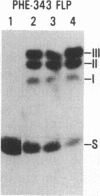
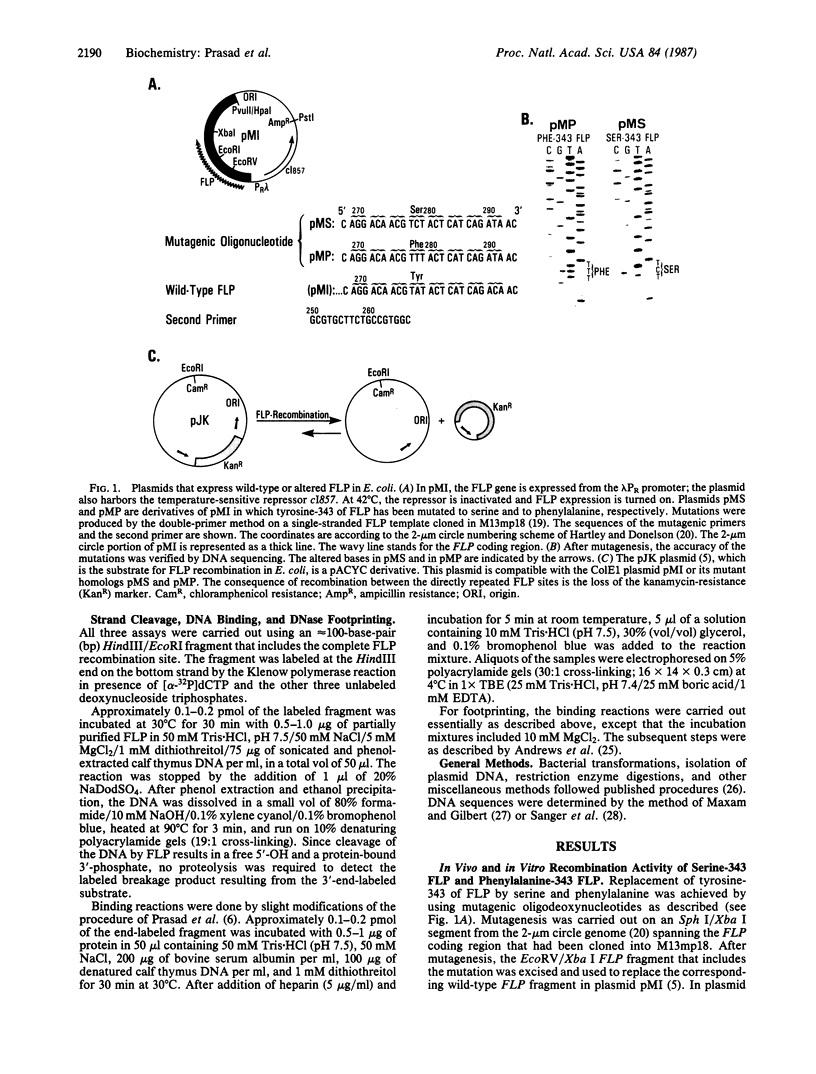
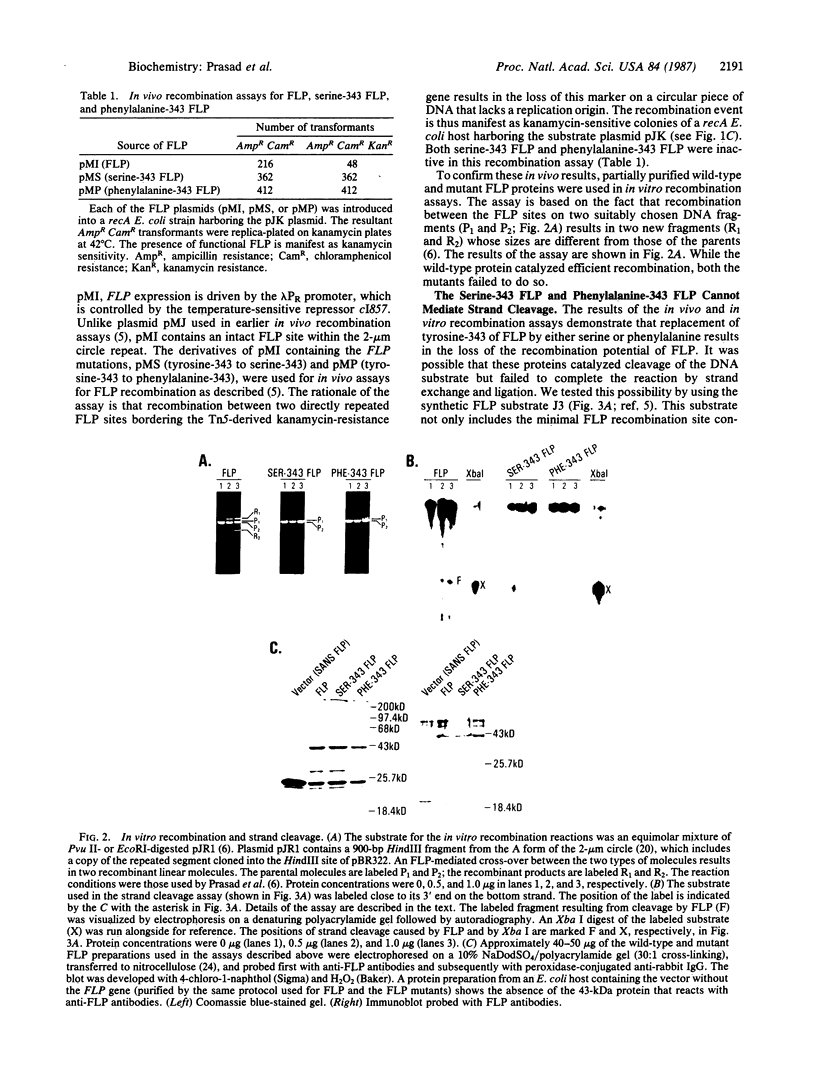
Abstract
The site-specific recombinase encoded by the yeast plasmid 2-microns circle (FLP) forms a transient covalent linkage with its substrate DNA via a tyrosine residue, which appears to be located near its COOH terminus. The homology of the COOH terminus of FLP with that of the Int family of recombinases suggests that tyrosine-343 of FLP could be involved in forming the DNA-protein bridge. We have mutated tyrosine-343 to a phenylalanine or serine. We demonstrate that the binding of each of the two mutant proteins to its substrate is indistinguishable from that of wild-type FLP. However, both mutant proteins are incapable of catalyzing strand cleavage and recombination.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Hoess R., Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983 Apr;32(4):1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- Andrews B. J., McLeod M., Broach J., Sadowski P. D. Interaction of the FLP recombinase of the Saccharomyces cerevisiae 2 micron plasmid with mutated target sequences. Mol Cell Biol. 1986 Jul;6(7):2482–2489. doi: 10.1128/mcb.6.7.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babineau D., Vetter D., Andrews B. J., Gronostajski R. M., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP protein of the 2-micron plasmid of yeast. Purification of the protein from Escherichia coli cells expressing the cloned FLP gene. J Biol Chem. 1985 Oct 5;260(22):12313–12319. [PubMed] [Google Scholar]
- Bruckner R. C., Cox M. M. Specific contacts between the FLP protein of the yeast 2-micron plasmid and its recombination site. J Biol Chem. 1986 Sep 5;261(25):11798–11807. [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. CAP and RNA polymerase interactions with the lac promoter: binding stoichiometry and long range effects. Nucleic Acids Res. 1983 Jan 11;11(1):141–158. doi: 10.1093/nar/11.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski R. M., Sadowski P. D. The FLP recombinase of the Saccharomyces cerevisiae 2 microns plasmid attaches covalently to DNA via a phosphotyrosyl linkage. Mol Cell Biol. 1985 Nov;5(11):3274–3279. doi: 10.1128/mcb.5.11.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hatfull G. F., Grindley N. D. Analysis of gamma delta resolvase mutants in vitro: evidence for an interaction between serine-10 of resolvase and site I of res. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5429–5433. doi: 10.1073/pnas.83.15.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Jayaram M. Two-micrometer circle site-specific recombination: the minimal substrate and the possible role of flanking sequences. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5875–5879. doi: 10.1073/pnas.82.17.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- McLeod M., Craft S., Broach J. R. Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2 microns circle. Mol Cell Biol. 1986 Oct;6(10):3357–3367. doi: 10.1128/mcb.6.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Leon L., Senecoff J. F., Bruckner R. C., Cox M. M. Site-specific genetic recombination promoted by the FLP protein of the yeast 2-micron plasmid in vitro. Cold Spring Harb Symp Quant Biol. 1984;49:797–804. doi: 10.1101/sqb.1984.049.01.090. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Weisberg R., Enquist L., Mizuuchi M., Buraczynska M., Foeller C., Hsu P. L., Ross W., Landy A. Structure and function of the phage lambda att site: size, int-binding sites, and location of the crossover point. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):429–437. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- Norris K., Norris F., Christiansen L., Fiil N. Efficient site-directed mutagenesis by simultaneous use of two primers. Nucleic Acids Res. 1983 Aug 11;11(15):5103–5112. doi: 10.1093/nar/11.15.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Van de Putte P. Genetic switches by DNA inversions in prokaryotes. Biochim Biophys Acta. 1984 Jun 16;782(2):111–119. doi: 10.1016/0167-4781(84)90013-7. [DOI] [PubMed] [Google Scholar]
- Prasad P. V., Horensky D., Young L. J., Jayaram M. Substrate recognition by the 2 micron circle site-specific recombinase: effect of mutations within the symmetry elements of the minimal substrate. Mol Cell Biol. 1986 Dec;6(12):4329–4334. doi: 10.1128/mcb.6.12.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. R., Grindley N. D. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981 Sep;25(3):721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Moser C. D. Resolvase-mediated recombination intermediates contain a serine residue covalently linked to DNA. Cold Spring Harb Symp Quant Biol. 1984;49:245–249. doi: 10.1101/sqb.1984.049.01.028. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Sadowski P. Site-specific recombinases: changing partners and doing the twist. J Bacteriol. 1986 Feb;165(2):341–347. doi: 10.1128/jb.165.2.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecoff J. F., Cox M. M. Directionality in FLP protein-promoted site-specific recombination is mediated by DNA-DNA pairing. J Biol Chem. 1986 Jun 5;261(16):7380–7386. [PubMed] [Google Scholar]
- Simon M., Zieg J., Silverman M., Mandel G., Doolittle R. Phase variation: evolution of a controlling element. Science. 1980 Sep 19;209(4463):1370–1374. doi: 10.1126/science.6251543. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Putte P., Cramer S., Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage mu. Nature. 1980 Jul 17;286(5770):218–222. doi: 10.1038/286218a0. [DOI] [PubMed] [Google Scholar]