Abstract
Acinetobacter baumannii is an important nosocomial pathogen that causes a high morbidity and mortality rate in infected patients, but pathogenic mechanisms of this microorganism regarding the secretion and delivery of virulence factors to host cells have not been characterized. Gram-negative bacteria naturally secrete outer membrane vesicles (OMVs) that play a role in the delivery of virulence factors to host cells. A. baumannii has been shown to secrete OMVs when cultured in vitro, but the role of OMVs in A. baumannii pathogenesis is not well elucidated. In the present study, we evaluated the secretion and delivery of virulence factors of A. baumannii to host cells via the OMVs and assessed the cytotoxic activity of outer membrane protein A (AbOmpA) packaged in the OMVs. A. baumannii ATCC 19606T secreted OMVs during in vivo infection as well as in vitro cultures. Potential virulence factors, including AbOmpA and tissue-degrading enzymes, were associated with A. baumannii OMVs. A. baumannii OMVs interacted with lipid rafts in the plasma membranes and then delivered virulence factors to host cells. The OMVs from A. baumannii ATCC 19606T induced apoptosis of host cells, whereas this effect was not detected in the OMVs from the ΔompA mutant, thereby reflecting AbOmpA-dependent host cell death. The N-terminal region of AbOmpA22-170 was responsible for host cell death. In conclusion, the OMV-mediated delivery of virulence factors to host cells may well contribute to pathogenesis during A. baumannii infection.
Introduction
Acinetobacter baumannii is an important nosocomial pathogen that causes a variety of human infections, particularly in severely ill patients [1], [2]. Multi-drug resistance or pan-drug resistance to clinically available antimicrobial agents in this organism induces serious therapeutic issues [3], [4]. A. baumannii is generally regarded as a low virulent pathogen [2], [3], but the full genome sequencing shows that this organism harbors a remarkable number of putative virulence-associated genes and elements homologous to the Legionella/Coxiella type IV secretion apparatus [5]. Several virulence determinants, such as biofilm formation [6], [7], adherence and ability to invade host cells [8], [9], as well as iron acquisition [10] and host cell death [11], have been assessed in previous studies. In prior studies, we determined that the bacterial molecules secreted from A. baumannii were directly responsible for host cell death [12]. Among the variety of bacterial molecules, outer membrane protein A of A. baumannii (AbOmpA) was identified as a potential virulence factor to induce host cell death via both mitochondrial and nuclear targeting [11], [13]. However, the secretion and delivery of AbOmpA to host cells remain to be thoroughly elucidated.
A wide variety of Gram-negative bacterial species have been demonstrated to secrete outer membrane vesicles (OMVs) during bacterial growth [14]. OMVs are spherical nanovesicles with an average diameter of 20–200 nm and are composed of lipopolysaccharides (LPS), proteins, lipids, and DNA or RNA [15]–[18]. Moreover, OMVs produced by pathogenic Gram-negative bacteria harbor toxins and specific virulence factors, including the heat-labile toxins of enterotoxigenic Escherichia coli (ETEC) [19], [20], the Shiga toxin of E. coli O157:H7 [21], the cytolethal distending toxin of Campylobacter jejuni [22], and the Cif protein of Pseudomonas aeruginosa [23]. Upon the delivery of virulence factors to host cells, OMVs perform an important function in bacterial pathogenesis without the direct interaction between the pathogens and the host cells.
We demonstrated recently that a clinical isolate of A. baumannii DU202 secreted OMVs into the extracellular milieu during in vitro growth; additionally, several putative virulence factors were identified in the OMVs of A. baumannii DU202 [24], thereby suggesting that A. baumannii OMVs may serve to deliver virulence factors to host cells. In this study, we evaluated the direct effects of A. baumannii OMVs on host cells, particularly, the secretion of OMVs from A. baumannii during in vivo infection, the delivery of virulence factors to host cells via OMVs and subsequent cytotoxicity. Here we report that A. baumannii OMVs represent a vehicle for the delivery of bacterial molecules to host cells and also that a potential virulence factor AbOmpA enriched in the OMVs contributes directly to host cell death.
Results
Secretion of OMVs from A. baumannii
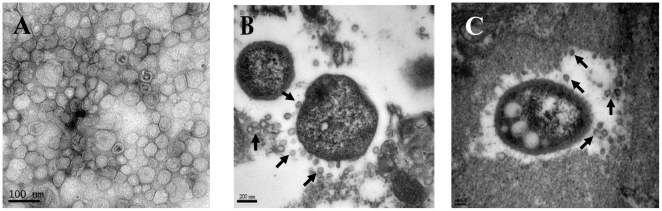
We first analyzed the secretion of OMVs from A. baumannii ATCC 19606T during in vitro culture. Bacteria were cultured in Luria-Bertani (LB) broth and OMVs were collected from the culture supernatants. Transmission electron microscopy (TEM) demonstrated that A. baumannii secreted spherical vesicles into the extracellular milieu (Fig. 1A). Next, in order to evaluate the secretion of OMVs from A. baumannii during in vivo infection, immunocompromised mice were infected intratracheally with 1×107 CFU of A. baumannii ATCC 19606T and sacrificed 48 h after bacterial injection. The histological examination demonstrated hemorrhage, necrosis, and infiltration of polymorphonuclear cells in both lung tissues (data not shown). TEM analysis revealed the budding of spherical nanovesicles from bacterial surfaces in the infected lung tissues (Fig. 1B and 1C). These results indicate that A. baumannii secretes OMVs during in vivo infection and the secreted OMVs are likely to interact with host cells without any direct contact between the pathogens and the host cells.
Figure 1. Secretion of OMVs from A. baumannii during in vitro culture and in vivo infection.
(A) Transmission electron micrograph of OMVs prepared from A. baumannii ATCC 19606T cultured in LB broth for 24 h. (B and C) Secretion of OMVs from A. baumannii ATCC 19606T in a murine pneumonia model. Mice were infected with 1×107 CFU of bacteria intratracheally and sacrificed 48 h after bacterial injection. Arrows indicate the OMVs secreted from A. baumannii.
Virulence factors packaged in A. baumannii OMVs
Proteomic analysis was conducted to identify proteins packaged in the OMVs from A. baumannii ATCC 19606T. Ultimately, a total of 113 proteins were identified (Table S1). A putative outer membrane protein (A1S_0884) with a molecular mass of 22.5 kDa was detected in the highest abundance, followed by AbOmpA (A1S_2840) with a molecular mass of 38.4 kDa. The cellular localization of proteins identified in the A. baumannii OMVs was predicted to occur in the extracellular space (n = 1), outer membrane (n = 26), periplasmic space (n = 8), inner membrane (n = 4), cytosol (n = 17), and unknown sites (n = 57). Virulence-associated proteins, including AbOmpA, CsuA/B (A1S_2218), CsuC (A1S_2215), CsuD (A1S_2214), putative hemolysin (A1S_1321), putative serine protease (A1S_2525), Cu/Zn superoxide dismutase (A1S_3143), fimbrial protein (A1S_1510), bacterioferritin (A1S_3175), RND superfamily transporter (A1S_0116), putative RND type efflux pump (A1S_0009), and putative protease (A1S_2470), were found in the OMVs of A. baumannii ATCC 19606T.
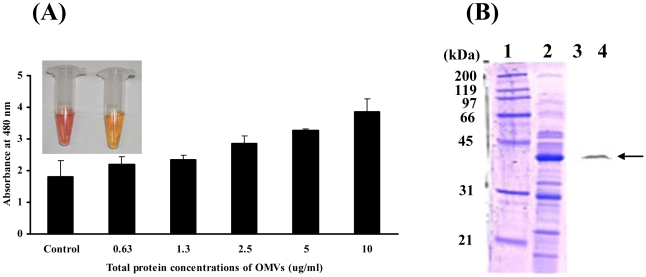
In order to determine whether AmpC β-lactamase (A1S_2367) identified in the A. baumannii OMVs were biologically active, the prepared OMVs were incubated with the β-lactamase substrate, nitrocefin. This showed that the β-lactamases associated with A. baumannii OMVs degraded nitrocefin, as demonstrated by the colorization of nitrocefin (Fig. 2A). Moreover, the results of Western blot analysis demonstrated that the full length of AbOmpA with a molecular mass of 38.4 kDa was detected in the A. baumannii OMVs (Fig. 2B). These results indicate that A. baumannii OMVs harbor biologically active virulence factors and enzymes, which can perform diverse biologic processes in host cells.
Figure 2. A. baumannii OMVs contain biologically active proteins.
(A) A. baumannii OMVs were incubated with 100 mM nitrocefin in PBS at 30°C for 30 min. Hydrolysis of nitrocefin was monitored at 480 nm using a UV spectrophotometer. Inlet figure: left, brown colorization of nitrocefin in A. baumannii OMVs; right, negative control of nitrocefin in PBS. (B) SDS-PAGE of proteins packaged in the OMVs from A. baumannii ATCC 19606T (lanes 1 and 2) and its Western blot analysis (lanes 3 and 4). The samples were immunoblotted with a rabbit anti-AbOmpA immune serum. Lanes 1 and 3, molecular weight maker; 2 and 4, OMV fraction. Arrows indicate AbOmpA.
Delivery of virulence factors to host cells via OMVs
Based on the results of proteome analysis showing that A. baumannii packaged multiple virulence factors into OMVs, we assessed the delivery of virulence factors to host cells via the OMVs. Three human cell lines, HeLa cell, HEp-2 cells, and U937 cells, were treated with OMVs acquired from A. baumannii ATCC 19606T for 24 h and the cellular distribution of AbOmpA, a documented OMV protein, was analyzed via confocal microscopy. The cells were treated with 4′,6-diamidino-2-phenyllindole dihydrochloride (DAPI) for nuclear staining and anti-AbOmpA polyclonal antibody, followed by Alexa Fluor® 488 or 568 for AbOmpA. Green or red fluorescence was noted principally in the cytosolic compartments of the cells, but some fluorescence was noted within the nuclei (Fig. 3A). Western blot analysis showed that the full length of AbOmpA appeared within the cells at 30 min and remained there for more than 12 h (Fig. 3B).
Figure 3. A. baumannii OMVs deliver virulence factor AbOmpA into host cells.
(A) HeLa and HEp-2 cells were treated with A. baumannii OMVs (5 µg/ml of protein concentrations) for 12 h. The cells were fixed, permeabilized, and stained with anti-rabbit AbOmpA antibody, followed by Alexa Fluor® 568-conjugated rabbit IgG (red). DAPI was used to stain the nuclei (blue). Magnification: ×400. The U937 cells were treated with 5 µg/ml of A. baumannii OMVs for 4 h and then stained with anti-rabbit AbOmpA antibody, followed by Alexa Fluor® 488-conjugated rabbit IgG (green). Magnification: ×1,260. (B) Western blot analysis of cell lysates. The differentiated U937 cells were treated with A. baumannii OMVs (20 µg/ml of protein concentrations) for the indicated times. Cell lysates were separated on 12% SDS-PAGE, transferred to membranes, and immunoblotted with a rabbit anti-AbOmpA immune serum and β-actin antibody.
The OMVs from E. coli and P. aeruginosa bind to host cells via lipid rafts, after which bacterial effector molecules are translocated into the cytosolic compartment [20], [23]. In an effort to determine whether or not A. baumannii OMVs delivered virulence factors to host cells via lipid rafts, HeLa cells were pretreated with a cholesterol-destroying agent, methyl-β-cyclodextrin (MβCD), and subsequently treated with OMVs for 4 h. We used HeLa cells to exclude the involvement of caveolins in the interactions of host cells with OMVs because these cells did not harbor caveolins in the cytoplasmic membrane [25]. AbOmpA was detected in the cytosol of HeLa cells treated with A. baumannii OMVs (Fig. 4A), whereas pretreatment of MβCD completely inhibited the cellular localization of AbOmpA in HeLa cells treated with OMVs (Fig. 4B), thereby indicating that a cholesterol-rich membrane microdomain is required for the delivery of virulence factors packaged in the A. baumannii OMVs to host cells.
Figure 4. A. baumannii OMVs interact with plasma membrane of host cells through a cholesterol-rich membrane microdomain.
(A) HeLa cells were treated with OMVs (5 µg/ml of protein concentrations) for 4 h. (B) HeLa cells were preterated with 10 mM MβCD for 45 min and then treated with OMVs for 4 h. The cells were fixed, permeabilized, and stained with anti-rabbit AbOmpA antibody, followed by Alexa Fluor® 488-conjugated rabbit IgG (green). DAPI was used to stain the nuclei (blue). Magnification: ×630.
Cytotoxicity of A. baumannii OMVs
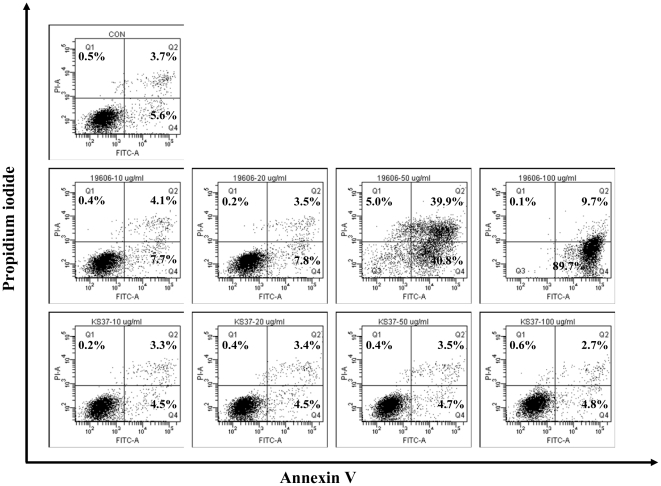
In order to determine whether A. baumannii OMVs induced host cell damage, macrophages were treated with various concentrations of OMVs for 24 h and then stained with Annexin V and propidium iodide (PI). The U937 cells were used in this study because macrophages had low threshold concentrations of AbOmpA for cell death as compared with epithelial cells (HEp-2 cells) and fibroblast cells (Cos-7 cells) [13]. Flow cytometric analysis demonstrated no cell death at ≤20 µg/ml (protein concentrations) of OMVs, but 50 and 100 µg/ml of OMVs did induce host cell death (Fig. 5, middle panel). Based on previous studies demonstrating that AbOmpA directly induced apoptotic cell death, OMVs were prepared from the ΔompA mutant and their ability to induce cytotoxicity was compared to that of the OMVs from wild-type A. baumannii. The OMVs from the ΔompA mutant did not induce cell death evenly at a concentration of 100 µg/ml (Fig. 5, lower panel). These results suggest that AbOmpA associated with A. baumannii OMVs is directly responsible for host cell death.
Figure 5. Flow cytometric analysis of cell death induced by the OMVs from A. baumannii ATCC 19606T and the ΔompA mutant.
The differentiated U937 cells were treated with various concentrations (0, 10, 20, 50, and 100 µg/ml) of OMVs and stained with Annexin V and PI. Upper panel, control cells without OMVs for 24 h. Middle panel, the cells were treated with OMVs from A. baumannii ATCC 19606T for 24 h. Lower panel, the cells were treated with OMVs from the ΔompA mutant for 24 h. Representative data from three independent experiments are shown. In the graph, cells in right upper and lower parts are apoptotic cells and cells in left upper part are necrotic cells.
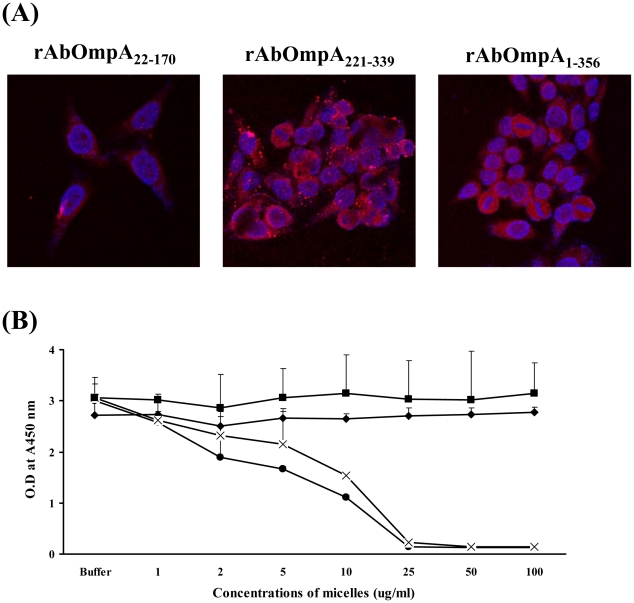
In order to determine which AbOmpA domains were directly responsible for host cell death, three recombinant proteins, including rAbOmpA22-170, rAbOmpA221-339, and rAbOmpA1-356, were generated and then micelles harboring each rAbOmpA fragment were constructed to insert rAbOmpA proteins into the membrane or lumen according to their hydrophobicity. These micelles were treated to HeLa cells for 24 h, after which the cellular distribution of rAbOmpA was assessed. Three different rAbOmpA proteins were internalized by host cells (Fig. 6A). Next, the cytotoxicity of different rAbOmpA proteins was assessed in the macrophages. Micelles composed of rAbOmpA22-170 and rAbOmpA1-356 induced cell death at a concentration of ≥5 µg/ml, whereas rAbOmpA221-339 did not induce cell death at a concentration of ≤100 µg/ml (Fig. 6B). These results indicate that the N-terminal resides of AbOmpA are responsible for host cell death.
Figure 6. Host cell entry and cytotoxicity of rAbOmpA fragments.
(A) HeLa cells were treated with different forms of micelles containing rAbOmpA fragments for 24 h. The cells were fixed, permeabilized, and stained with anti-rabbit AbOmpA antibody, followed by Alexa Fluor® 568-conjugated rabbit IgG (red). DAPI was used to stain the nuclei (blue). The subcellular localization of AbOmpA was observed by confocal microscopy. Magnification: ×400. (B) The differentiated U937 cells were treated with different forms of micelles containing rAbOmpA fragments for 24 h. Cell viability was determined by a WST-1 assay. Untreated control cells (⧫), rAbOmpA22-170 (•), rAbOmpA1-356 (X), and rAbOmpA221-339 (▪).
Discussion
In our previous studies to identify the virulence factors of A. baumannii, we demonstrated that AbOmpA induced host cell death via both mitochondrial and nuclear targeting [11], [13]. However, the secretion of AbOmpA from bacteria and its delivery to host cells have yet to be characterized. AbOmpA is a porin that allows for the passing of small solutes in the outer membrane and is one of the most abundant proteins in culture supernatants [11], [24]. Thus, we hypothesized that A. baumannii packaged AbOmpA into OMVs and that the OMV-mediated delivery of AbOmpA to host cells induced cytotoxicity. In this study, we identified A. baumannii OMVs as an important vehicle for the delivery of AbOmpA to host cells, after which AbOmpA packaged in OMVs induced cytotoxicity. These results help to establish that the previously uncharacterized secretion and delivery pathways of AbOmpA induce host cell death.
Proteome analysis of OMVs from A. baumannii ATCC 19606T and a clinical isolate of A. baumannii DU202 identified more than 110 proteins derived from the outer membrane, periplasmic space, inner membrane, cytosol, and unknown sites [24]. Twenty-one inner membrane and cytosolic proteins were identified in the OMVs, although OMVs were not likely to contain inner membrane and cytosolic components during the vesicle biogenesis [14], [15]. However, several proteomic studies demonstrated that some inner membrane and cytosolic proteins were also present in the OMV preparations [17], [18], [26]–[28]. A putative outer membrane protein (A1S_0884) with a molecular mass of 22.5 kDa was the most abundant in the proteomic analysis of OMVs (Table S1), but 1-dimentional SDS-PAGE showed the most dense band in the molecular mass of 35–40 kDa (Fig. 2B). This difference is due to the presence of several proteins with a similar molecular mass, such as putative competence protein (ComL) (40.6 kDa), hypothetical protein A1S_1691 (39.2 kDa), AbOmpA (38.4 kDa), β-lactamase (37.3 kDa), hypothetical protein A1S_0505 (36.5 kDa), and putative RND type efflux pump (35.6 kDa). The protein composition of OMVs varied between A. baumannii ATCC 19606T and a clinical isolate DU202. Virulence-associated proteins, including AbOmpA, putative RND efflux pump (A1S_0009), bacterioferritin (A1S_3175), and several putative outer membrane proteins, were identified commonly in both A. baumannii strains, whereas CsuA/B, CsuC, CsuD, fimbrial protein, and putative hemolysin were identified only in the OMVs from A. baumannii ATCC 19606T (Table S1). The protein compositions of the OMVs prepared from a clinical P. aeruginosa isolate and from laboratory strain PAO1 differed significantly [29], [30], thus eliciting a different pro-inflammatory cytokine response in macrophages [31]. Accordingly, the strain-specific protein compositions or virulence factor profiles in A. baumannii OMVs may result in differences in the virulence of A. baumannii strains.
The OMV content is delivered to host cells via either receptor-mediated endocytic pathway or the fusion with host cell plasma membranes [32], [33]. The results of this study demonstrated that a cholesterol-rich membrane microdomain was required for the delivery of AbOmpA packaged in A. baumannii OMVs to the cytosol of host cells, thus reflecting the lipid raft-dependent endocytosis of OMVs. Toxins and other vesicular membrane components of OMVs were shown to bind specifically to the receptors within lipid rafts [20], [31]. The heat-labile toxins in ETEC OMVs were bound to monosialoganglioside (GM1) [20]. Ellis et al. [31] reported that vesicular LPS and proteins were responsible for the binding of P. aeruginosa OMVs to macrophage surfaces and cellular internalization, respectively. Since AbOmpA played a pivotal role in adherence to and invasion of A. baumannii in epithelial cells [9], we attempted to determine whether AbOmpA played a role in the interaction of A. baumannii OMV with host cells. The ΔompA mutant of A. baumannii ATCC 19606T did not express AbOmpA in the outer membrane [11], but the results of proteomic analysis demonstrated the existence of a truncated AbOmpA in the OMVs from the ΔompA mutant (data not shown), thereby suggesting that a truncated AbOmpA may be located in the lumen of OMVs. When HeLa cells were treated with OMVs from the ΔompA mutant, a truncated AbOmpA was detected in the cytosol of host cells. These results indicate that the interaction of A. baumannii OMVs with host cells occurred independently of AbOmpA. Future studies will clearly be necessary to identify the bacterial molecules associated with lipid rafts. Moreover, the subsequent trafficking pathway of OMVs following the entry should be clarified to elucidate A. baumannii pathogenesis occurring by way of OMV-mediated cytotoxicity.
We previously demonstrated that AbOmpA purified from A. baumannii ATCC 19606T and rAbOmpA from E. coli caused death in host cells [11], [13], but the cytotoxic domains have yet to be clearly identified. The results of the present study showed that AbOmpA was the most potent cytotoxic molecule in the A. baumannii OMVs and the N-terminal 170 residues of AbOmpA were required for the induction of host cell death. A 50 µg/ml (protein concentrations) of OMVs induced host cell death. Because molecular percentage of AbOmpA was determined to be 9.08 based on the proteomic analysis of A. baumannii OMVs (Table S1), an approximately 4.5 µg/ml of AbOmpA in the 50 µg/ml of OMVs induced host cell death. Based on the prediction of the tertiary structure of AbOmpA (http://www.pymol.org/), the N-terminal 220 residues of AbOmpA traversed the outer membrane with eight anti-parallel β-sheet segments and four external loops exposed on the bacterial surface. Thus, the N-terminal residues of AbOmpA, which may traverse the vesicular membrane and form external loops in OMVs, induce cytotoxicity. However, the translocation of AbOmpA located in the vesicular membrane to the cytosolic compartment has yet to be characterized.
The contribution of OMVs to bacterial pathogenesis have previously been determined in a variety of pathogenic Gram-negative bacteria, including ETEC [20], uropathogenic E, coli [34], Helicobacter pylori [35], [36], Actinobacillus actinomycetemcomitans [37], Vibrio cholerae [38], and P. aeruginosa [29]. In this study, we demonstrate that OMVs are an important secretory vehicle for the delivery of virulence factors to host cells in A. baumannii. The OMV-mediated cytotoxicity occurring via AbOmpA is a crucial regulator in the induction of host cell death. Further studies of the virulence attributes of each virulence factor packaged in OMVs are expected to provide insights into the association of A. baumannii pathogenesis and alterations of host cell biology.
Materials and Methods
Bacterial strains and cell culture
A. baumannii ATCC 19606T and isogenic ΔompA mutant (KS37) were employed to prepare the OMVs [11]. E. coli BL21 (DE3) was employed in the preparation of the rAbOmpA proteins. The organisms were maintained on blood agar plates or MacConkey agar plates, and cultivated in LB broth. HEp-2 cells from laryngeal epithelial cells, HeLa cells from cervical carcinoma, and U937 cells from monocytes were employed in this study. HEp-2 cells and HeLa cells were grown in Dulbecco's modified Eagle medium (HyClone) supplemented with 10% fetal bovine serum (FBS, HyClone), 2 mM L-glutamine, 1000 U/ml penicillin G, and 50 µg/ml streptomycin at 37°C in 5% CO2. U937 monocytes were differentiated into macrophages for three or four days and matured via the addition of 500 ng/ml of phorbol 12-myristate 13-acetate (Sigma-Aldrich). Macrophages were cultured in RPMI-1640 (Gibco BRL) supplemented with 10% FBS and 2 mM L-glutamine at 37°C in 5% CO2.
Purification of OMVs from bacterial culture supernatants
The OMVs from A. baumannii were prepared as previously described [18], [39]. In brief, bacteria were grown in 500 ml of LB broth until the optical density at 600 nm (OD600) reached 1.0 at 37°C with shaking. Bacterial cells were removed via 15 min of centrifugation at 6,000 x g at 4°C. The supernatants were filtered through a 0.2 µm vacuum filter to remove residual cellular debris. The samples were concentrated via ultrafiltration with a QuixStand Benchtop System (GE Healthcare) using a 100 kDa hollow fiber membrane (GE Healthcare), which could exclude molecules with a molecular mass of 100 kDa in the samples. The OMVs were collected via 3 h of ultracentrifugation at 150,000 x g at 4°C and resuspended in phosphate-buffered saline (PBS). The protein concentration was then determined using a modified BCA assay (Thermo Scientific). The purified OMVs were checked for sterility and stored at −80°C.
Production of rAbOmpA and micelle formation
The ompA gene was amplified from A. baumannii ATCC 19606T and cloned into the pET28a expression vector (Novagen) [13], [40]. The recombinant proteins were overexpressed in E. coli BL21 (DE3) and loaded onto a HiTrap™ FF column (Amersham Biosciences) to elute His-tagged rAbOmpA. The rAbOmpA was incubated with endotoxin removal resin (Sigma-Aldrich) for the removal of LPS. The protein concentration was determined using a modified BCA assay (Thermo Scientific). In order to construct micelles, the purified rAbOmpA proteins, rAbOmpA1-356 and rAbOmpA22-170, were incubated with 05% n-dodecyl-N,N-dimethylamine-N-oxide in a buffer consisting of 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. The hydrophilic rAbOmpA221-339 was mixed with 20 mM Tris-HCl (pH 6.8) and 100 mM NaCl.
Identification of proteins in A. baumannii OMVs
One-dimensional electrophoresis-liquid chromatography-tandem mass spectrometry (1-DE-LC-MS/MS) was performed to identify proteins packaged in the A. baumannii OMVs as described previously [24], [41]. Proteins were separated via 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and in-gel digested. The protein digests were resolved in 15 µl of 0.02% formic acid in 0.5% acetic acid, and the samples were concentrated on a MGU30-C18 trapping column (LC Packings) and a nano-column (C18 reverse-phase column, Proxeon) at a flow rate of 120 nl/min. The peptides were eluted by 0–65% acetonitrile for 80 min. All MS and MS/MS spectra in the LCQ-Deca ESI ion trap mass spectrometer were acquired in data-dependent mode. The MS/MS spectra were searched using MASCOT software (Matrix Science, Inc.) using the genome data of A. baumannii ATCC 17978T from NCBInr (http://www.ncbi.nlm.nih.gov/) and the decoy sequence database. The exponentially modified protein abundance index (emPAI) was generated using MASCOT software [42].
Western blot analysis
Cells were treated with OMVs (20 µg/ml of protein concentrations) for the indicated time periods and then were lysed in lysis buffer (10 mM Tris pH 7.4, 5 mM EDTA, 130 mM NaCl, 1% Triton X-100, 10 mg/ml PMSF, 10 mg/ml aprotinin, 10 mg/ml leupeptin, 5 mM phenanthroline, and 28 mM benzamidine-HCl) for 30 min on ice. The lysates were clearly centrifugation and then quantified using a modified BCA assay (Thermo Scientific). Each sample was separated with 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by electrotransfer onto the nitrocellulose membranes (Hybond-ECL; Amersham Pharmacia Biotech). The blots were blocked in 5% non-fat skim milk and incubated with a rabbit anti-AbOmpA immune serum and β-actin antibodies (Santa Cruz Biotechnology). The membranes were incubated with a secondary antibody coupled to horseradish peroxidase and developed using an enhanced chemiluminescence system (ECL plus; Amersham Pharmacia Biotech).
Cell proliferation assay
The growth of cell treated with different rAbOmpA proteins was measured with a Premix WST1 cell proliferation assay system (TaKaRa Shuzo) [11]. The cells were seeded at a concentration of 2.0×105/ml in a 96-well microplate. After treating with different concentrations of micelles for 24 h, cell growth was measured at 450 nm 3 h after treatment with WST1.
Flow cytometric analysis
Cells were treated with OMVs for 24 h and stained with FITC-conjugated Annexin V and PI (BD Pharmingen) according to the manufacturer's instructions. The 1×106 cells were stained with FITC-conjugated Annexin V in Annexin V binding buffer for 15 min. PI was added to determine alterations in cell membrane integrity. The samples were immediately analyzed by the flow cytometry and CellQuest Pro software (BD Biosciences). For each sample, 5,000 or 104 cells were acquired for data analysis.
Confocal microscopy
The cultured cells were seeded at a density of 5×104 on glass coverslips the day before the assay. After treating the OMVs, the cells were washed in PBS, fixed in 4% paraformaldehyde and permeabilized for 10 min in PBS containing 0.25% Triton X-100. The OMVs were labeled with a polyclonal anti-rabbit AbOmpA antibody, followed by Alexa-488- or Alexa-568-conjugated goat anti-rabbit IgG antibody (Molecular Probes). The nuclei of the cells were stained with DAPI (Molecular Probes). HeLa cells were treated with 10 mM MβCD (Sigma-Aldrich) to disrupt cholesterol-rich membrane domains for 45 min in serum-free medium at 37°C in a CO2 incubator. After washing with PBS, cells were treated with OMVs. The samples were observed with a Carl-Zeiss confocal fluorescent microscope.
Determination of β-lactamase activity
The chromogenic cephalosporin nitrocefin (Oxoid, U. K) was used to determine β-lactamase activity [43]. The assay was performed at 30°C with 100 mM nitrocefin in PBS (pH 7.4). Hydrolysis was monitored at 480 nm using a UV spectrophotometer (Shimadzu, Japan).
TEM analysis
After the resuspension of OMV preparations in PBS, the samples were applied to copper grids and stained with 2% uranyl acetate. The lung tissues removed from the infected mice were fixed with 2.5% glutaraldehyde and post-fixed in 1% osmium tetroxide. The samples were subsequently dehydrated in a series of ethanol concentrations and embedded in Epon. Thin sections were cut with an ultramicrotome (RMC Boeckeler Instruments) equipped with a diamond knife and stained with 3% uranyl acetate and lead citrate. The samples were then visualized with a TEM (Hitachi H-7500, Hitachi, Japan) operated at 120 kV.
Mouse pneumonia model
Eight-week-old female C57BL/6 mice were maintained under specific pathogen free conditions. Immunocompromised mice were infected with A. baumannii ATCC 19606T [9]. Neutropenic mice were induced via intraperitoneal injections of cyclophosphamide (150 mg/Kg) on days -4 and -3 before bacterial injection [44]. The mice were anesthetized with pentobarbital and then 100 µl of 1×108 cfu/ml of bacteria were administrated intratracheally. The control mice were injected with 100 µl of PBS (pH 7.4). The mice were sacrificed two days after bacterial challenge and the lungs were removed. All procedures involving animals were approved by the Animal Care Committee of Kyungpook National University (KNU2010-40).
Supporting Information
Proteins identified in the OMV fraction of A. baumannii ATCC 19606T using 1-DE and LC-MS/MS analysis.
(XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MEST) (Project No. R01-2008-000-11083-0). SI Kim & SO Kwon were supported by a grant (K-MeP T30100) from the Korea Basic Science Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon MC, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents. 2010;35:219–226. doi: 10.1016/j.ijantimicag.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Falagas ME, Bliziotis IA, Kasiakou SK, Samonis G, Thanassopoulou P, et al. Outcome of infections due to pandrug-resistant (PDR) Gram-negative bacteria. BMC Infect Dis. 2005;5:24–30. doi: 10.1186/1471-2334-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HW, Koh YM, Kim J, Lee JC, Lee YC, et al. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect. 2008;14:49–54. doi: 10.1111/j.1469-0691.2007.01842.x. [DOI] [PubMed] [Google Scholar]
- 7.Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun. 2009;77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JC, Koerten H, van den Broek P, Beekhuizen H, Wolterbeek R, et al. Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res Microbiol. 2006;157:360–366. doi: 10.1016/j.resmic.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Choi CH, Lee JS, Lee YC, Park TI, Lee JC. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 2008;8:216. doi: 10.1186/1471-2180-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimbler DL, Penwell WF, Gaddy JA, Menke SM, Tomaras AP, et al. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals. 2009;22:23–32. doi: 10.1007/s10534-008-9202-3. [DOI] [PubMed] [Google Scholar]
- 11.Choi CH, Lee EY, Lee YC, Park TI, Kim HJ, et al. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol. 2005;7:1127–1138. doi: 10.1111/j.1462-5822.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee JC, Oh JY, Kim KS, Jeong YW, Park JC, et al. Apoptotic cell death induced by Acinetobacter baumannii in epithelial cells through caspase-3 activation. APMIS. 2001;109:679–684. doi: 10.1034/j.1600-0463.2001.d01-132.x. [DOI] [PubMed] [Google Scholar]
- 13.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, et al. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol. 2008;10:309–319. doi: 10.1111/j.1462-5822.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 14.Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 16.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 17.Galka F, Wai SN, Kusch H, Engelmann S, Hecker M, et al. Characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infect Immun. 2008;76:1825–1836. doi: 10.1128/IAI.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee EY, Choi DS, Kim KP, Gho YS. Proteomics in Gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev. 2008;27:535–555. doi: 10.1002/mas.20175. [DOI] [PubMed] [Google Scholar]
- 19.Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem. 2000;275:12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23:4538–4549. doi: 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:1843–1848. doi: 10.1128/aem.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindmark B, Rompikuntal PK, Vaitkevicius K, Song T, Mizunoe Y, et al. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 2009;9:220. doi: 10.1186/1471-2180-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, et al. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon SO, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett. 2009;297:150–156. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 25.Lim JS, Choy HE, Park SC, Han JM, Jang IS, et al. Caveolae-mediated entry of Salmonella typhimurium into senescent nonphagocytic host cells. Aging Cell. 2010;9:243–251. doi: 10.1111/j.1474-9726.2010.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee EY, Bang JY, Park GW, Choi DS, Kang JS, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 27.Berlanda Scorza F, Doro F, Rodríguez-Ortega MJ, Stella M, Liberatori S, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli ΔtolR IHE3034 mutant. Mol Cell Proteomics. 2008;7:473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Sidhu VK, Vorhölter FJ, Niehaus K, Watt SA. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol. 2008;8:87. doi: 10.1186/1471-2180-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauman SJ, Kuehn MJ. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 2006;8:2400–2408. doi: 10.1016/j.micinf.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauman SJ, Kuehn MJ. Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. BMC Microbiol. 2009;9:26. doi: 10.1186/1471-2180-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellis TN, Leiman SA, Kuehn MJ. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both LPS and protein components. Infect Immun. 2010;78:3822–3831. doi: 10.1128/IAI.00433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouokam JC, Wai SN, Fällman M, Dobrindt U, Hacker J, et al. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect Immun. 2006;74:2022–2030. doi: 10.1128/IAI.74.4.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ismail S, Hampton MB, Keenan JI. Helicobacter pylori outer membrane vesicles modulate proliferation and interleukin-8 production by gastric epithelial cells. Infect Immun. 2003;71:5670–5675. doi: 10.1128/IAI.71.10.5670-5675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keenan JI, Davis KA, Beaugie CR, McGovern JJ, Moran AP. Alterations in Helicobacter pylori outer membrane and outer membrane vesicle-associated lipopolysaccharides under iron-limiting growth conditions. Innate Immun. 2008;14:279–290. doi: 10.1177/1753425908096857. [DOI] [PubMed] [Google Scholar]
- 37.Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32:1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 38.Kondo K, Takade A, Amako K. Release of the outer membrane vesicles from Vibrio cholerae and Vibrio parahaemolyticus. Microbiol Immunol. 1993;37:149–152. doi: 10.1111/j.1348-0421.1993.tb03192.x. [DOI] [PubMed] [Google Scholar]
- 39.Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 40.Lee JS, Lee JC, Lee CM, Jung ID, Jeong YI, et al. Outer membrane protein A of Acinetobacter baumannii induces differentiation of CD4+ T cells toward a Th1 polarizing phenotype through the activation of dendritic cells. Biochem Pharmacol. 2007;74:86–97. doi: 10.1016/j.bcp.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Yun SH, Choi CW, Park SH, Lee JC, Leem SH, et al. Proteomic analysis of outer membrane proteins from Acinetobacter baumannii DU202 in tetracycline stress condition. J Microbiol. 2008;46:720–727. doi: 10.1007/s12275-008-0202-3. [DOI] [PubMed] [Google Scholar]
- 42.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.O'Callaghan CH, Morris A, Kirby SM, Shingler AH. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Faassen H, KuoLee R, Harris G, Zhao X, Conlan JW, et al. Neutrophils play an important role in host resistance to respiratory infection with Acinetobacter baumannii in mice. Infect Immun. 2007;75:5597–5608. doi: 10.1128/IAI.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins identified in the OMV fraction of A. baumannii ATCC 19606T using 1-DE and LC-MS/MS analysis.
(XLS)