Abstract
High-risk HPV E6 and E7 oncoproteins cooperate to subvert critical host cell cycle checkpoint control mechanisms in order to promote viral genome replication. This results not only in aberrant proliferation but also in host cellular changes that can promote genomic instability. The HPV-16 E7 oncoprotein was found to induce centrosome abnormalities thereby disrupting mitotic fidelity and increasing the risk for chromosome missegregation and aneuploidy. In addition, expression of the high-risk HPV E7 oncoprotein stimulates DNA replication stress as a potential source of DNA breakage and structural chromosomal instability. Proliferation of genomically unstable cells is sustained by several mechanisms including the accelerated degradation of claspin by HPV-16 E7 and the degradation of p53 by the high-risk HPV E6 oncoprotein. These results highlight the oncogenic potential of aberrant proliferation and opens new avenues for prevention of malignant progression, not only in HPV-associated cervical cancer but also in non-virally associated malignancies with disrupted cell cycle checkpoint control mechanisms.
1. Introduction
Genomic instability comprises both structural and numerical chromosomal abnormalities and is a defining phenotype found in many malignant tumors [1]. While such chromosomal alterations may give certain cells a growth advantage, they are likely to be lethal to the vast majority of cells. Nonetheless, cell populations with increased genomic plasticity may gain a selective growth advantage and ultimately promote cancer development and progression. High-risk human papillomavirus (HPV)-associated neoplasms are an excellent model system to study genomic instability. This is underscored by the fact that even pre-malignant lesions contain chromosomally unstable cells and that the expression of two viral oncoproteins, HPV E6 and E7, drive malignant progression. This opens the opportunity to understand causes and consequences of genomic instability by utilizing viral oncoproteins as tools [1].
HPV infection is the main cause of cervical cancer, the second most common cause of cancer mortality in women worldwide [1,2]. There are over 100 HPV genotypes which have been identified and these are classified into two major groups: cutaneous and mucosal HPV types. Infection with cutaneous HPV types, such as HPV-1 or -2, usually leads to benign diseases such as skin or plantar warts. However, certain cutaneous HPV types, such as HPV-5, have been associated with skin carcinoma in patients with the skin disease epidermodysplasia verruciformis (EV) as well as non-melanoma skin cancers associated with HPV infections in organ transplant patients [3,4].
Mucosal HPVs are further subdivided into low-risk and high-risk HPVs [5]. Low-risk types such as HPV-6 or -11 are associated with benign lesions including condylomata acuminata or laryngeal papillomas. High-risk HPV types such as HPV-16, -18, -31, -33, or -45 are associated with carcinomas of the oropharyngeal or anogenital tract, in particular cervical carcinoma [2]. Epidemiological and biological studies have shown that HPV-16, and -18 are the two most frequently detected oncogenic types within the high-risk group accounting for 50% and 20%, respectively, of cervical cancers [2]. HPV-16 is also the most commonly found type in HPV-positive head and neck squamous cell carcinoma [6].
High-risk HPVs express two oncoproteins, E6 and E7, which function to deregulate the host cell cycle in order to promote replication of the viral genome. Long-term expression of HPV E6 and E7 oncoproteins is known to both extend the life-span of primary human cells and facilitate their immortalization [7]. In line with this notion, expression of HPV E6 and E7 oncoproteins is consistently up-regulated in HPV-associated cancers due to integration of the viral DNA and de-regulation of the normal control of HPV E6 and E7 expression [1].
Despite the high prevalence of HPV infection in sexually active women, most HPV infections are self-limiting and hence transient. Progression to cancer is a result of both persistent infection with high-risk HPV types as well as co-factors, for example tobacco use and exogenous estrogen or UV exposure [8]. Mounting evidence suggests that genomic instability is also an important co-factor in promoting malignant progression. Support for this notion stems from the fact that patients with Fanconi Anemia (FA), an X-linked and autosomal recessive chromosomal instability syndrome, are at a significantly higher risk for developing HPV-associated cancerous lesions [9,10]. There is also compelling evidence suggesting that HPV oncoproteins can by themselves drive genomic instability [11,12].
Besides high-risk HPV oncoprotein-induced chromosomal instability, there is evidence suggesting that exogenous co-factors also exist and contribute to high-risk HPV-associated genomic instability. Exposure of cells expressing episomal HPV-16 genomes to increased levels of nitric oxide (NO), a free radical normally produced at infectious or inflammatory sites, ledtoenhanced high-risk HPV oncoprotein gene transcription, increased DNA double-strand breaks and chromosomal instability [13]. Supporting the role of NO in promoting high-risk HPV oncoprotein induced cervical carcinogenesis, it was recently found that nitric oxide synthase-dependent DNA damage was observed in biopsy specimens from patients with cervical dysplasia [14]. Other exogenous co-factors include the host cell response to HPV infection, which may lead to chronic inflammation. In line with this notion, HPV-16 E6 and E7 expressing cells treated with transforming growth factor β1 (TGF-β1), a known effector of the host-cell inflammatory response, promoted enhanced chromosomal instability [15].
Together, these findings substantiate the idea that disruption of host genome integrity by expression of high-risk HPV oncoproteins is a major driving force in supporting cellular transformation which ultimately promotes cervical carcinogenesis.
2. HPV Oncoproteins
High-risk human papillomaviruses are small double-stranded DNA viruses of approximately 8,000 base pairs with a tropism for keratinocytes. Their life-cycle is intimately linked to the differentiation state of the host cell. Oncogenic HPV types, such as HPV-16, express eight open reading frames (ORFs) transcribed as polycistronic mRNAs [16]. HPV gene products, functionally outlined in Table 1, are expressed in a temporal manner under control of the long control region (LCR), which harbors non-coding sequences [16]. The virus does not encode its own DNA replication enzymes and consequently relies on the host cell machinery to replicate its genome. Viral replication occurs in differentiating keratinocytes that are normally permanently withdrawn from the cell division cycle. To overcome this barrier, the HPV E6 and E7 oncoproteins have evolved to re-introduce an S-phase like milieu under differentiating conditions of the host cell in order to promote viral replication.
Table 1.
Function of HPV-16 proteins
| HPV protein | Function | Size (a.a.) | Temporal expression | Localization | Functional domains | Ref. |
|---|---|---|---|---|---|---|
| E1 | Viral replication, DNA helicase | 649 | Early* | Nucleus | Amino-terminal: DNA binding Carcoxy-terminal: enzymatic | [10,86] |
| E2 | Viral replication, origin binding, transcriptional activation and repression tethering of viral DNA to host chromosomes | 365 | Early | Nucleus | Amino-terminal: transcriptional activation and viral DNA tethering Carboxy-terminal: DNA binding/dimerization | [10] |
| E4 | Destabilization of cytokeratin network | 95 | Early and Late | Cytoplasm | DEAD-box protein binding motif, LLXLL motif | [10] |
| E5 | Mediates mitogenic signalling of growth factors | 83 | Early and Late | Endosomal membranes, Golgi, and cell membranes | Hydrophobic regions | [10] |
| E6 | Major oncoprotein | 158 | Early | Nucleus and cytoplasm | p53 binding domain, PDZ-binding domain, four Cys-X-X-Cys motifs | See text |
| E7 | Major oncoprotein | 98 | Early | Nucleus and cytoplasm | LXCXE motif, caesin kinase II phosphorylation site, two Cys-X-X-Cys motifs | See text |
| L1 | Major viral capsid protein | 531 | Late** | Diffuse nuclear | Multimerization domain, L2-binding domain | [87,88] |
| L2 | Minor viral capsid protein | 473 | Late | Diffuse nuclear | L1-binding domain | [87] |
expressed prior to productive viral replication
expressed at the time of productive viral replication
2.1 The HPV E7 oncoprotein
High-risk HPV E7 proteins are small phosphoproteins with no known human homologs [17]. HPV E7 oncoproteins contain two conserved domains (CR1 and CR2) which share sequence similarity to both adenovirus E1A and SV40 large T antigen [18]. High-risk HPV E7 inactivates the retinoblastoma tumor suppressor protein (pRB), and the related pocket protein family members p107 and p130, which are responsible for regulating E2F-mediated transcription of S-phase genes [17]. Specifically, HPV-16 E7 binds to and induces the proteasomal degradation of pRB by cullin 2-containing E3 ubiquitin ligases [19]. High-risk HPV E7 associates with pRB and its family members through a Leu-X-Cys-X-Glu (LXCXE) motif located within the CR2 homology domain [20]. Additional sequences located in the amino-terminal CR1 homology domain are necessary for pRB degradation [19]. High-risk HPV-16 E7 proteins bind with a higher efficiency to pRB than do low-risk HPV-6 E7 proteins. This difference maps to a single amino acid change within the pRB-binding domain which confers high-affinity binding [21]. High-risk HPV-16 E7 has also been shown to inactivate p600, a pRB-associated protein [22].
To further disrupt host gene expression control, HPV-16 and HPV-31 E7 oncoproteins also interact with histone deacetylases type -1 and -2 (HDAC-1, and -2) [23,24]. HDACs function as transcriptional repressors by reversing acetyl modifications of lysine residues on histones. The indirect association between oncogenic HPV-16 E7 and HDACs is mediated by Mi2β, a component of the NURD histone deacetylase complex [23]. This interaction is dependent on the integrity of two Cys-X-X-Cys motifs in the HPV E7 oncoprotein carboxy-terminus and results in increased E2F-mediated gene transcription from HDAC responsive promoters [23]. High-risk HPV-16 E7 also associates with histone acetyl transferases (HATs) such as, p300 and pCAF, which function to activate transcription and stimulate cellular proliferation [25,26].
In addition to chromatin remodeling, high-risk HPV-16 E7 can directly alter cellular transcription through interaction with E2F1. This interaction results in the pRB-independent enhancement of E2F-mediated gene transcription [27]. However, the promoter of E2F6, a transcriptional repressor responsible for directing cell cycle exit, is also E2F-responsive [28]. The HPV-16 E7 oncoprotein has therefore evolved to directly associate with E2F6 resulting in inactivation of its transcriptional repression and maintenance of an S-phase like environment [28].
Together, along with the ability of HPV-16 E7 to interact with cyclin/CDK complexes and its ability to overcome cellular growth arrest signals mediated by the cyclin dependent kinase (CDK) inhibitors p21Cip1 and p27Kip1, the HPV E7 oncoprotein profoundly disrupts the pRB-signaling axis to favor replication of the viral genome [29,30,31]. Why high-risk HPV E7 has evolved to target a multitude of G1/S checkpoint components to achieve this goal is currently unknown.
2.2 The HPV E6 oncoprotein
Disruption of the host cell cycle by HPV-16 E7 is likely to activate cellular stress responses and apoptotic signaling cascades. The HPV E6 oncoprotein has evolved to inhibit the host cell response to unscheduled cell cycle entry by mediating the degradation of p53. High-risk E6 degrades p53 by re-directing a host cell HECT domain containing E3 ubiquitin ligase, E6-associated protein (E6AP) [32]. Moreover, HPV-16 E6 binds the transcriptional co-activators CBP/p300 and decreases the ability to activate p53-responsive promoter elements [33]. High-risk HPV E6 has furthermore been suggested to switch p53-p300 from an activating to a repressor complex independently of E6AP [34].
High-risk HPV E6 has additional p53-independent functions that are important for cellular transformation and immortalization. Oncogenic HPV E6 contains a PDZ-domain binding motif, X-(S/T)-X-(V/I/L)-COOH, that is unique to high-risk HPV E6 and is not present in low-risk HPV E6 [35]. The HPV E6 oncoprotein binds PDZ-containing host proteins targeting them for degradation in both an E6AP-dependent and -independent manner [36,37,38]. Candidate PDZ-containing protein targets include hDlg, hScrib, MAGI1-3, and MUPP1 [36,37]. PDZ-containing proteins localize to membrane-cytoskeleton interfaces and have been implicated as molecular signaling scaffolds modulating cell growth, polarity and adhesion in response to cell contact. The targeted inactivation of these proteins by oncogenic HPV E6 may disrupt cell junctions, induce loss of cell polarity and promote cellular transformation [39].
The high-risk HPV-16 E6 oncoprotein promotes cellular immortalization through the transcriptional up-regulation of hTERT, the catalytic subunit of human telomerase, and can contribute to telomere maintenance [40]. High-risk HPV E6 can enhance hTERT transcription through several mechanisms including association with the transcriptional activator c-Myc and/or the E6AP-dependent degradation of a putative transcriptional repressor of the hTERT promoter, NFX1-91 as well as others [41,42].
2.3 HPV oncoproteins and genomic instability
Genomic instability is a defining phenotype of many malignant tumors including HPV-associated malignancies [1,43]. Over 100 years ago Theodor Boveri hypothesized that genomic instability and cancer can result from the presence of extra centrosomes and the subsequent formation of multipolar mitoses. Such a disruption of spindle polarity may consequently promote chromosome missegregation and ultimately aneuploidy [44]. Multipolar, specifically tri-polar, mitoses are a hallmark of high-risk HPV-associated carcinomas [1]. Furthermore, the frequency of aneuploidy increases with both malignant grade and tumor aggressiveness in HPV-associated lesions, which make HPV-associated neoplasms a suitable model system to test Boveri’s hypothesis [1,45].
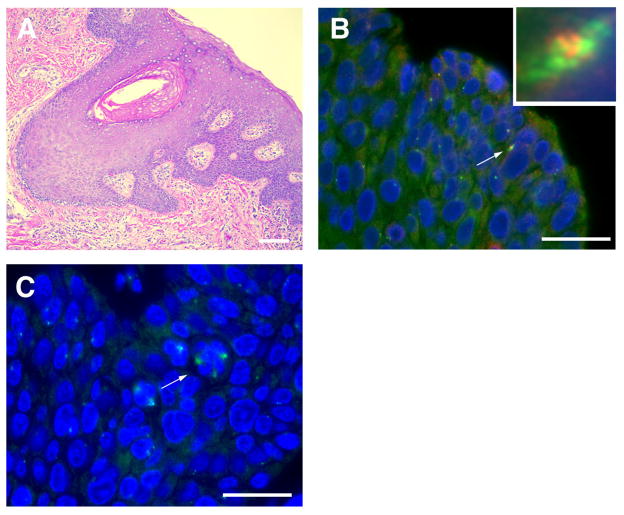
The major consequences of supernumerary centrosomes are polarity disturbances, such as multipolar mitoses, and/or merotelic kinetochore attachments, which can lead to lagging chromosomes during cell division [46]. However, extra centrosomes do not necessarily lead to cell division errors since centrosomes can cluster thereby preserving bipolarity of the mitotic spindle [47]. Nonetheless, centrosome abnormalities have been detected in a wide range of malignant tumors including breast, colon, and prostate cancer [1]. In the context of high-risk HPV, centrosome amplification is observed in cells expressing episomal HPV-16 genomes, which underscores that viral integration and overexpression of HPV E6 and HPV E7 oncoproteins is not required for the disruption of centrosome duplication control [48]. Studies in HPV-associated primary human tumors have demonstrated that centrosome overduplication correlates with the frequency of cell division errors, which lends important support to the notion that the presence of supernumerary centrosomes can promote these defects (Figure 1) [49].
Figure 1. High-risk HPV-associated neoplasms exhibit centrosome overduplication and multipolar mitoses.
(A) High-risk HPV-associated anal squamous cell carcinoma stained with hematoxylin and eosin (H&E). Scale bar indicates 100 μm (B) Co-immunofluorescence microscopy for the centrosomal marker γ-tubulin (green) co-stained with Cep170 (red), a marker of mature centrosomes in a tissue sample from (A). The arrow points to a cell with overduplicated centrosomes and the inset represents a high-powered view of this cell. Nuclei stained with DAPI. Scale bar indicates 50 μm. (C) Arrow indicates an example of cell division errors during metaphase in high-risk HPV-associated anal neoplasms detected by immunofluorescence microscopy for γ-tubulin (green). Nuclei stained with DAPI. Scale bar indicates 50 μm.
Besides aneuploidy, structural chromosomal instability is a critical factor for malignant progression. This is evidenced by the enhanced rate of tumor formation in patients with mutations in DNA repair pathway genes such as BRCA1, BRCA2 or the mismatch-repair (MMR) pathway [50]. Unrepaired, broken DNA can promote gene translocations or gene amplifications/deletions, which may provide a growth advantage to cells through gain of oncogenes or loss of tumor suppressors [50]. Several lines of evidence show that expression of HPV-16 E6 and E7 can independently induce structural chromosomal instability [51] (see section 2.6).
2.4 HPV-16 E7 disrupts centrosome duplication control
The centrosome consists of two centrioles embedded in a cloud of pericentriolar proteins, also known as pericentriolar material (PCM). In order to generate two spindle poles, the single centrosome of a non-dividing cell must duplicate precisely once prior to mitosis in order to ensure faithful cell division [52].
Centrosome duplication begins during late mitosis/early G1-phase of the cell division cycle, when the two pre-existing centrioles of the centrosome disengage through the action of polo kinase 1 (PLK1) and separase, and move into a near parallel position [53]. This step is followed by recruitment of polo-like kinase 4 (PLK4) to the wall of the maternal centriole at the site of daughter centriole synthesis [54]. Subsequently, structural proteins are recruited to the nascent pro-centriole to stabilize and elongate the new daughter centriole. Centrosome duplication is complete during late G2-phase, when the two fully formed centriole pairs separate to form the mitotic spindle poles [52].
There are two mechanisms by which centriole amplification in tumor cells may occur: centriole overduplication and centriole accumulation. These two phenotypes can be distinguished by immunostaining for markers of older, mature centrioles (Figure 1) [55]. Genuine centriole overduplication is characterized by the presence of one or two mature maternal centrioles and multiple immature daughter centrioles. In contrast, centriole accumulation is defined by the presence of multiple maternal centrioles with a normal ratio of daughter centrioles [55]. The distinction between centriole overduplication and accumulation is important because cells exhibiting centriole accumulation may arise due to abortive mitoses or cytokinesis errors and such cells may not be able to produce viable progeny. Conversely, cells which exhibit a genuine centriole overduplication defect are, in general, less altered and hence are more likely to give rise to genomically unstable daughter cells.
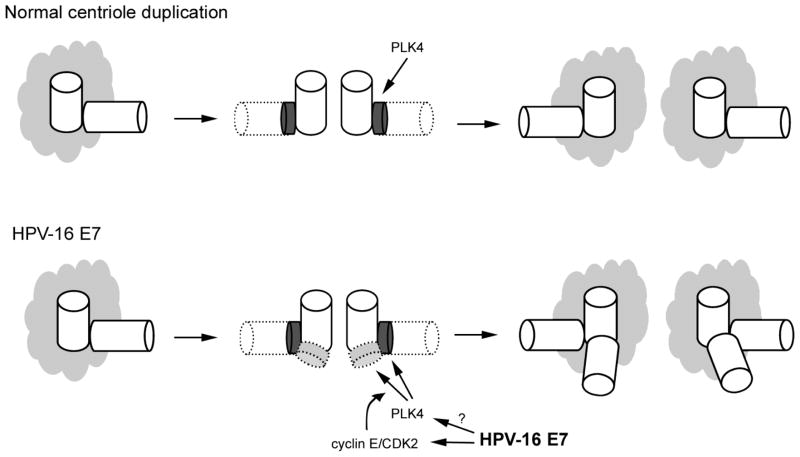
There are several lines of evidence that HPV-16 E7 oncoprotein expression disrupts genomic integrity by directly interfering with centriole duplication control. First, HPV-16 E7 expression produces abnormal centriole numbers in otherwise normal cells prior to the onset of genomic instability [56]. This is in contrast to HPV-16 E6 expressing cells which exhibit centrosome accumulation in cells which are already genomically unstable, often expressing markers of cellular senescence, and are unlikely to remain in the proliferative pool [56]. Secondly, supernumerary centrioles appear rapidly and within a single cell division cycle, suggesting they arise due to direct disruption of centriole duplication control [57]. This was initially difficult to reconcile with the prevailing model of centriole duplication described above, where a single maternal centriole initiates the synthesis of only a single daughter centriole. Further analysis of HPV-16 E7 induced centriole abnormalities led to the discovery that the HPV E7 oncoprotein rapidly induces centriole overduplication through stimulation of a novel centriole duplication pathway, referred to as centriole multiplication (Figure 2) [57]. This pathway is characterized by a single maternal centriole initiating the simultaneous synthesis of two or more daughter centrioles. This phenotype had not previously been observed in the context of an oncogenic stimulus relevant for a major human cancer [57].
Figure 2. HPV-16 E7 expression subverts normal centriole duplication control.
During normal centriole duplication, PLK4 is recruited to the wall of the maternal centriole to initiate the synthesis of a single daughter centriole per mother. Following HPV-16 E7 expression, centriole duplication control is disrupted which can result in the simultaneous synthesis of more than one daughter centriole per maternal centriole (centriole multiplication). Overexpression of cyclin E/CDK2 complexes has been shown to lead to the aberrant recruitment of PLK4 to maternal centrioles [58]. This, along with deregulation of PLK4 protein expression promotes centriole multiplication. HPV-16 E7 is known to deregulate cyclinE/CDK2 activity, whether the HPV E7 oncoprotein interferes with PLK4 protein levels and/or kinase activity is currently unknown.
CDK2, cyclin E, and PLK4 were found to be necessary factors for centriole multiplication [57]. Ectopic expression of CDK2/cyclin E complexes alone, however, was not sufficient to induce centriole multiplication. This only occurred when PLK4 was upregulated, suggesting that PLK4 protein levels are rate-limiting for centriole multiplication [58]. High-risk HPV-16 E7 protein expression is known to deregulate cyclin E/CDK2 complexes which may help to aberrantly recruit PLK4 to maternal centrioles [30,58,59]. Whether HPV-16 E7 also deregulates steady-state PLK4 protein expression awaits further clarification.
An HPV-16 E7 deletion mutant lacking amino acid residues 21–24 and hence unable to bind and degrade pRB (HPV-16 E7 Δ21–24) was found to also be unable to induce centriole overduplication in both normal and pRB-family deficient mouse embryo fibroblasts [60]. In contrast, full-length wild-type HPV-16 E7 was able to induce centriole abnormalities even in pRB/p107/p130-deficient cells [60]. These results suggest that pRB-degradation is not the only mechanism by which oncogenic HPV E7 induces centriole overduplication. One possible mechanism for pRB-independent induction of supernumerary centrioles is through the ability of HPV-16 E7 to interact with γ-tubulin, a component of the PCM important for microtubule nucleation. This interaction, which is pRB-independent, relies on an intact LXCXE motif [61]. It has been suggested that disruption of γ-tubulin plays a role in the regulation of centrosome duplication and this mechanism may hence contribute to overduplication induced by HPV-16 E7 [62].
2.5 Additional mechanisms of HPV-16 E7 induced disruption of mitotic fidelity
Besides centriole overduplication, the HPV-16 E7 oncoprotein has been found to disrupt mitotic fidelity through several additional mechanisms. Remarkably, the proportion of multipolar metaphase cells in HPV-16 E7 oncoprotein-expressing populations was found to be significantly higher than the proportion of multipolar ana- or telophase cells [49]. This suggests that the majority of HPV-16 E7-expressing cells exhibiting multipolar spindle poles in metaphase are unable to complete mitosis.
Eukaryotic cells can be arrested at the metaphase-anaphase transition by activation of the spindle-assembly checkpoint (SAC). The SAC is activated in response to aberrant microtubule-kinetochore attachments but not necessarily due to the presence of supernumerary centrosomes or multiple spindle poles [63,64]. Once the SAC is activated several outcomes may occur. Cells may resolve the microtubule-kinetochore attachments and proceed through a normal mitosis, they may be unable to resolve the mitotic dysfunction and activate apoptotic cascades leading to mitotic catastrophe, or they may adapt to prolonged SAC activation, decondense their chromosomes and enter a G1-like state with tetraploid DNA content [63]. The fate of high-risk HPV E7-expressing cells that have multiple spindle poles in metaphase and do not proceed through mitosis is currently unknown. Since the presence of aberrant spindle poles per se may not trigger SAC activation, it is possible that altered DNA structure may contribute to the over-representation of multipolar metaphase cells.
However, both HPV-16 E6 and HPV-16 E7 have independently been shown to overcome the SAC and promote the accumulation of polyploid cell populations [65,66]. The ability of HPV-16 E6 to abrogate the SAC was not unexpected, due to the fact that activation of the SAC is thought to be controlled through p53-dependent pathways [63]. However, it was surprising that HPV-16 E7 expression alone bypassed the SAC. Whether this activity is due to direct interaction with SAC proteins or pRB-inactivation, which can lead to Mad2 deregulation, needs to be determined in detail [67]. Both mechanisms are not mutually exclusive.
High-risk HPV-16 E7 has been reported to delocalize dynein from mitotic spindles in a pRB-independent manner [68]. Dynein targets nuclear apparatus protein 1 (NuMA) to the mitotic spindle poles where it is responsible for spindle organization and stabilization as well as chromosome alignment. Subsequent studies showed that HPV-16 E7 and low-risk HPV-6b and HPV-11 E7 physically associate with NuMA [69]. The ability of the HPV-16 E7 protein to delocalize dynein and interact with NuMA mapped to the same carboxy-terminal sequences of the protein. The ability of HPV-16 E7 to delocalization dynein and interact with NuMA resulted in a mitotic delay and defects in chromosome alignment [69].
Genomically unstable high-risk HPV oncoprotein expressing cells may also progress through the cell division cycle by disruption of the post-mitotic spindle checkpoint. The post-mitotic checkpoint is triggered following adaptation to a prolonged SAC activation, during which cells may exit mitosis, bypass cytokinesis and progress into a G1-like state with tetraploid DNA content [70]. These cells are then prevented from continuing through the cell cycle and replicating their DNA through a p53- and pRb-dependent post-mitotic checkpoint [71]. High-risk E6 and E7 oncoprotein expression individually has been shown to abrogate the post-mitotic checkpoint promoting polyploidization possibly predisposing cells to aneuploidy [71,72].
2.6 HPV-16 E7 and host cellular DNA damage
There is convincing evidence that HPV oncoproteins promote not only mitotic defects but also structural chromosomal alterations and DNA damage. Cytogenic analyses of HPV-associated lesions revealed recurring patterns of chromosome gains and losses. In particular, gain of chromosome 3q has been linked to the transition to invasiveness in high-risk HPV-associated lesions [73]. Several other common structural alterations observed in HPV-associated neoplasms include gains of genetic material on chromosomes 1q, 5p, 6p and 20q and losses mapped to chromosomes 2q, 3p, 4, 8p and 13q [74,75]. A further reflection of structural instabilities seen in high-risk HPV-associated lesions include an increased incidence of anaphase bridges. Anaphase bridges may form through chromosome fusions at telomeres or double-stranded DNA breaks [1].
Supporting the hypothesis that high-risk HPV oncoproteins can promote DNA breakage in host cells, both HPV-16 E6 and E7 expression has been shown to enhance mutation frequency in primary human keratinocytes [76]. Additionally, cells independently expressing high-risk but not low-risk HPV E6 and E7 oncoproteins have an increased ability to integrate foreign DNA [77]. It has furthermore been shown that both the high-risk HPV oncoproteins independently induce DNA breakage using the comet assay [51]. Lastly, nuclear foci of phosphorylated H2AX (y-H2AX) appear following HPV-16 E7 expression, indicating induction of DNA double-strand break repair pathways [51]. Despite the well-known function of HPV-16 E7 to induce DNA damage, the precise source of DNA double-strand breaks remains poorly understood.
Aberrant expression of S-phase specific cyclins has been suggested to result in anomalous DNA replication, increased stalling of replication forks, and chromosomal instability [78]. Using the Fanconi Anemia (FA) DNA damage response pathway as a surrogate marker, it was found that HPV-16 E7 expression triggers host cell replication stress [79].
Upon activation, the core complex of FA proteins mediates the monoubiquitination of two other FA proteins, FANCD2 and FANCI [80]. This step is necessary for translocation of FANCD2 and FANCI to the site of the stalled replication forks [80]. Monoubiquitinated FANCD2, along with several other interacting proteins, mediates replication fork stabilization and restart. Using FANCD2 nuclear foci and the recruitment of FANCD2 and FANCDI/BRCA2 as markers for FA pathway activation, Spardy, et al showed that primary human keratinocytes stably expressing high-risk HPV-16 E7, but not low-risk HPV-6 E7, have an activated FA pathway [79]. Stable expression of HPV-16 E6 in primary human keratinocytes did not induce a significant increase in FANCD2 foci, however co-expression of HPV-16 E6 and E7 did enhance the formation of foci over HPV-16 E7 expression alone [79]. It was further demonstrated that cells deficient for the FA pathway were prone to accelerated high-risk HPV-associated chromosomal breakage [79]. These findings suggest that FA pathway activation is an early host cell response to HPV-infection and that genetic or epigenetic inactivation of the FA pathway contributes to increased genomic instability and malignant progression [81].
Importantly, these experimental results are mirrored by clinical findings where patients suffering from the cancer susceptibility syndrome Fanconi Anemia have a 500- to 700-fold increase in the incidence of squamous cell carcinomas (SCCs) at sites of HPV infection [9]. One analysis of head and neck SCCs from FA patients found over 80% of the tumors contained high-risk HPV DNA [9]. SCCs develop at much younger ages in FA-deficient individuals suggesting a role for the FA pathway in suppressing malignant progression [9]. Besides genomic inactivation of FA genes, the FANCF promoter has been shown to be hypermethylated in approximately 30% of advanced stage invasive cervical SCCs [82]. Interestingly, patients younger than 45 years of age showed a significantly higher frequency of FANCF promoter hypermethylation than patients older than 45 years of age [82]. This suggests that FA pathway inactivation or deficiency may result in increased risk for the development of cancer in younger age group patients. Finally, single-nucleotide polymorphisms in the FANCA gene have been associated with an increased risk for development of HPV-associated cervical cancer [83]. Taken together, these observations suggest that disruption of the FA pathway may promote a cellular environment which enhances the rate of HPV-associated malignant progression.
HPV-16 E7 expression also modulates additional host cell DNA damage response proteins. Both HPV-16 E6 and HPV-16 E7 have been shown to bind the breast cancer 1 (BRCA1) tumor suppressor through their zinc-finger domains which may inhibit its function [84]. HPV-16 E6 has been shown to interact with the DNA repair factor XRCC1 possibly interfering with single-strand DNA break repair [85]. The ability of HPV oncoproteins to impair the cellular DNA damage response may be a further source of chromosomal breakage seen in HPV oncoprotein-expressing cells [51]. The manipulation of DNA damage response pathways by virally encoded proteins in order to facilitate the viral life-cycle has been well-established for viruses [86]. The question whether manipulation of the host cell DNA damage response may contribute to HPV genome integration remains to be answered.
2.7 HPV-16 E7, DNA damage and host cellular immortalization
Expression of high-risk HPV-16 E6 and E7 are known to individually extend the lifespan of primary human cells and facilitate their immortalization when co-expressed [7]. Cellular immortalization ultimately requires the activation of pathways that help to prevent critical shortening of telomeres. The ability of high-risk HPV E6 to promote cellular immortalization has been suggested to depend on its ability to activate telomerase [40]. How the high-risk HPV E7 oncoprotein facilitates cellular immortalization independent of the HPV E6 oncoprotein is not understood, but alternative lengthening of telomeres (ALT) has previously been suggested [87]. ALT is a homologous recombination (HR)-based mechanism of telomere maintenance that uses, for example, sister chromatids as templates [88].
More detailed insights into the question whether the HPV E7 oncoprotein can trigger ALT were attained recently by the discovery that HPV-16 E7 causes an increase in the formation of ALT-associated promeolytic leukemia bodies (APBs) [88]. The onset of ALT immortalization coincides with the appearance of APBs and their formation is often used as a surrogate marker for ALT activity [89]. APBs are known to contain telomeric DNA, telomere-binding proteins, and several proteins involved in HR [88]. High-risk HPV-16 E7 was found to induce APBs that contained these components as well as previously unknown components including FANCD2, BRCA2 and MUS81 [88]. FANCD2 was found to be critical for the maintenance of telomere homeostasis in ALT-positive cells and the occurrence of FANCD2-positive APBs correlated with HPV-16 E7-induced extension of life span in primary human keratinocytes population [88].
This function of HPV-16 E7 was found to be dependent on its ability to degrade pRB and hence suggests altered replication of telomeric DNA as a trigger for APB formation and stimulation of ALT [88]. The correlation that was discovered between APB formation and sustained proliferation in early passage primary human keratinocytes suggests that APB-positive cells may be endowed with a growth advantage at early stages of immortalization [88].
2.8 HPV-16 E7 and continued proliferation in the presence of DNA damage
How HPV-16 E7-expressing cells maintain proliferative capacity despite the presence of DNA damage and DNA damage checkpoint activation is not understood in detail. Recently, it has been shown that HPV-16 E7 expression attenuates DNA damage checkpoints through enhancement of the proteolytic turnover of claspin [90]. Claspin is a critical regulator of the ATR/CHK1 signaling axis and DNA damage checkpoint recovery in the G2 phase of the cell cycle [91]. ATR is activated by stalled replication forks and requires claspin to phosphorylate the downstream kinase Chk1 in order to arrest the cell allowing time for DNA repair [91]. Degradation of claspin plays a critical role in recovery from DNA damage checkpoint activation and is necessary to promote mitotic entry [91].
Paradoxically, HPV-16 E7 oncoprotein expressing cells actually express a higher baseline level of claspin than control cells, possibly promoting efficient DNA replication during S-phase [90]. High-risk HPV-16 E7 then accelerates the degradation of claspin as cells approach the G2-phase of the cell division cycle, in part through up-regulation of E2F-responsive components of the claspin degradation machinery [90]. This, together with the HPV E6 oncoprotein-mediated degradation of p53, allows cells that contain DNA damage to aberrantly enter mitosis, maintain proliferative capacity and may ultimately contribute to the propagation of structural chromosomal abnormalities seen in high-risk HPV-associated neoplasms.
3. Outlook
HPV oncoproteins have a plethora of functions which converge on host cell cycle checkpoints to promote DNA replication in cells that are at the same time undergoing differentiation and are normally withdrawn from the cell division cycle. Research over the past several years has shown that aberrant proliferation can set the stage for genomic instability and malignant progression by disrupting centrosome duplication control and normal DNA replication dynamics (Figure 3). The identification of critical pathways involved in aberrant centrosome duplication such as CDK2 and PLK4 may help to design innovative approaches for prevention of malignant progression in early HPV-associated lesions. In addition, a better understanding of the DNA damage response in high-risk HPV-associated malignancies may be exploited to develop novel therapies for advanced stage lesions, for example through synthetic lethality. Besides this translational potential, understanding the molecular basis of genomic instability in the context of high-risk HPV oncoproteins has yielded unexpected insights into some important aspects of cell biology. These and future findings will ultimately help to develop a systems biology approach to our understanding of cancer formation and progression.
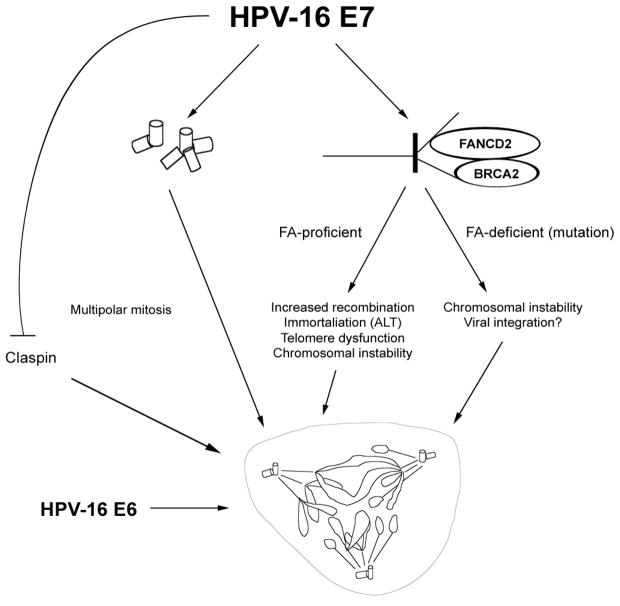
Figure 3. A comprehensive view of HPV-16 E7-induced genomic instability.
There is compelling evidence that disruption of host cell cycle checkpoint control by HPV-16 E7 promotes numerical and structural chromosomal instability through disruption of centrosome duplication control and altered DNA replication dynamics, respectively. Activation of the Fanconi Anemia (FA) pathway by HPV-16 E7 can have different outcomes depending on the presence of a functional FA pathway (see text for details). HPV-16 E7 has been shown to facilitate entry into mitosis by inducing accelerated claspin degradation. Whereas the exact fate of altered metaphase cells in the context of HPV-16 E7 remains to be determined, it is clear that the ability of HPV-16 E6 to attenuate p53-mediated host cell responses contributes ultimately to host cell viability and continued proliferation.
Acknowledgments
We would like to thank all members of the Duensing laboratory for helpful discussions. Work on HPV in the authors’ laboratory is supported by the NIH/NCI (R01 CA112598) and the American Cancer Society (RSG-07-075-01-MBC).
Footnotes
Conflict of Interest Statement
The authors declare no competing confict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duensing S, Munger K. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int J Cancer. 2004;109:157–162. doi: 10.1002/ijc.11691. [DOI] [PubMed] [Google Scholar]
- 2.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 3.Orth G. Epidermodysplasia verruciformis: a model for understanding the oncogenicity of human papillomaviruses. Ciba Found Symp. 1986;120:157–174. doi: 10.1002/9780470513309.ch11. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout RJ, Bouwes Bavinck JN, ter Schegget J. Persistence of human papillomavirus DNA in benign and (pre)malignant skin lesions from renal transplant recipients. J Clin Microbiol. 2000;38:2087–2096. doi: 10.1128/jcm.38.6.2087-2096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 6.Paz IB, Cook N, Odom-Maryon T, Xie Y, Wilczynski SP. Human papillomavirus (HPV) in head and neck cancer. An association of HP 16 with squamous cell carcinoma of Waldeyer’s tonsillar ring. Cancer. 1997;79:595–604. doi: 10.1002/(sici)1097-0142(19970201)79:3<595::aid-cncr24>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 7.Halbert CL, Demers GW, Galloway DA. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J Virol. 1992;66 :2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kjellberg L, Hallmans G, Ahren AM, Johansson R, Bergman F, Wadell G, Angstrom T, Dillner J. Smoking, diet, pregnancy and oral contraceptive use as risk actors for cervical intra-epithelial neoplasia in relation to human papillomavirus infection. Br J Cancer. 2000;82 :1332–1338. doi: 10.1054/bjoc.1999.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kutler DI, Wreesmann VB, Goberdhan, Ben-Porat L, Satagopan J, Ngai I, Huvos AG, Giampietro P, Levran O, Pujara K, Diotti R, Carlson D, Huryn LA, Auerbach AD, Singh B. Human papillomavirus DNA and p53 polymorphisms in squamous cell carcinomas from Fanconi anemia patients. J Natl Cancer Inst. 2003;95 :1718–1721. doi: 10.1093/jnci/djg091. [DOI] [PubMed] [Google Scholar]
- 10.Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43 :223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.zur Hausen H. Papillomavirus infections--a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 12.White AE, Livanos EM, Tlsty TD. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8 :666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 13.Wei L, Gravitt PE, Song H, Maldonado AM, Ozbun MA. Nitric oxide induces early viral transcription coincident with increased DNA damage and mutation rates in human papillomavirus-infected cells. Cancer Res. 2009;69 :4878–4884. doi: 10.1158/0008-5472.CAN-08-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiraku Y, Tabata T, Ma N, Murata M, Ding X, Kawanishi S. Nitrative and oxidative DNA damage in cervical intraepithelial neoplasia associated with human papilloma virus infection. Cancer Sci. 2007;98 :964–972. doi: 10.1111/j.1349-7006.2007.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Tsao SW, Kwok YK, Wong E, Huang XR, Liu S, Tsang CM, Ngan HY, Cheung AN, Lan HY, Guan XY, Cheung AL. Transforming growth actor beta1 promotes chromosomal instability in human papillomavirus 16 E6E7-infected cervical epithelial cells. Cancer Res. 2008;68 :7200–7209. doi: 10.1158/0008-5472.CAN-07-6569. [DOI] [PubMed] [Google Scholar]
- 16.Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68 :362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaughlin-Drubin ME, Munger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384 :335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vousden KH, Jat PS. Functional similarity between HPV16E7, SV40 large T and adenovirus E1a proteins. Oncogene. 1989;4 :153–158. [PubMed] [Google Scholar]
- 19.Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, Harper JW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81 :9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66 :6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heck DV, Yee CL, Howley PM, Munger K. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc Natl Acad Sci U S A. 1992;89 :4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Munger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated actor, p600. Proc Natl Acad Sci U S A. 2005;102 :11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18 :2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longworth MS, Laimins LA. The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential or the life cycle of human papillomavirus type 31. J Virol. 2004;78 :3533–3541. doi: 10.1128/JVI.78.7.3533-3541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avvakumov N, Torchia J, Mymryk JS. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene. 2003;22 :3833–3841. doi: 10.1038/sj.onc.1206562. [DOI] [PubMed] [Google Scholar]
- 26.Bernat A, Avvakumov N, Mymryk JS, Banks L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene. 2003;22 :7871–7881. doi: 10.1038/sj.onc.1206896. [DOI] [PubMed] [Google Scholar]
- 27.Hwang SG, Lee D, Kim J, Seo T, Choe J. Human papillomavirus type 16 E7 binds to E2F1 and activates E2F1-driven transcription in a retinoblastoma protein-independent manner. J Biol Chem. 2002;277 :2923–2930. doi: 10.1074/jbc.M109113200. [DOI] [PubMed] [Google Scholar]
- 28.McLaughlin-Drubin ME, Huh KW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J Virol. 2008;82 :8695–8705. doi: 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen CL, Munger K. Direct association of the HPV16 E7 oncoprotein with cyclin A/CDK2 and cyclin E/CDK2 complexes. Virology. 2008;380 :21–25. doi: 10.1016/j.virol.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11 :2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DL, Alani RM, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11 :2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75 :495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 33.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18 :5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005;17 :251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci U S A. 1997;94 :11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SS, Glaunsinger B, Mantovani F, Banks L, Javier RT. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J Virol. 2000;74 :9680–9693. doi: 10.1128/jvi.74.20.9680-9693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardiol D, Kuhne C, Glaunsinger B, Lee SS, Javier R, Banks L. Oncogenic human papillomavirus E6 proteins target the disc large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18 :5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- 38.Massimi P, Shai A, Lambert P, Banks L. HPV E6 degradation of p53 and PDZ containing substrates in an E6AP null background. Oncogene. 2008;27 :1800–1804. doi: 10.1038/sj.onc.1210810. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen ML, Nguyen MM, Lee D, Griep AE, Lambert PF. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required or E6’s induction of epithelial hyperplasia in vivo. J Virol. 2003;77 :6957–6964. doi: 10.1128/JVI.77.12.6957-6964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380 :79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 41.Gewin L, Myers H, Kiyono T, Galloway DA. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004;18 :2269–2282. doi: 10.1101/gad.1214704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA. NFX1-123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes. J Virol. 2009;83 :6446–6456. doi: 10.1128/JVI.02556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Knebel Doeberitz M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer. 2002;38 :2229–2242. doi: 10.1016/s0959-8049(02)00462-8. [DOI] [PubMed] [Google Scholar]
- 44.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci. 2008;121(Suppl 1 ):1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 45.Crum CP, Ikenberg H, Richart RM, Gissman L. Human papillomavirus type 16 and early cervical neoplasia. N Engl J Med. 1984;310 :880–883. doi: 10.1056/NEJM198404053101403. [DOI] [PubMed] [Google Scholar]
- 46.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460 :278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307 :127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 48.Duensing S, Duensing A, Flores ER, Do A, Lambert PF, Munger K. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J Virol. 2001;75 :7712–7716. doi: 10.1128/JVI.75.16.7712-7716.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duensing A, Chin A, Wang L, Kuan SF, Duensing S. Analysis of centrosome overduplication in correlation to cell division errors in high-risk human papillomavirus (HPV)-associated anal neoplasms. Virology. 2008;372 :157–164. doi: 10.1016/j.virol.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khanna KK, Jackson SP. DNA double-strand breaks: signaling repair and the cancer connection. Nat Genet. 2001;27 :247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 51.Duensing S, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002;62 :7075–7082. [PubMed] [Google Scholar]
- 52.Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120 :2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- 53.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17 :344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7 :1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 55.Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16 :1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duensing S, Duensing A, Crum CP, Munger K. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 2001;61 :2356–2360. [PubMed] [Google Scholar]
- 57.Duensing A, Liu Y, Perdreau SA, Kleylein-Sohn J, Nigg EA, Duensing S. Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene. 2007;26 :6280–6288. doi: 10.1038/sj.onc.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korzeniewski N, Zheng L, Cuevas R, Parry J, Chatterjee P, Anderton B, Duensing A, Munger K, Duensing S. Cullin 1 functions as a centrosomal suppressor of centriole multiplication by regulating polo-like kinase 4 protein levels. Cancer Res. 2009;69 :6668–6675. doi: 10.1158/0008-5472.CAN-09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin LG, Demers GW, Galloway DA. Disruption of the G1/S transition in human papillomavirus type 16 E7-expressing human cells is associated with altered regulation of cyclin E. J Virol. 1998;72 :975–985. doi: 10.1128/jvi.72.2.975-985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duensing S, Munger K. Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J Virol. 2003;77 :12331–12335. doi: 10.1128/JVI.77.22.12331-12335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen CL, Eichwald C, Nibert ML, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the centrosomal component gamma-tubulin. J Virol. 2007;81 :13533–13543. doi: 10.1128/JVI.01669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10 :322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sluder G, Thompson EA, Miller FJ, Hayes J, Rieder CL. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J Cell Sci. 1997;110(Pt 4):421–429. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- 65.Thomas JT, Laimins LA. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J Virol. 1998;72 :1131–1137. doi: 10.1128/jvi.72.2.1131-1137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson DA, Belinsky G, Chang TH, Jones DL, Schlegel R, Munger K. The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene. 1997;15 :3025–3035. doi: 10.1038/sj.onc.1201495. [DOI] [PubMed] [Google Scholar]
- 67.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430 :797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen CL, McLaughlin-Drubin ME, Munger K. Delocalization of the microtubule motor Dynein from mitotic spindles by the human papillomavirus E7 oncoprotein is not sufficient or induction of multipolar mitoses. Cancer Res. 2008;68 :8715–8722. doi: 10.1158/0008-5472.CAN-08-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen CL, Munger K. Human papillomavirus E7 protein deregulates mitosis via an association with nuclear mitotic apparatus protein 1. J Virol. 2009;83 :1700–1707. doi: 10.1128/JVI.01971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18 :1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heilman SA, Nordberg Y, Liu JJ, Sluder G, Chen JJ. Abrogation of the postmitotic checkpoint contributes to polyploidization in human papillomavirus E7-expressing cells. J Virol. 2009;83 :2756–2764. doi: 10.1128/JVI.02149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Southern SA, Lewis MH, Herrington CS. Induction of tetrasomy by human papillomavirus type 16 E7 protein is independent of pRb binding and disruption of differentiation. Br J Cancer. 2004;90 :1949–1954. doi: 10.1038/sj.bjc.6601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci U S A. 1996;93 :479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hashida T, Yasumoto S. Induction of chromosome abnormalities in mouse and human epidermal keratinocytes by the human papillomavirus type 16 E7 oncogene. J Gen Virol. 1991;72(Pt 7):1569–1577. doi: 10.1099/0022-1317-72-7-1569. [DOI] [PubMed] [Google Scholar]
- 75.Wilting SM, Snijders PJ, Meijer GA, Ylstra B, van den Ijssel PR, Snijders AM, Albertson DG, Coffa J, Schouten JP, van de Wiel MA, Meijer CJ, Steenbergen RD. Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol. 2006;209 :220–230. doi: 10.1002/path.1966. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Han S, Baluda MA, Park NH. HPV-16 oncogenes E6 and E7 are mutagenic in normal human oral keratinocytes. Oncogene. 1997;14 :2347–2353. doi: 10.1038/sj.onc.1201078. [DOI] [PubMed] [Google Scholar]
- 77.Kessis TD, Connolly DC, Hedrick L, Cho KR. Expression of HPV16 E6 or E7 increases integration of foreign DNA. Oncogene. 1996;13 :427–431. [PubMed] [Google Scholar]
- 78.Tanaka S, Diffley JF. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev. 2002;16 :2639–2649. doi: 10.1101/gad.1011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spardy N, Duensing A, Charles D, Haines N, Nakahara T, Lambert PF, Duensing S. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J Virol. 2007;81 :13265–13270. doi: 10.1128/JVI.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7 :249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 81.Hoskins EE, Morris TA, Higginbotham JM, Spardy N, Cha E, Kelly P, Williams DA, Wikenheiser-Brokamp KA, Duensing S, Wells SI. Fanconi anemia deficiency stimulates HPV-associated hyperplastic growth in organotypic epithelial raft culture. Oncogene. 2009;28 :674–685. doi: 10.1038/onc.2008.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Narayan G, Arias-Pulido H, Nandula SV, Basso K, Sugirtharaj DD, Vargas H, Mansukhani M, Villella J, Meyer L, Schneider A, Gissmann L, Durst M, Pothuri B, Murty VV. Promoter hypermethylation of FANCF: disruption of Fancoi Anemia-BRCA pathway in cervical cancer. Cancer Res. 2004;64 :2994–2997. doi: 10.1158/0008-5472.can-04-0245. [DOI] [PubMed] [Google Scholar]
- 83.Wang SS, Bratti MC, Rodriguez AC, Herrero R, Burk RD, Porras C, Gonzalez P, Sherman ME, Wacholder S, Lan ZE, Schiffman M, Chanock SJ, Hildesheim A. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. J Infect Dis. 2009;199 :20–30. doi: 10.1086/595563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Fan S, Meng Q, Ma Y, Katiyar P, Schlegel R, Rosen EM. BRCA1 interaction with human papillomavirus oncoproteins. J Biol Chem. 2005;280 :33165–33177. doi: 10.1074/jbc.M505124200. [DOI] [PubMed] [Google Scholar]
- 85.Iftner T, Elbel M, Schopp B, Hiller T, Loizou JI, Caldecott KW, Stubenrauch F. Interference of papillomavirus E6 protein with single-strand break repair by interaction with XRCC1. EMBO J. 2002;21:4741–4748. doi: 10.1093/emboj/cdf443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15:119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 87.Stoppler H, Hartmann DP, Sherman L, Schlegel R. The human papillomavirus type 16 E6 and E7 oncoproteins dissociate cellular telomerase activity from the maintenance of telomere length. J Biol Chem. 1997;272:13332–13337. doi: 10.1074/jbc.272.20.13332. [DOI] [PubMed] [Google Scholar]
- 88.Spardy N, Duensing A, Hoskins EE, Wells SI, Duensing S. HPV-16 E7 reveals a link between DNA replication stress, fanconi anemia D2 protein, and alternative lengthening of telomere-associated promyelocytic leukemia bodies. Cancer Res. 2008;68:9954–9963. doi: 10.1158/0008-5472.CAN-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 90.Spardy N, Covella K, Cha E, Hoskins EE, Wells SI, Duensing A, Duensing S. Human papillomavirus 16 E7 oncoprotein attenuates DNA damage checkpoint control by increasing the proteolytic turnover of claspin. Cancer Res. 2009;69:7022–7029. doi: 10.1158/0008-5472.CAN-09-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chini CC, Chen J. Claspin, a regulator of Chk1 in DNA replication stress pathway. DNA Repair (Amst) 2004;3:1033–1037. doi: 10.1016/j.dnarep.2004.03.001. [DOI] [PubMed] [Google Scholar]