), which directly phosphorylates TonEBP/OREBP-T135. Inhibition of CDK5 activity reduces the rapid high NaCl–induced nuclear localization of TonEBP/OREBP but does not affect its transactivating activity. High NaCl induces nuclear localization of TonEBP/OREBP faster (≤2 h) than it increases its overall protein abundance (≥6 h). Inhibition of CDK5 reduces the increase in TonEBP/OREBP transcriptional activity that has occurred by 4 h after NaCl is raised, associated with less nuclear TonEBP/OREBP at that time, but does not reduce either activity or nuclear TonEBP/OREBP after 16 h. Thus high NaCl–induced increase of the overall abundance of TonEBP/OREBP, by itself, eventually raises its effective level in the nucleus, but its rapid CDK5-dependent nuclear localization accelerates the process, speeding transcription of osmoprotective target genes.
INTRODUCTION
Hypertonicity, caused by elevated levels of NaCl and other poorly permeating solutes, perturbs cells and can kill them by apoptosis (Burg et al., 2007). Protection is provided by tonicity-responsive enhancer–binding protein/osmotic response element–binding protein (TonEBP/OREBP) (Miyakawa et al., 1999; Ko et al., 2000), often called NFAT5, a rel family transcription factor whose activation by hypertonicity increases enzymes and transporters that elevate intracellular organic osmolytes and heat shock proteins (Burg et al., 2007). High NaCl activates TonEBP/OREBP by increasing its abundance (Miyakawa et al., 1999; Ko et al., 2000), phosphorylation (Dahl et al., 2001), nuclear localization (Miyakawa et al., 1999; Ko et al., 2000), and transactivating activity (Ferraris et al., 2002). Numerous proteins, including kinases and phosphatases, contribute to the activation of TonEBP/OREBP (Burg et al., 2007). For example, concurrent high NaCl–induced activation of c-Abl kinase (Gallazzini et al., 2010) and inhibition of SHP-1 phosphatase (Zhou et al., 2010) increase phosphorylation of TonEBP/OREBP on tyrosine 143, resulting in increased TonEBP/OREBP transcriptional activity, nuclear localization, and transactivating activity, dependent on association of phospholipase C (PLC)–γ1 with TonEBP/OREBP at phospho-Y143 (Irarrazabal et al., 2010). Also, high NaCl–induced activation of ataxia telangiectasia mutated (ATM) kinase contributes to the subsequent increase of TonEBP/OREBP nuclear localization (Zhang et al., 2005) as well as increase of transcriptional and transactivating activity, dependent on TonEBP/OREBP-S1274 and -S1367 (Irarrazabal et al., 2004).
In the present studies we used protein mass spectrometry to search for additional phosphorylation sites in TonEBP/OREBP, and we tested for possible roles of those phosphorylation events in regulation of TonEBP/OREBP activity.
RESULTS
Discovery of additional phosphorylation sites in TonEBP/OREBP
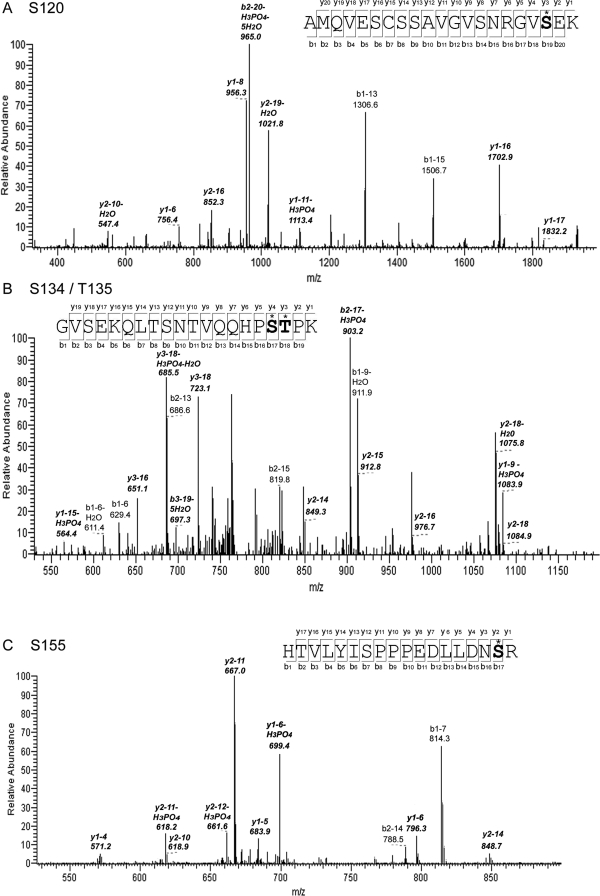
Native TonEBP/OREBP is not sufficiently abundant in human embryonic kidney (HEK) 293 cells for analysis by protein mass spectrometry. Therefore, in order to discover amino acids that are phosphorylated, we stably expressed TonEBP/OREBP-1–547-V5 in the cells (Chen et al., 2007) and immunoprecipitated it from cytoplasmic and nuclear extracts of cells bathed in medium kept at 300 mOsm/kg or changed for 2 h to 200 or 500 mOsm/kg by adjusting NaCl. This region of TonEBP/OREBP contains nuclear localization, DNA binding, and dimerization domains (Burg et al., 2007). To maximize coverage of phosphorylation sites by mass spectrometry, we used two different proteolytic enzymes, trypsin and endoproteinase Arg C. We used the SEQUEST (Eng et al., 1994) algorithm for initial identification of phosphorylated amino acids, and Ascore (Beausoleil et al., 2006) for confirmation. Ascore (http://Ascore.med.harvard.edu) estimates the probability of correct identification of phosphorylation sites from the presence and intensity of site-determining ions in tandem mass spectometry (MS/MS) spectra. Ascore > 19 predicts >99% probability of correct identification of phosphorylation sites. We found numerous peptides from TonEBP/OREBP in both nuclear and cytoplasmic fractions—up to 12 unique peptides in a single sample. We identified four likely phosphorylation sites in three different phosphopeptides (Table 1). We found phospho-S120 in nuclear extracts at 200 and 500 mOsm/kg and cytoplasmic extracts at 500 mOsm/kg. We found phospho-S134 and -T135, always together, in nuclear extracts at 500 mOsm/kg. We found phospho-S155 in cytoplasmic extracts at 200 mOsm/kg. Each peptide was detected in several independently prepared samples. Representative MS2 spectra of the phosphopeptides are shown in Figure 1.
Table 1:
Phosphorylation sites in TonEBP/OREBP
| Site | Enzyme | Charge | Xcorr | Ascore |
|---|---|---|---|---|
| S120 K.AMQVESCSSAVGVSNRGVS*EK.Q | Trypsin | 2 | 2.4 | 9.3 |
| S134 R.GVSEKQLTSNTVQQHPS*T*PK.R | Endoproteinase Arg-C | 3 | 3.1 | 21.7 |
| T135 R.GVSEKQLTSNTVQQHPS*T*PK.R | Endoproteinase Arg-C | 3 | 3.1 | 19.5 |
| S155 R.HTVLYISPPPEDLLDNS*R.M | Endoproteinase Arg-C | 3 | 5.1 | 174.4 |
FIGURE 1:
MS2 spectra showing phosphorylation of TonEBP/OREBP at (A) S120, (B) S134 and T135, and (C) S155. Ions that are determining for the specific phosphorylation sites are indicated by italics.
Effect on nuclear localization of TonEBP/OREBP of mutating the phosphorylation sites to alanine
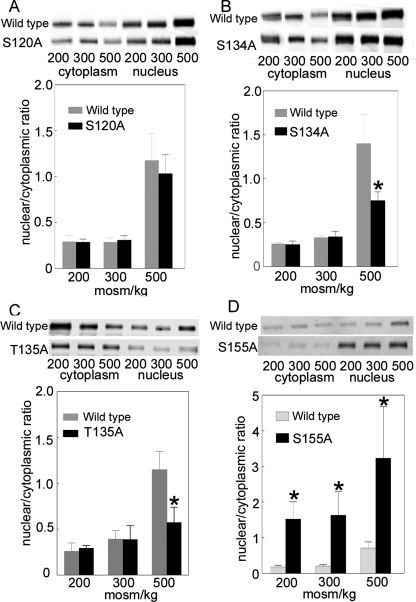
We measured the nuclear to cytoplasmic ratio of TonEBP/OREBP after changing osmolality from 300 to 200, 300, or 500 mOsm/kg for 1 h by altering NaCl. Raising NaCl increases nuclear to cytoplasmic ratio of wild-type (WT) TonEBP/OREBP-V5 approximately fourfold (Figure 2). Mutation of the amino acids in TonEBP/OREBP that are sites of phosphorylation has varying effects. Mutating S120 to alanine has no significant effect at any osmolality (Figure 2A). Mutating S134 (Figure 2B) or T135 (Figure 2C) to alanine decreases nuclear to cytoplasmic ratio at 500 mOsm/kg but has no effect at 200 or 300 mOsm/kg. Mutating S155 to alanine increases nuclear to cytoplasmic ratio at all osmolalities (Figure 2D).
FIGURE 2:
Effect on its nuclear to cytoplasmic ratio of mutating to alanine TonEBP/OREBP-V5- (A) S120, (B) S134, (C) T135, or (D) S155. HEK293 cells were transiently transfected with wild-type TonEBP/OREBP-V5 or a mutant at 300 mOsm/kg, and then the osmolality was changed to 200, 300, or 500 mOsm/kg by varying NaCl for 1 h. The relative amounts of TonEBP/OREBP-V5 in the cytoplasmic and nuclear fractions and the nuclear/cytoplasmic ratio were calculated from Western blots with anti-V5 antibody (mean ± SEM, *P ≤ 0.05, n = 3).
Because mutation of S120 to alanine does not affect nuclear to cytoplasmic ratio, we elected not to study it further at this time and turned our attention to S134 and T135. For this purpose, we produced phosphospecific antibodies against each of them. The antibody against TonEBP/OREBP–phospho-S134 detects a peptide (TonEBP/OREBP-130–139) containing phosphorylated, but not nonphosphorylated, TonEBP/OREBP-S134 (Supplemental Figure 1A). Unfortunately, it does not recognize phosphorylation at that site if T135 is also phosphorylated (Supplemental Figure 1A), as occurs in peptides that we identify by mass spectrometry (Figure 1 and Table 1). Further, it recognizes TonEBP/OREBP-S134A-V5, in which the amino acid at that site (alanine) cannot be phosphorylated (Supplemental Figure 2). Because we were able to produce an antibody against TonEBP/OREBP–phospho-T135 that does not suffer from those limitations, we elected to study the role of phosphorylation at that site.
High-NaCl induces phosphorylation of TonEBP/OREBP at threonine 135, catalyzed by CDK5 kinase
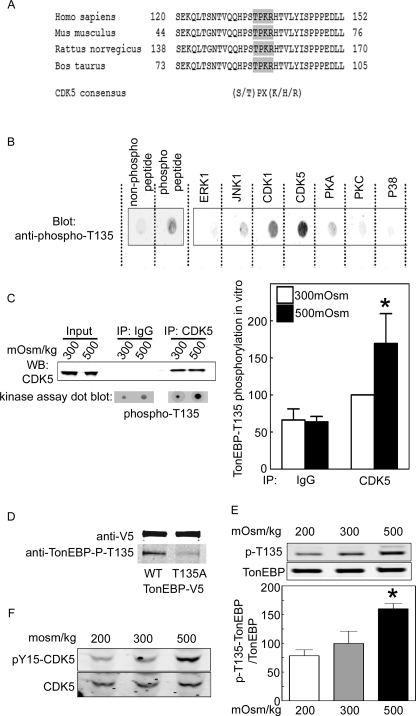
TonEBP/OREBP-T135 is within a consensus cyclin-dependent kinase 5 (CDK5) phosphorylation site that is conserved among mammalian species (Figure 3A). We used a specific anti-TonEBP/OREBP–phospho-T135 antibody to test for phosphorylation of TonEBP/OREBP-T135. The antibody cross-reacts with a TonEBP/OREBP peptide containing T135 (QQHPSTPKRH) only when the T135 is phosphorylated (Figure 3B, left panel, and Supplemental Figure 1B). To test whether CDK5 catalyzes phosphorylation of TonEBP/OREBP-T135, we performed in vitro kinase assays using the TonEBP/OREBP peptide containing T135 as substrate (Figure 3B, right panel) and various recombinant kinases. The peptide is strongly phosphorylated by CDK5, less by CDK1, and not at all by other serine/threonine kinases: extracellular signal–regulated kinase 1 (ERK1), c-Jun N-terminal kinase 1 (JNK1), protein kinase A (PKA), protein kinase C (PKC), and p38. It is not surprising that CDK1 phosphorylates this peptide in vitro because CDK1 and CDK5 consensus phosphorylation sites are similar. However, CDK1 is inhibited by high NaCl (Dmitrieva et al., 2002) and is thus unlikely to be the kinase of interest. We also tested the kinase activity of native CDK5 immunoprecipitated from HEK293 cells. The cells were exposed to medium kept at 300 or increased to 500 mOsm/kg (NaCl added) for 1 h. The immunoprecipitate containing CDK5 phosphorylates TonEBP/OREBP-T135, but the control immunoglobulin G (IgG) immunoprecipitate does not (Figure 3C, left panel). Also, native CDK5 kinase activity is greater when the cells from which it is immunoprecipitated have been exposed to high NaCl (Figure 3C, right panel). Further, in HEK293 cells high NaCl increases phosphorylation at T135 of recombinant WT TonEBP/OREBP-V5 (Figure 3D) and of native TonEBP/OREBP (Figure 3E). Because CDK5 is activated by phosphorylation on tyrosine 15 (Zukerberg et al., 2000), we tested for phosphorylation at that site, using a phosphospecific antibody. High NaCl increases phosphorylation of CDK5-Y15 (Figure 3F). We conclude that high NaCl increases CDK5 kinase activity, leading to increased phosphorylation of TonEBP/OREBP-T135.
FIGURE 3:
High NaCl induces phosphorylation of TonEBP/OREBP on threonine 135. (A) TonEBP/OREBP-T135 is in a consensus CDK5 phosphorylation site, conserved across mammalian species. (B) Left, specificity of the anti–phospho-T135 TonEBP/OREBP antibody. The antibody recognizes TonEBP/OREBP peptide containing T135 (QQHPSTPKRH) only when T135 is phosphorylated. Right, recombinant CDK1 and CDK5 phosphorylate TonEBP/OREBP-T135, but other recombinant serine/threonine kinases do not. In vitro kinase assay using the TonEBP/OREBP peptide containing T135 as substrate and detecting phosphorylation with the anti–phospho-T135-TonEBP/OREBP antibody. (C) High NaCl increases CDK5 kinase activity. HEK293 cells were transiently transfected with recombinant CDK5, and then osmolality was increased to 500 mOsm/kg (NaCl added) for 1 h before immunoprecipitating CDK5 for in vitro assay of its kinase activity, using TonEBP/OREBP peptide containing T135 as substrate and phospho-T135 TonEBP/OREBP antibody. Left, representative Western blot of immunoprecipitated CDK5 (top) and dot-blot result of the in vitro kinase assay (bottom) (mean ± SEM, *P ≤ 0.05, n = 3). (D) Anti–TonEBP/OREBP-phospho-T135 antibody recognizes wild-type (WT) TonEBP/OREBP-V5 but not TonEBP/OREBP-T135A-V5. HEK293 cells were transiently transfected with TonEBP/OREBP constructs at 300 mOsm/kg and then increased to 500 mOsm/kg NaCl for 1 h (NaCl added). (E) High NaCl increases phosphorylation of native TonEBP/OREBP on T135. HEK293 cells were maintained at 300 mOsm/kg or exposed to 500 mOsm/kg (NaCl added) for 1 h, and then whole cell extracts were analyzed by Western blot, using anti-TonEBP/OREBP or anti–TonEBP/OREBP-phospho-T135 antibodies (mean ± SEM, *P ≤ 0.05, n = 3). (F) High NaCl increases phosphorylation of CDK5 at tyrosine 15. HEK293 cells were exposed to medium at 300 or 500 mOsm/kg (NaCl added) for 1 h. Western blots used anti-CDK5 or anti–CDK5-phospho-Y15 antibodies. Representative of three experiments.
CDK5 physically associates with TonEBP/OREBP, and high NaCl increases the association
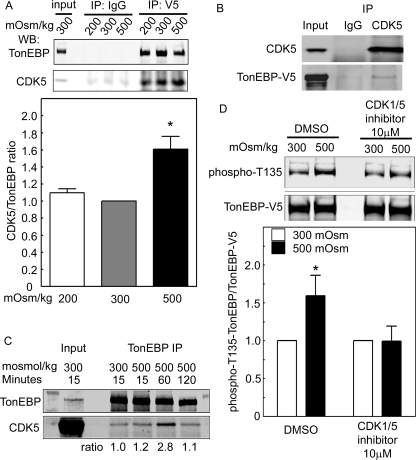
We transfected HEK293 cells with TonEBP/OREBP-V5 and then changed osmolality by varying NaCl for 1 h before immunoprecipitating with anti-V5 antibody or IgG (as control). Anti-V5 immunoprecipitates from cells at all the osmolalities contain CDK5 (Figure 4A, top). Increasing osmolality to 500 mOsm/kg by adding NaCl increases association of CDK5 with TonEBP/OREBP (Figure 4A, bottom). Also, TonEBP-V5 coimmunoprecipitates with CDK5 (Figure 4B), and native TonEBP/OREBP coimmunoprecipitates with native CDK5, reaching a maximum at 60 min (Figure 4C). We conclude that CDK5 physically associates with TonEBP/OREBP and that high NaCl increases the association.
FIGURE 4:
(A) CDK5 coimmunoprecipitates with TonEBP/OREBP-V5. HEK293 cells were transiently transfected with recombinant TonEBP/OREBP-V5 and then were exposed to media at 200, 300, or 500 mOsm/kg (NaCl varied) for 1 h. TonEBP/OREBP-V5 was immunoprecipitated (IP) with an anti-V5 antibody (IP: V5). Top, CDK5 coimmunoprecipitates with TonEBP/OREBP-V5 at all osmolalities. Immunoprecipitation with IgG is the control for specificity of the coimmunoprecipitation (IP: IgG). Bottom, high NaCl increases the coimmunoprecipitation of CDK5 with TonEBP/OREBP-V5 (mean ± SEM, *P ≤ 0.05, n = 3). (B) TonEBP-V5 coimmunoprecipitates with CDK5. As in (A) except that cells were exposed only to medium at 300 or 500 mOsm/kg and immunoprecipitation was with anti-CDK5. (C) Native CDK5 coimmunoprecipitates with native TonEBP. Medium bathing HEK293 cells was increased from 300 mOsm/kg to 500 mOsm/kg for 15, 60, or 120 min or left at 300 mOsm/kg for 15 min. Immunoprecipitation from whole cell extracts was performed using anti-TonEBP/OREBP antibody (TonEBP IP). Western blots were probed with anti-TonEBP/OREBP (TonEBP) or anti-CDK5 antibodies (CDK5). (D) Inhibition of CDK5 prevents the high NaCl–induced increase of phosphorylation of TonEBP/OREBP at T135. HEK293 cells were incubated with CDK1/5 inhibitor or DMSO and then maintained at 300 mOsm/kg or increased to 500 mOsm/kg (NaCl added) for 1 h. Western analysis of cell extracts was performed using anti–TonEBP/OREBP-phospho-T135 and anti–TonEBP/OREBP-V5 antibodies. Top, representative Western blot. Bottom, quantification of phosphorylation (mean ± SEM, *P < 0.05, n = 3).
Inhibition of CDK5 activity reduces high NaCl–induced phosphorylation of TonEBP/OREBP on T135
HEK293 cells were transfected with TonEBP/OREBP-V5 and then were incubated with dimethyl sulfoxide (DMSO) (control) or CDK1/5 inhibitor before increasing osmolality for 1 h by adding NaCl. CDK1/5 inhibitor prevents the 50% increase of phosphorylation of TonEBP/OREBP at T135 observed in the control (Figure 4D). This result is consistent with CDK5 kinase activity leading to increased phosphorylation at TonEBP/OREBP-T135.
CDK5 activity contributes to the rapid high NaCl–induced nuclear localization of TonEBP/OREBP
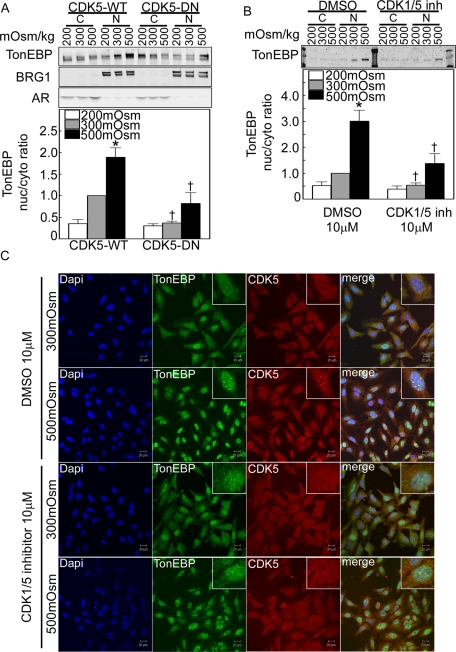
Within 2 h after NaCl is increased, the nuclear to cytoplasmic ratio of TonEBP/OREBP increases two- to threefold in HEK293 cells (Figure 5, A and B). Possible cross contamination between the nuclear and cytoplasmic extracts is excluded by clear separation of cytoplasmic (aldose reductase) and nuclear (Brg1) markers (Figure 5A). We inhibited CDK5 activity by overexpression of dominant-negative CDK5 kinase dead (CDK5-DN) (Figure 5A) or by addition of CDK1/5 inhibitor (Figure 5, B and C). Both ways of inhibiting CDK5 activity reduce the rapid increase of nuclear to cytoplasmic ratio of TonEBP/OREBP (Figure 5, A and B). Roscovitine, which is a less specific inhibitor of CDK5, also reduces the high NaCl–induced increase of TonEBP/OREBP nuclear localization (Supplemental Figure 3). Interestingly, under control conditions (DMSO) when osmolality is increased from 300 to 500 mOsm/kg by adding NaCl, bright foci of CDK5 appear in nuclei, colocalized with native TonEBP/OREBP. CDK1/5 inhibitor prevents formation of the foci (Figure 5C).
FIGURE 5:
CDK5 contributes to regulation of TonEBP/OREBP nuclear localization. (A) and (B) Inhibition of CDK5 kinase activity abolishes high NaCl–induced nuclear localization of TonEBP/OREBP. Western analysis was performed on proteins extracted from cytoplasm and nucleus, using antibodies against TonEBP/OREBP. (A) HEK293 cells transiently transfected with CDK5 wild-type (CDK5-WT) or dominant-negative kinase dead (CDK5-DN) were incubated for 2 h at 200, 300, or 500 mOsm/kg (NaCl varied). Top, representative Western blot. Bottom, summary data. Cytoplasmic (aldose reductase) and nuclear (Brg1) markers rule out important cross-contamination between the respective extracts. (B) HEK293 cells were incubated for 2 h at 200, 300, or 500 mOsm/kg (NaCl varied), with or without 10 μM CDK1/5 inhibitor (CDK1/5 inh). Top, representative Western blot. Bottom, summary data (mean ± SEM, n = 3, *P < 0.05 vs. 300 mOsm/kg control, †P < 0.05 control vs. experimental. (C) HeLa cells were preincubated for 1 h at 300 mOsm/kg with DMSO or with 10 μM CDK1/5 inhibitor, and then osmolality was increased to 500 mOsm/kg (NaCl added) for 1 h or kept at 300 mOsm/kg in the continued presence of CDK1/5 inhibitor or vehicle. Cells were fixed and stained with anti-TonEBP/OREBP and anti-CDK5 antibodies. Under control (DMSO) conditions at 300 mOsm/kg, TonEBP/OREBP is present in the cytoplasm, where it colocalizes with CDK5. Under control conditions at 500 mOsm/kg, TonEBP/OREBP moves into the nucleus, where it colocalizes in speckles with CDK5. The CDK1/5 inhibitor decreases both nuclear localization of TonEBP/OREBP at 500 mOsm/kg and its colocalization with CDK5 in the nucleus.
CDK5 contributes to the rapid increase of TonEBP/OREBP transcriptional activity 4 h after NaCl is raised
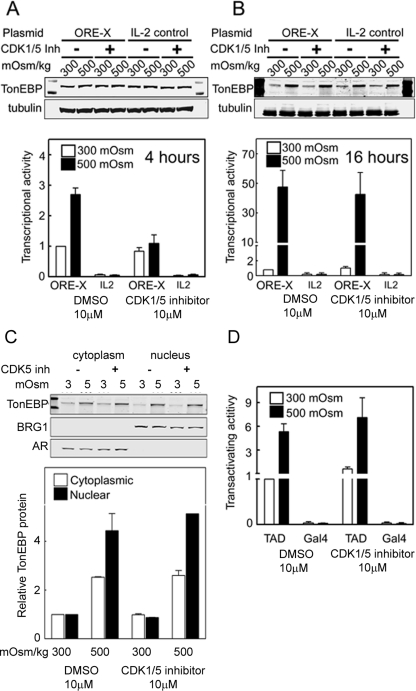
We measured TonEBP/OREBP transcriptional activity in HEK293 cells stably expressing an ORE-X reporter. Four hours after osmolality is increased from 300 to 500 mOsm/kg by adding NaCl, the transcriptional activity increases 2.7-fold (Figure 6A, bottom). CDK1/5 inhibitor prevents the increase (Figure 6A, bottom).
FIGURE 6:
CDK5 contributes to high NaCl–induced regulation of TonEBP/OREBP transcriptional activity but not to regulation of its transactivating activity. Osmolality bathing HEK293 cells stably expressing a TonEBP/OREBP transcription reporter (ORE-X) was increased from 300 to 500 mOsm/kg (NaCl added). (A) Transcriptional activity increases at 4 h under control conditions (DMSO), but not in the presence of 10 μM CDK1/5 inhibitor (CDK1/5). TonEBP protein expression does not change during the 4 h that osmolality is increased (top). However, after 16 h (B) the inhibitor no longer reduces TonEBP/OREBP transcriptional activity and TonEBP protein expression has increased, also not affected by the inhibitor (top). These effects do not occur with a control reporter that lacks ORE elements (IL2) (mean ± SEM, *P < 0.05, n = 3). (C) Inhibition of CDK5 does not affect the increase of TonEBP/OREBP in the nucleus 16 h after NaCl is raised. HEK293 cells were incubated with CDK1/5 inhibitor (CDK1/5 inh) or vehicle and then maintained at 300 mOsm/kg or increased to 500 mOsm/kg (NaCl added) for 16 h. Western analysis of nuclear and cytoplasmic extracts was performed using anti-TonEBP/OREBP antibody. Aldose reductase (AR, cytoplasmic marker) and BRG1 (nuclear marker) are controls for the purity of the cytoplasmic and nuclear extracts. Top, representative Western blot. Bottom, TonEBP/OREBP relative abundance (mean ± range, n = 2). (D) High NaCl does not affect TonEBP/OREBP transactivating activity. Osmolality bathing HEK293 cells stably expressing a dual luciferase reporter of TonEBP/OREBP transactivating activity (TAD) was increased from 300 to 500 mOsm/kg (NaCl added) for 16 h. An otherwise identical reporter lacking the TonEBP/OREBP transactivation domain (GAL4) serves as control (mean ± SEM, n = 3).
CDK5 does not contribute to the increase of TonEBP/OREBP transcriptional activity 16 h after NaCl is raised
At 16 h after osmolality is increased from 300 to 500 mOsm/kg by adding NaCl, the transcriptional activity increases 45-fold (Figure 6B, bottom). In contrast to the result at 4 h (Figure 6A), however, the increase at 16 h is not prevented by CDK1/5 inhibitor (Figure 6B). As a control, with an otherwise identical reporter lacking the ORE-X DNA element (IL2), high NaCl and CDK1/5 inhibitor are without effect. Note that although high NaCl has increased TonEBP/OREBP protein expression after 16 h (Figure 6B, top), it has not increased by 4 h (Figure 6A, top). Further, after 16 h, the amount of TonEBP/OREBP protein in the nucleus is unaffected by inhibition of CDK5 (CDK1/5 inh, Figure 6C). We conclude that the contribution of CDK5 to the increase of TonEBP/OREBP transcriptional activity 4 h after NaCl is elevated occurs because CDK5 contributes to its increased nuclear localization, but that after 16 h, CDK5 no longer contributes to increased TonEBP/OREBP transcriptional activity because it no longer contributes to regulation of TonEBP/OREBP nuclear localization.
CDK5 does not contribute to the high NaCl–induced increase of TonEBP/OREBP transactivating activity
We measured TonEBP transactivating activity with a dual luciferase reporter. At 16 h after increasing osmolality from 300 to 500 mOsm/kg by adding NaCl, the transactivating activity increases fivefold (DMSO; Figure 6D). CDK1/5 inhibitor does not affect the increase of transactivating activity (CDK1/5 inh; Figure 6D).
TonEBP/OREBP-T135 phosphorylation varies with osmolality in the rat renal inner medulla in vivo
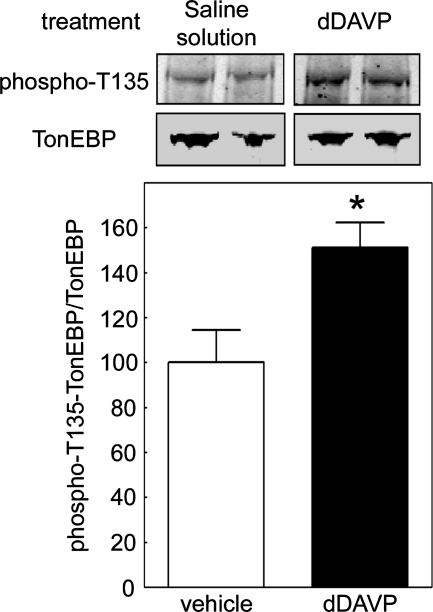
Brattleboro rats, being deficient in vasopressin, do not maintain a high renal medullary interstitial osmolality, nor do they concentrate their urine. One hour after injection of 2 nmol 1-desamino-8-d-arginine vasopressin (dDAVP), their urine osmolality increases from 203 to 575 mOsm/kg (Gallazzini et al., 2010). We find that phosphorylation of TonEBP/OREBP at T135 is significantly higher in renal inner medullas of Brattleboro rats 1 h after injection with dDAVP, compared with control rats injected with saline (Figure 7). This is consistent with high NaCl–induced increase of phosphorylation of TonEBP/OREBP on T135 in the renal inner medulla in vivo as well as in cell culture. We cannot exclude direct effects of vasopressin in vivo, but elevated vasopressin evidently does not contribute to the effect of high NaCl on HEK293 cells because it is not added in those experiments.
FIGURE 7:
Phosphorylation of TonEBP/OREB-T135 is regulated by osmolality in the rat renal inner medullary cells in vivo. Osmolality of urine and renal inner medullary interstitial fluid is low in Brattleboro rats because of their congenital lack of vasopressin but is rapidly restored following administration of dDAVP. Injection of 2 nmol dDAVP (dDAVP) increases phosphorylation of TonEBP/OREBP at T135 in the renal inner medulla compared with control injection (vehicle). Top, representative Western blots from inner medullas of two rats from each group, using anti-TonEBP/OREBP and anti–TonEBP/OREBP-phospho-T135 antibodies. Bottom, TonEBP/OREBP–phospho-T135/TonEBP (mean ± SEM, n = 3, *P < 0.05).
High NaCl–induced phosphorylation of TonEBP/OREBP-T135 does not depend on c-Abl or Fyn kinase
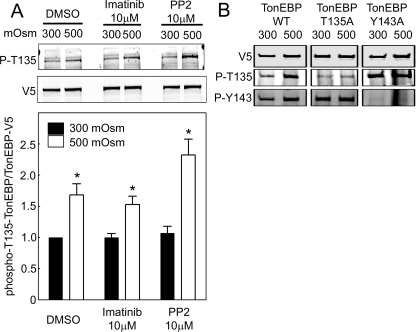
CDK5 activity can be modulated by c-Abl kinase (Lee et al., 2008; Cancino et al., 2009) and Fyn kinase (Okada et al., 2008). High NaCl activates c-Abl (Gallazzini et al., 2010) and Fyn (Ko et al., 2002), which contribute to activation of TonEBP/OREBP. With this in mind, we tested the effect of inhibiting c-Abl and Fyn on the high NaCl–induced increase of phosphorylation of TonEBP/OREBP-T135. A total of 10 μM of imatinib (Gallazzini et al., 2010) or protein phosphatase 2 (PP2) (Ko et al., 2002) was previously shown to inhibit TonEBP/OREBP transcriptional activity. However, neither inhibition of c-Abl kinase activity by imatinib (Druker and Lydon, 2000) nor inhibition of Fyn kinase activity by PP2 (Hanke et al., 1996) reduces the CDK5-mediated phosphorylation of TonEBP/OREBP (Figure 8A), indicating that they are not likely to be involved in the high NaCl–induced activation of CDK5. Further, phosphorylation of TonEBP/OREBP-Y135 does not depend on phosphorylation of TonEBP/OREBP-Y134 and vice versa. When NaCl is high (500 mOsm/kg total), phosphorylation of TonEBP/OREBP-Y143 is not reduced in the TonEBP/OREBP-Y135A mutant, and phosphorylation of TonEBP/OREBP-Y135 is not reduced in the TonEBP/OREBP-Y143A mutant (Figure 8B).
FIGURE 8:
(A) High NaCl–induced phosphorylation of TonEBP/OREBP at threonine 135 does not depend on c-Abl or Fyn kinase activity. HEK293 cells transfected with TonEBP/OREBP-V5 were incubated with c-Abl inhibitor (imatinib), Fyn inhibitor (PP2), or vehicle for 1 h and then maintained at 300 mOsm/kg or increased to 500 mOsm/kg (NaCl added) for 1 h. Western analysis of cell extracts was performed using anti–TonEBP/OREBP-phospho-T135 and anti–TonEBP/OREBP-V5 antibodies. Top, representative Western blot. Bottom, quantification of phosphorylation (mean ± SEM, *P < 0.05, n = 3). (B) High NaCl–induced phosphorylation of TonEBP/OREBP-T135 does not require phosphorylation of TonEBP/OREBP-Y143 and vice versa. HEK293 cells transfected with wild-type TonEBP/OREBP-V5 (TonEBP WT), TonEBP/OREBP-T135A (TonEBP T135A), or TonEBP/OREBP-Y143A (TonEBP Y135A) were exposed to high NaCl (500 mOsm/kg, NaCl added) for 1 h, and then whole cell extracts were analyzed by Western blot, using anti-V5, anti–TonEBP/OREBP-phospho-T135 (P-T135), or anti–TonEBP/OREBP-phospho-Y143 antibodies.
DISCUSSION
CDK5
Cyclin-dependent kinases (Cdks) are small serine/threonine protein kinases that play essential roles in regulating cell cycle progression. Their molecular partners, the cyclins, activate and confer specificity to Cdks. Although CDK5 (Rosales and Lee, 2006; Dhariwala and Rajadhyaksha, 2008; Lalioti et al., 2010) has homology to CDK1 and 2, its function is vastly different because it does not regulate cell cycle and it is activated by noncyclin proteins, p35 and p39. While CDK5 is expressed ubiquitously in mammalian tissues and phosphorylates a variety of proteins, its activator proteins are predominantly expressed in postmitotic neurons and its activity has been studied mainly in the central nervous system. Cellular processes previously known to be regulated by CDK5 include cytoskeletal dynamics, transcription, apoptosis, and senescence. Interestingly, some of these same processes are also altered in osmotically stressed cells, as will be noted.
Cytoskeletal dynamics and the proteins that regulate them are among the major known targets of CDK5. CDK5 controls the reorganization of the cytoskeleton involved in cell propulsion and in expansion and guidance of cell outgrowths. Rho GTPases are among preferred targets of CDK5 and are critical elements in the complex pathways that regulate actin and microtubule dynamics in the cytoskeleton. Rho GTPases are also involved in cytoskeletal changes associated with restoration of cell volume following changes in tonicity (Hoffmann and Pedersen, 2006), but it is not clear what role, if any, CDK5 has in this control of cell volume.
CDK5 also regulates other transcription factors and events besides the high NaCl–induced increase of TonEBP/OREBP activity identified in the present studies. CDK5 activates signal transducer and activator of transcription 3 by phosphorylating it on Ser-727 (Fu et al., 2004), resulting in increased transcription of many genes, including c-fos, junB, and fibronectin. CDK5 is expressed in insulin-producing β cells of the pancreas. Elevation of glucose increases p35/CDK5 activity, which stimulates transcription of the insulin gene (Ubeda et al., 2004). CDK5 also increases transcription of the acetylcholine receptor (Fu et al., 2001). Further, CDK5 is involved in gene regulation that promotes apoptosis. Thus it promotes apoptosis by inhibiting myocyte enhancer factor 2 (Gong et al., 2003). It also stabilizes the transcription factor, p53, by phosphorylating it on Ser-15, Ser-33, and Ser-46 in response to genotoxic and oxidative stresses. The phosphorylation prevents association of p53 with Mdm2, an association that would otherwise facilitate ubiquitination and degradation of p53 (Lee et al., 2007). Thus the p53 phosphorylated by CDK5 is stabilized, contributing to the induction of proapoptotic gene expression. Hypertonicity activates p53, but this activation protects against apoptosis, rather than promoting it (Dmitrieva et al., 2000; Dmitrieva et al., 2001). The role, if any, of CDK5 in hypertonicity-induced apoptosis and activation of p53 has not been investigated.
CDK5 has a role in cellular senescence. Senescent cells are enlarged and flattened and express senescence-associated β-galactosidase (SA-β-Gal). CDK5 plays a central role in these changes by reducing Rac1 activity (Alexander et al., 2004). CDK5 activity increases in cells induced to senesce by a variety of stimuli, and inhibition of CDK5 activity reduces the morphological changes and the expression of SA-β-Gal. Repression of Rac1 activity by CDK5 is necessary for expression of the senescent phenotype, particularly for the actin polymerization accompanying senescent morphology. Interestingly, we previously found that high NaCl accelerates senescence both in cell culture and in the renal inner medulla in vivo (Dmitrieva and Burg, 2007), and we now find that it increases CDK5 activity (Figure 3F). Therefore it is plausible that activation of CDK5 contributes to high NaCl–induced cellular senescence.
Regulation of TonEBP/OREBP transcriptional activity by its nuclear localization
One factor determining the effectiveness of transcription factors like TonEBP/OREBP is how much of them is available in the nucleus. The amount available in the nucleus depends on both the total amount in the cell and its nuclear localization. High salt rapidly increases nuclear localization of TonEBP/OREBP (Figure 5), but not the total amount of it (Figure 6A). Later, both increase (Figure 6, B and C). Inhibition of CDK5 rapidly reduces high NaCl–induced nuclear localization of TonEBP/OREBP (Figure 5), but not its total abundance (Figure 6A), and this reduction is associated with decreased transcriptional activity (Figure 6A). In contrast, after 16 h of high NaCl, inhibition of CDK5 does not affect either the amount of TonEBP/OREBP in the nucleus (Figure 6C) or its transcriptional activity (Figure 6B). We propose that the reduction of high NaCl–induced increase of TonEBP/OREBP transcriptional activity at 4 h caused by inhibition of CDK5 is due to the reduction of the amount of TonEBP/OREBP in the nucleus. The effect disappears by 16 h (Figure 6B) because the amount of TonEBP/OREBP in the nucleus is no longer affected by inhibition of CDK5 (Figure 6C). Nuclear localization of TonEBP/OREBP apparently is a significant determinant of its transcriptional activity under these conditions. The CDK5-dependent, rapid nuclear localization of TonEBP/OREBP serves to accelerate transcription of its osmoprotective target genes.
CDK5 does not affect TonEBP/OREBP transactivating activity
Transactivating activity is another determinant of its transcriptional activity. Although the assay of TonEBP/OREBP transcriptional activity depends on the amount of native TonEBP/OREBP available in the nucleus, the assay of transactivating activity does not because the reporter of TonEBP/OREBP transactivating activity is a dual GAL4-based system that locates constitutively in the nucleus and is driven by a recombinant TonEBP/OREBP transactivating domain that is independent of native TonEBP/OREBP (Figure 6D) (Ferraris et al., 2002).
Functional consequences of phosphorylation of TonEBP/OREBP-S155
S155 was previously found to be phosphorylated in HeLa cells and to affect nuclear localization (Xu et al., 2008). TonEBP/OREBP-132–581 plasmid produces a truncated TonEBP/OREBP protein devoid of its nuclear export signal, so it is preferentially localized to the nucleus at 300 mOsm/kg. However, it is exported to the cytoplasm at 250 mOsm/kg (Tong et al., 2006). In contrast, the S155A mutant remains in the nucleus at 250 mOsm/kg (Xu et al., 2008). Results for osmolality greater than 300 mOsm/kg were not reported. Our finding that nuclear localization of TonEBP/OREBP-S155A is enhanced at all osmolalities, including 500 mOsm/kg (Figure 2D), is consistent with the previous finding that phosphorylation at TonEBP/OREBP-S155 contributes to its nuclear export (Xu et al., 2008). Therefore we elected not to study it further at this time.
No apparent toxicity from inhibition of CDK5
HEK293 cells survive well in media with NaCl added to a total concentration up to 600 mOsm/kg. At higher levels, toxicity is apparent from altered cell appearance and reduced number. Because inhibition of CDK5 did not result in any apparent toxicity, we did not further investigate this possibility. Lack of toxicity is not surprising considering that the effects of CDK5 on TonEBP/OREBP activity are partial and transient.
Relation between nuclear localization of TonEBP/OREBP and its transactivating activity
In this study we found that CDK5 contributes to high NaCl–induced activation of TonEBP/OREBP by accelerating its nuclear accumulation without changing its transactivating activity. This is the first instance that we are aware of in which nuclear localization of TonEBP/OREBP increases its high NaCl–induced activity without an accompanying increase of its transactivating activity. There are regulators of TonEBP/OREBP transcriptional activity that affect only its transactivating activity (Ferraris et al., 2002) without affecting its nuclear localization, namely, MDC1, a DNA damage response protein (Kunin et al., 2010); HSP90, a chaperone and coactivator (Chen et al., 2007); Fyn, a tyrosine kinase (Ko et al., 2002); and p38, a mitogen-activated protein kinase (Ko et al., 2002). Four additional proteins regulate both nuclear localization and transactivating activity, namely, c-Abl (Gallazzini et al., 2010), PLC-γ1 (Irarrazabal et al., 2010), ATM (Irarrazabal et al., 2004; Zhang et al., 2005), and phosphatidylinositol 3-kinase IA (PI3K-IA) (Irarrazabal et al., 2006). CDK5, however, is unique in that it regulates transcriptional activity by affecting nuclear localization without affecting transactivating activity. Considering the modular composition of TonEBP/OREBP, it has seemed surprising that any individual protein could affect both nuclear localization and transactivating activity. TonEBP/OREBP’s nuclear localization and export domains are located within its N-terminal one-third and its transactivating domain is located in the C-terminal two-thirds, as are the sequences involved in modulating it (Burg et al., 2007). The explanation for the combined actions may lie in dual activities of PLC-γ1 (Irarrazabal et al., 2010), plus its network connections. PLC-γ1 signals both through its lipase activity and through association with other proteins via its SH3 domain (Irarrazabal et al., 2010). The lipase activity is necessary for its effect on nuclear localization of TonEBP/OREBP and the SH3 domain for its effect on transactivation, possibly by guanine nucleotide exchange factor activity (Irarrazabal et al., 2010). Its probable network connections include c-Abl (Plattner et al., 2003), PI3K-IA (Ye et al., 2002; Ye, 2005), and ATM (Khanna et al., 1997), explaining how they could participate in activating both the nuclear localization and transactivating activity of TonEBP/OREBP.
MATERIALS AND METHODS
Cell culture and treatment
HEK293 cells in passages 38–48 were cultured at 300 mOsm/kg in Eagle’s minimal essential medium (American Type Culture Collection, Manassas, VA) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT) as previously described (Irarrazabal et al., 2004). Osmolality of the control medium was 300 mOsm/kg, and at experiment-specific time points, medium was replaced with ones at 300 mOsm/kg, 200 mOsm/kg (NaCl added to NaCl-free medium; Biofluids, Rockville, MD), or 500 mOsm/kg (NaCl added). CDK5 inhibitors roscovitine and CDK1/5 inhibitor were from EMD Chemicals (Gibbstown, NJ). Inhibitors were added to medium bathing HEK293 cells 60 min before changing the osmolality and were added in any subsequent changes of medium. All cells were cultured at 37°C in 5% CO2 and were studied while subconfluent. Anti-TonEBP/OREBP, anti–aldose reductase, anti-BRG1, anti-CDK5 and anti–CDK5-phospho-Y15 antibodies were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA) and anti-tubulin from Rockland (Gilbertsville, PA). Anti-V5 was from AbD Serotec (Raleigh, NC). The rabbit anti–TonEBP/OREBP-phospho-T135 and –phospho-S134 were produced by PhosphoSolutions (Aurora, CO). HEK293 cells stably expressing TonEBP/OREBP-1–547-V5-His were previously described (Chen et al., 2007).
Plasmids and transfection
Human TonEBP/OREBP cDNA clone KIAA0827 was a gift from Takahiro Nagase (Kazusa DNA Research Institute, Chiba, Japan). Sequences coding for amino acids 1–547 or 1–1531 of KIAA0827 were cloned into expression vector pcDNA6V5-His (Invitrogen, Carlsbad, CA) to generate 1–547- or 1–1531-V5-His, as previously described (Irarrazabal et al., 2004; Zhang et al., 2005). Mutants S120A, S134A, T135A, and S155A of 1–1531-V5 were prepared by site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA). All constructs were generated using standard cloning procedures and verified by restriction enzyme digestion and DNA sequencing. HEK293 cells were transfected with Lipofectamine 2000 (Invitrogen), according to the supplier’s instructions. CDK5 WT or kinase dead dominant negative (KD) were kindly provided by Harish C. Pant (Laboratory of Neurochemistry, NINDS, Bethesda, MD). The ORE-X luciferase reporter construct contains two copies of human ORE-X element (Ferraris et al., 1996) in a plasmid with a minimal IL2 promoter (hTonE-GL3, a gift from S. N. Ho, University of California at San Diego, La Jolla, CA) (Trama et al., 2000). The binary GAL4/TAD reporter system was previously described (Ferraris et al., 2002).
Immunoprecipitation
After washing, cells were collected in buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, protease inhibitor cocktail tablet (Roche, Indianapolis, IN), and phosphatase cocktail inhibitor I and II (Sigma-Aldrich, St. Louis, MO). Cell extracts were precleared with mouse IgG plus Protein A/G PLUS-Agarose (Santa Cruz Biotechnologies) at 4ºC for 1 h and then incubated with mouse anti-V5 antibody or anti-CDK5 antibody plus Protein A/G PLUS-Agarose at 4ºC overnight. The agarose beads were washed gently once with the lysis buffer. Proteins were eluted with the SDS loading buffer, boiled for 5 min, then subjected to Western analysis or kept at 4°C for kinase assay.
Sample preparation for mass spectrometry
HEK 293 cells stably transfected with TonEBP/OREBP-1–547-V5-His were grown in 15-cm dishes. Osmolality was increased to 500 mOsm/kg by adding NaCl for 2 h, and then nuclear extracts were prepared with NE-PER (Pierce, Rockford, IL), according to the supplier’s instructions, with added protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail I and II (Sigma-Aldrich). Extracts were precleared for 1 h with rabbit IgG, biotin conjugated (Santa Cruz Biotechnology), on Dynabeads (Invitrogen) for 1 h. Precleared supernatants were incubated overnight with rabbit anti-V5, biotin conjugated (ICL Lab, Newberg, OR), on Dynabeads (Invitrogen). Beads were washed three times with buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1% Triton X-100 and three times with phosphate-buffered saline (PBS) containing 1% Triton X-100. Both buffers included phosphatase and protease inhibitor cocktails. The beads were resuspended in 6 M guanidine-HCl/50mM NH4HCO3 to denature the proteins and elute them from the beads. Protein samples were then reduced with 100 mM dithiothreitol (DTT) for 1 h at 56°C and alkylated using 100 mM iodoacetamide for 1 h at room temperature in the dark. The sample buffer was exchanged to 50 mM NH4HCO3 (for trypsin digestion) or to 100 mM Tris-HCl, 10 mM CaCl2, pH 7.6 (for endoproteinase Arg-C digestion). Amicon Ultra Centrifugal Filter Devices (Millipore, Billerica, MA) were used for buffer exchange. Proteins were digested with trypsin (Promega, Madison, WI) or endoproteinase Arg-C (Roche Applied Science, Mannheim, Germany) in a ratio of 1:50 wt/wt overnight at 37°C. Peptide samples were desalted using a 1-ml hydrophilic-lipophilic–balanced cartridge (Oasis, Milford, MA), followed by volume reduction in vacuo. Samples were resuspended in 50 μl of 5% acetic acid, pH 2.5–3.0 and then loaded onto an IMAC column (Pierce) for phosphopeptide enrichment. Peptides were incubated with this Ga3+ resin for 20 min agitating gently every 5 min and then washed and eluted according to the supplier’s protocol. Samples were then dried in vacuo, resuspended in 1% formic acid, and desalted with C18 Ziptips (Millipore) before analysis by MS/MS.
Phosphopeptides identification and validation
IMAC eluates and flow-through fractions were both analyzed on an Agilent 1100 nanoflow system (Agilent Technologies, Palo Alto, CA) LC connected to a Finnigan LTQ mass spectrometer (Thermo Electron, San Jose, CA) equipped with a nanoelectrospray ion source. MS spectra were analyzed with BIOWORKS software (Thermo Electron), using the SEQUEST search algorithm for peptide identification. Peak masses were searched against the most current version of the Human RefSeq Database (National Center for Biotechnology Information) and its reversed complement with the following parameters: fixed carbamidomethylation of Cys; variable phosphorylation of Ser, Thr, and Tyr when searching MS2; and use of a preliminary exclusion filter to remove poor-quality spectra. The filter removed any spectrum with the following values for the charge state of the peptide and the associated Xcorr score: +1 ≤ 1.5; +2 ≤ 2.0; and +3 ≤ 2.5. Xcorr takes into account the number of peaks matched between actual and theoretical spectra and is directly proportional to spectral quality. We used Ascore, a probability-based score (Beausoleil et al., 2006), to estimate the probability of correct phosphorylation site localization.
Kinase assays
Beads containing CDK5 immunoprecipitated from cell extracts were suspended in Kinase Buffer (Cell Signaling Technology, Danvers, MA) and then incubated with 1 μg TonEBP/OREBP-T135 peptide (QQHPSTPKRH) in assay buffer supplemented with 200 μM ATP (Cell Signaling Technology) at 30°C for 30 min. Supernatants were analyzed by dot blot for phosphorylated TonEBP/OREBP-T135. The relative amounts of CDK5 present on the beads were measured by Western analysis after decanting the supernatant. For in vitro kinase assay, the following recombinant kinases were purchased from Cell Signaling Technology: ERK1, JNK1, PKAc, PKC, p38, CDK1, and CDK5; 100 ng of recombinant kinase was incubated in kinase buffer containing 200 μM ATP and 1 μg TonEBP-T135 peptide in 30 μl final volume for 30 min at 30°C and analyzed by dot blot for phosphorylated TonEBP/OREBP-T135.
Phosphospecific antibodies
Antibodies against TonEBP/OREBP–phospho-S134 and -T135 were prepared by PhosphoSolutions. To verify their specificity, dilutions of the peptide containing TonEBP/OREBP amino acids 130–139 (QQHPSTPKRH), unphosphorylated or phosphorylated at S134, T135, or both, in 50 μl PBS were spotted on nitrocellulose membranes with a manifold suction device (Bio-Rad Laboratories, Hercules, CA). Nonspecific binding was blocked by incubating membranes at room temperature for 1 h with Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE) diluted 1:1 with PBS. Membranes were incubated with rabbit anti–TonEBP/OREBP phospho-S134 or anti–TonEBP/OREBP phospho-T135 at 4°C overnight. After washing with 0.1% Tween-20 in PBS, blots were incubated with Alexa Fluor 680 goat anti–rabbit IgG (Molecular Probes, Carlsbad, CA) for 1 h in the dark. Blots were visualized and quantified using an Odyssey Infrared Imager (LI-COR Biosciences).
Western analysis
Western analysis was performed as previously described (Irarrazabal et al., 2010). Briefly, cells were lysed with buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1% Triton X-100 (for whole-cell extracts) or with NE-PER (Pierce) for separate nuclear and cytoplasmic fractions, according to the supplier’s instructions. Protease inhibitor mixture (Roche Diagnostics, Alameda, CA) and phosphatase inhibitor cocktails I (Sigma-Aldrich) were included in the lysis buffers. Total protein concentration was measured in cell extracts by BCA assay (Pierce), and proteins were separated on 4–12% gradient Novex Bis-Tris or 3–8% gradient Tris-Acetate gels and transferred electrophoretically to nitrocellulose membranes (Invitrogen). Nonspecific binding was blocked by incubating membranes for 1 h at room temperature with Odyssey Blocking Buffer (LI-COR Biosciences) diluted 1:1 with PBS. Membranes were then incubated with mouse anti-V5 (Invitrogen) or rabbit anti–TonEBP/OREBP phospho-S134 or rabbit anti–TonEBP/OREBP phospho-T135 at 4°C overnight. After washing with 0.1% Tween-20 in PBS, blots were incubated with Alexa Fluor 680 goat anti–rabbit IgG or Alexa Fluor 780 or 800 goat anti–mouse IgG (Molecular Probes) for 1 h in the dark. Blots were visualized and quantified using an Odyssey Infrared Imager (LI-COR Biosciences).
Nuclear to cytoplasmic ratio
Nuclear to cytoplasmic ratio was determined as previously described (Ferraris and Burg, 2007). Cytoplasmic and nuclear proteins were extracted separately by using NE-PER (Pierce). Specific proteins in each fraction were measured by Western analysis. The relative amounts of TonEBP/OREBP in the cytoplasmic and nuclear fractions and the nuclear/cytoplasmic ratio were calculated from the relative concentrations of the respective protein in each cytoplasmic or nuclear extract and the relative volumes of the extracts.
Immunofluorescence studies of HeLa cells
The cells were fixed for immunofluorescence with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 PBS, and then blocked with 1% bovine serum albumin. Primary antibodies were rabbit anti-TonEBP/OREBP and mouse anti-CDK5. The secondary antibodies were conjugated to Alexa 488 or 633 fluorophores (Invitrogen), respectively. Images were acquired using a Zeiss LSM 510 META microscope with a 40× NA = 1.3, oil immersion objective (Carl Zeiss MicroImaging, Thornwood, NY).
Luciferase assays
Reporter experiments were performed as previously described. In brief, for assay of the transcriptional activity of native TonEBP/OREBP, HEK293 cells expressing stably transfected ORE-X IL2 min GL3_Bsd or IL2 promoter only (control) reporters were used (Irarrazabal et al., 2004). For assay of TonEBP/OREBP transactivating activity, cells expressing the stably transfected GAL4 reporter (pFR-Luc) and GAL4dbd-548–1531, which contain the recombinant TonEBP/OREBP transactivation domain, were used (Ferraris et al., 2002). HEK293 cells were preincubated with CDK1/5 inhibitor for 1 h followed by 4 or 16 h at either 300 or 500 mOsm/kg (NaCl added).
Animal studies
Pathogen-free male Sprague-Dawley rats (Taconic Farms, Germantown, NY) or homozygous Brattleboro rats (Harlan Sprague-Dawley, Indianapolis, IN) weighing 150–200 g were used. All experiments were conducted in accord with animal protocol H-0110R1 approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute. Homogenates were prepared from the inner medullas, in Laemmli buffer (1.5% SDS, 10 mM Tris, pH 6.8), containing a protease/phosphatase inhibitor cocktail (Pierce). Following determination of total protein concentration (Pierce BCA, Rockland, IL), samples were heated to 60°C for 15 min and then stored in 6% glycerol plus 40 mM DTT for Western analysis.
Statistical analysis
Data were compared by analysis of variance (ANOVA), followed by a Student–Newman–Keuls posttest. Normalized data were transformed before ANOVA. Results are expressed as means ± standard error of the mean (SEM) (n = number of independent experiments). Differences were considered significant for P < 0.05.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute.
Abbreviations used:
- ANOVA
analysis of variance
- ATM
ataxia telangiectasia mutated
- CDK
cyclin-dependent kinase
- dDAVP
1-desamino-8-d-arginine vasopressin
- DMSO
dimethyl sulfoxide
- ERK
extracellular signal–regulated kinase
- HEK
human embryonic kidney
- IgG
immunoglobulin G
- IP
immunoprecipitation
- JNK
c-Jun N-terminal kinase
- MS/MS
tandem mass spectometry
- OREBP
osmotic response element–binding protein
- PBS
phosphate-buffered saline
- PI3K
phosphatidylinositol 3-kinase
- PKA
protein kinase A
- PKC
protein kinase C
- PLC
phospholipase C
- PP2
protein phosphatase
- SA-β-Gal
senescence-associated β-galactosidase
- TonEBP
tonicity-responsive enhancer–binding protein
- WT
wild-type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0681) on January 5, 2011.
REFERENCES
- Alexander K, Yang HS, Hinds PW. Cellular senescence requires CDK5 repression of Rac1 activity. Mol Cell Biol. 2004;24:2808–2819. doi: 10.1128/MCB.24.7.2808-2819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiol Rev. 2007;87:1441–1474. doi: 10.1152/physrev.00056.2006. [DOI] [PubMed] [Google Scholar]
- Cancino GI, Perez de Arce K, Castro PU, Toledo EM, von Bernhardi R, Alvarez AR. c-Abl tyrosine kinase modulates tau pathology and Cdk5 phosphorylation in AD transgenic mice. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.07.007. in press) [DOI] [PubMed] [Google Scholar]
- Chen Y, Schnetz MP, Irarrazabal CE, Shen RF, Williams CK, Burg MB, Ferraris JD. Proteomic identification of proteins associated with the osmoregulatory transcription factor TonEBP/OREBP: functional effects of Hsp90 and PARP-1. Am J Physiol Renal Physiol. 2007;292:F981–F992. doi: 10.1152/ajprenal.00493.2005. [DOI] [PubMed] [Google Scholar]
- Dahl SC, Handler JS, Kwon HM. Hypertonicity-induced phosphorylation and nuclear localization of the transcription factor TonEBP. Am J Physiol Cell Physiol. 2001;280:C248-C253. doi: 10.1152/ajpcell.2001.280.2.C248. [DOI] [PubMed] [Google Scholar]
- Dhariwala FA, Rajadhyaksha MS. An unusual member of the Cdk family: Cdk5. Cell Mol Neurobiol. 2008;28:351–369. doi: 10.1007/s10571-007-9242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieva N, Kultz D, Michea L, Ferraris J, Burg M. Protection of renal inner medullary epithelial cells from apoptosis by hypertonic stress-induced p53 activation. J Biol Chem. 2000;275:18243–18247. doi: 10.1074/jbc.M000522200. [DOI] [PubMed] [Google Scholar]
- Dmitrieva N, Michea L, Burg MB. p53 tumor suppressor protein protects renal inner medullary cells from hypertonic stress by restricting DNA replication. Am J Physiol Renal Physiol. 2001;281:F522–F530. doi: 10.1152/ajprenal.2001.281.3.F522. [DOI] [PubMed] [Google Scholar]
- Dmitrieva NI, Bulavin DV, Fornace AJ Jr, Burg MB. Rapid activation of G2/M checkpoint after hypertonic stress in renal inner medullary epithelial (IME) cells is protective and requires p38 kinase. Proc Natl Acad Sci USA. 2002;99:184–189. doi: 10.1073/pnas.231623498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieva NI, Burg MB. High NaCl promotes cellular senescence. Cell Cycle. 2007;6:3108–3113. doi: 10.4161/cc.6.24.5084. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Lydon NB. Lessons learned from the development of an Abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest. 2000;105:3–7. doi: 10.1172/JCI9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Amer Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Ferraris JD, Burg MB. Tonicity-regulated gene expression. Methods Enzymol. 2007;428:279–296. doi: 10.1016/S0076-6879(07)28016-4. [DOI] [PubMed] [Google Scholar]
- Ferraris JD, Williams CK, Jung KY, Bedford JJ, Burg MB, Garcia-Perez A. ORE, a eukaryotic minimal essential osmotic response element. The aldose reductase gene in hyperosmotic stress. J Biol Chem. 1996;271:18318–18321. doi: 10.1074/jbc.271.31.18318. [DOI] [PubMed] [Google Scholar]
- Ferraris JD, Williams CK, Persaud P, Zhang Z, Chen Y, Burg MB. Activity of the TonEBP/OREBP transactivation domain varies directly with extracellular NaCl concentration. Proc Natl Acad Sci USA. 2002;99:739–744. doi: 10.1073/pnas.241637298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu AK, Fu WY, Cheung J, Tsim KW, Ip FC, Wang JH, Ip NY. Cdk5 is involved in neuregulin-induced AChR expression at the neuromuscular junction. Nat Neurosci. 2001;4:374–381. doi: 10.1038/86019. [DOI] [PubMed] [Google Scholar]
- Fu AK, Fu WY, Ng AK, Chien WW, Ng YP, Wang JH, Ip NY. Cyclin-dependent kinase 5 phosphorylates signal transducer and activator of transcription 3 and regulates its transcriptional activity. Proc Natl Acad Sci USA. 2004;101:6728–6733. doi: 10.1073/pnas.0307606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallazzini M, Yu MJ, Gunaratne R, Burg MB, Ferraris JD. c-Abl mediates high NaCl-induced phosphorylation and activation of the transcription factor TonEBP/OREBP. FASEB J. 2010;24:4325–4335. doi: 10.1096/fj.10-157362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, Pollok BA, Connelly PA. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- Hoffmann EK, Pedersen SF. Sensors and signal transduction pathways in vertebrate cell volume regulation. Contrib Nephrol. 2006;152:54–104. doi: 10.1159/000096318. [DOI] [PubMed] [Google Scholar]
- Irarrazabal CE, Burg MB, Ward SG, Ferraris JD. Phosphatidylinositol 3-kinase mediates activation of ATM by high NaCl and by ionizing radiation: role in osmoprotective transcriptional regulation. Proc Natl Acad Sci USA. 2006;103:8882–8887. doi: 10.1073/pnas.0602911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irarrazabal CE, Gallazzini M, Schnetz MP, Kunin M, Simons BL, Williams CK, Burg MB, Ferraris JD. Phospholipase C-g1 is involved in signaling the activation by high NaCl of the osmoprotective transcription factor TonEBP/OREBP. Proc Natl Acad Sci USA. 2010;107:906–911. doi: 10.1073/pnas.0913415107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irarrazabal CE, Liu JC, Burg MB, Ferraris JD. ATM, a DNA damage-inducible kinase, contributes to activation by high NaCl of the transcription factor TonEBP/OREBP. Proc Natl Acad Sci USA. 2004;101:8809–8814. doi: 10.1073/pnas.0403062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Yan J, Watters D, Hobson K, Beamish H, Spring K, Shiloh Y, Gatti RA, Lavin MF. Defective signaling through the B cell antigen receptor in Epstein-Barr virus-transformed ataxia-telangiectasia cells. J Biol Chem. 1997;272:9489–9495. doi: 10.1074/jbc.272.14.9489. [DOI] [PubMed] [Google Scholar]
- Ko BC, Lam AK, Kapus A, Fan L, Chung SK, Chung SS. Fyn and p38 signaling are both required for maximal hypertonic activation of the OREBP/TonEBP. J Biol Chem. 2002;277:46085–46092. doi: 10.1074/jbc.M208138200. [DOI] [PubMed] [Google Scholar]
- Ko BC, Turck CW, Lee KW, Yang Y, Chung SS. Purification, identification, and characterization of an osmotic response element binding protein. Biochem Biophys Res Commun. 2000;270:52–61. doi: 10.1006/bbrc.2000.2376. [DOI] [PubMed] [Google Scholar]
- Kunin M, Dmitrieva NI, Gallazzini M, Shen RF, Wang G, Burg MB, Ferraris JD. Mediator of DNA damage checkpoint 1 (MDC1) contributes to high NaCl-induced activation of the osmoprotective transcription factor TonEBP/OREBP. PLoS One. 2010;5:e12108. doi: 10.1371/journal.pone.0012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioti V, Pulido D, Sandoval IV. Cdk5, the multifunctional surveyor. Cell Cycle. 2010;9:284–311. doi: 10.4161/cc.9.2.10466. [DOI] [PubMed] [Google Scholar]
- Lee JH, Jeong MW, Kim W, Choi YH, Kim KT. Cooperative roles of c-Abl and Cdk5 in regulation of p53 in response to oxidative stress. J Biol Chem. 2008;283:19826–19835. doi: 10.1074/jbc.M706201200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim HS, Lee SJ, Kim KT. Stabilization and activation of p53 induced by Cdk5 contributes to neuronal cell death. J Cell Sci. 2007;120:2259–2271. doi: 10.1242/jcs.03468. [DOI] [PubMed] [Google Scholar]
- Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a Rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, et al. CDK5-dependent phosphorylation of the Rho family GTPase TC10(alpha) regulates insulin-stimulated GLUT4 translocation. J Biol Chem. (2008;283:35455–35463. doi: 10.1074/jbc.M806531200. [DOI] [PubMed] [Google Scholar]
- Plattner R, Irvin BJ, Guo S, Blackburn K, Kazlauskas A, Abraham RT, York JD, Pendergast AM. A new link between the c-Abl tyrosine kinase and phosphoinositide signalling through PLC-gamma1. Nat Cell Biol. 2003;5:309–319. doi: 10.1038/ncb949. [DOI] [PubMed] [Google Scholar]
- Rosales JL, Lee KY. Extraneuronal roles of cyclin-dependent kinase 5. Bioessays. 2006;28:1023–1034. doi: 10.1002/bies.20473. [DOI] [PubMed] [Google Scholar]
- Tong EH, Guo JJ, Huang AL, Liu H, Hu CD, Chung SS, Ko BC. Regulation of nucleocytoplasmic trafficking of transcription factor OREBP/TonEBP/NFAT5. J Biol Chem. 2006;281:23870–23879. doi: 10.1074/jbc.M602556200. [DOI] [PubMed] [Google Scholar]
- Trama J, Lu Q, Hawley RG, Ho SN. The NFAT-related protein NFATL1 (TonEBP/NFAT5) is induced upon T cell activation in a calcineurin-dependent manner. J Immunol. 2000;165:4884–4894. doi: 10.4049/jimmunol.165.9.4884. [DOI] [PubMed] [Google Scholar]
- Ubeda M, Kemp DM, Habener JF. Glucose-induced expression of the cyclin-dependent protein kinase 5 activator p35 involved in Alzheimer’s disease regulates insulin gene transcription in pancreatic β-cells. Endocrinology. 2004;145:3023–3031. doi: 10.1210/en.2003-1522. [DOI] [PubMed] [Google Scholar]
- Xu SX, Wong CC, Tong EH, Chung SS, Yates JR III, Yin YB, Ko BC. Phosphorylation by casein kinase 1 regulates tonicity-induced OREBP/TonEBP nucleocytoplasmic trafficking. J Biol Chem. 2008;283:17624–17634. doi: 10.1074/jbc.M800281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K. PIKE/nuclear PI 3-kinase signaling in preventing programmed cell death. J Cell Biochem. 2005;96:463–472. doi: 10.1002/jcb.20549. [DOI] [PubMed] [Google Scholar]
- Ye K, Aghdasi B, Luo HR, Moriarity JL, Wu FY, Hong JJ, Hurt KJ, Bae SS, Suh PG, Snyder SH. Phospholipase Cγ1 is a physiological guanine nucleotide exchange factor for the nuclear GTPase PIKE. Nature. 2002;415:541–544. doi: 10.1038/415541a. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ferraris J, Irarrazabal CE, Dmitireva NI, Park JH, Burg MB. Ataxia-telangiectasia mutated (ATM), a DNA damage-inducible kinase, contributes to high NaCl-induced nuclear localization of the transcription factor TonEBP/OREBP. Am J Physiol Renal Physiol. 2005;289:F506–F511. doi: 10.1152/ajprenal.00417.2004. [DOI] [PubMed] [Google Scholar]
- Zhou X, Gallazzini M, Burg MB, Ferraris JD. Contribution of SHP-1 protein tyrosine phosphatase to osmotic regulation of the transcription factor TonEBP/OREBP. Proc Natl Acad Sci USA. 2010;107:7072–7077. doi: 10.1073/pnas.1002795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukerberg LR, Patrick GN, Nikolic M, Humbert S, Wu CL, Lanier LM, Gertler FB, Vidal M, Van Etten RA, Tsai LH. Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron. 2000;26:633–646. doi: 10.1016/s0896-6273(00)81200-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.