Abstract
We report the complete amino acid sequence of callinectin, a 32 amino acid, proline-, arginine-rich AMP with four cysteines and having the sequence WNSNRRFRVGRPPVVGRPGCVCFRAPCPCSNY-amide. The primary structure of callinectin is highly similar to arasins, AMPs recently identified in the small spider crab (Hyas araneus). Callinectin exists in three isomers that vary in the functional group on the tryptophan (W) residue. The most prevalent isomer had a hydroxy-N-formylkynurenine group, while the other two isomers had either N-formylkynurenine or hydroxy-tryptophan. Using a sequence highly similar to native callinectin, we chemically synthesized a peptide which we called callinectin-like peptide (CLP). Via immunoelectron microscopy, affinity-purified rabbit antibodies raised to CLP successfully localized the site of callinectin in blue crab hemocytes to the large electron-dense granules that are found primarily in large granule hemocytes.
Keywords: antimicrobial peptide, cystine, proline, arginine, hydroxylated tryptophan
1. INTRODUCTION
The blue crab (Callinectes sapidus) is a keystone species in many estuaries of the western Atlantic Ocean and the Gulf of Mexico (Anonymous, 1999; Hines, 2007). It is a member of the family Portunidae (swimming crabs), which also includes other ecologically and commercially important species, such as the mud crabs (Scylla spp.), of the Indo-Pacific region. Wild blue crabs have frequently been affected by infectious disease epidemics, especially bacteria and parasites (Noga et al., 1998; Lee and Frischer, 2004; Shields and Overstreet, 2007). Cultured blue crabs, such as those held in shedding operations for the production of “soft-shelled” crabs, have also been plagued by disease (Noga et al., 1998).
While disease causes serious losses, we know little about the mechanisms blue crabs use to protect themselves from infection. Antimicrobial peptides (AMPs) are typically potent, broad-spectrum chemical defenses. In recent years, a number of AMPs have been identified in various crab species, including a proline-rich 6.5 kDa peptide from the shore crab (Carcinus maenas) (Schnapp et al., 1996), crustins from green crab (Carcinus maenas) (Relf et al., 1999) and mud crab (Scylla paramamosain) (Imjongjirak et al., 2009), antilipopolysaccharide factors from mud crab (Imjongjirak et al., 2007), mangrove mud crab (S. serrata ) (Yedery and Reddy, 2009), Chinese mitten crab (Eriocheir sinensis) (Li et al., 2008) and swimming crab (Portunus trituberculatus) (Yue et al., 2010), as well as arasin (proline-arginine-rich peptide) and hyastatin (glycine-rich multidomain peptide) from the small spider crab (Hyas araneus) (Stensvåg et al., 2008, Sperstad et al., 2009).
Blue crabs also synthesize an AMP, callinectin, in the hemocyte fraction of hemolymph (Khoo et al., 1999), which was one of the first AMPs identified from any crab. However, only a partial sequence (about 2/3 of this peptide) was determined. The purpose of our present study was to elucidate the entire primary structure of callinectin. In addition, we wished to use these data to develop an antibody probe that would identify the cellular location of callinectin in blue crab immunocytes.
2. MATERIALS AND METHODS
2.1 Sequence Determination of Callinectin
Native callinectin was purified as described previously (Khoo et al., 1999). The intact mass of callinectin was initially performed using Matrix Assisted Laser Desorption Mass spectrometry (MALDI-MS) on a Micromass TofSpec-SE in linear mode. Four μl (~ 4 pmoles) of HPLC-purified, non-alkylated callinectin (see below) was diluted into 6 μl of 70% formic acid. One μl was removed and mixed with one μl of alpha-cyano-4-hydroxy cinnamic acid matrix in 0.1% trifluoroacetic acid (TFA), 50% acetonitrile, 50% water, with one μl of internal calibrants. The internal calibrants used (to maintain better than 100 ppm mass accuracy) were 25 fmol bradykinin (protonated monoisotopic mass of 1060.57 Da) and ACTH clip (protonated monoisotopic mass of 2465.2 Da). The average protonated mass of callinectin was determined to be 3647 Da with 3 masses at +16 Da from this parent mass (indicating sites of oxidation) with the most intense mass at 3695.2 Da.
Approximately 400 pmoles (two μl) of purified callinectin, dissolved in 10 μl of 8 M urea/0.4 M ammonium bicarbonate, was reduced by adding five μl of 45 mM dithiothreitol and incubating at 37°C for 20 min. The sample was then alkylated by adding five μl of 100 mM iodoacetamide and incubating at room temperature in the dark for 20 min. HPLC purification of the alkylated callinectin, and a non-alkylated aliquot (25 pmols) of callinectin, were done on a Hewlett Packard 1090 system equipped with a 1 mm × 25 cm Vydac C18 (5 micron particle size, 300 Å pore size) reverse phase column equilibrated with 98% Buffer A (0.06% TFA) and 2% Buffer B (0.052% TFA, 80% acetonitrile). Callinectin was eluted at 50 μl/min with the following gradient program: 0-60 min (2-37% Buffer B), 60-90 min (37-75% Buffer B) and 90-105 min (75-98% Buffer B), with detection at 210 nm. Edman chemical degradation was performed on 75% of this alkylated callinectin using an Applied Biosystems Procise cLC Peptide/Protein sequencer. Thirty two cycles were sequenced, including 4 carboxamidomethylated cysteines.
Protease treatment was used to determine the C-terminal amino acid, and to identify residue 8. Approximately 125 pmols of callinectin was speedvacuumed dry, dissolved in 25 μl of 10% acetonitrile/10 mM ammonium bicarbonate, and reduced using 5.5 μl of 4.5 mM dithiothreitol. After incubating at 37°C for 20 min, the sample was alkylated by adding 5.5 μl of 10 mM iodoacetamide, with incubation in the dark at room temperature for 20 min. After speedvac drying, the sample was dissolved in 35 μl of 1.0 μg/ml trypsin (Promega) in 10% acetonitrile/10 mM ammonium bicarbonate, and incubated at 37°C for 18 hr. MALDI-MS was performed on one μl of the digest after mixing with matrix as described above.
Additional ESI mass spectrometry was performed on a one pmol aliquot of the tryptic digest. The sample was first desalted on a C18 ZipTip® (Millipore); the tip (P10 size) was washed five times with 10 μl of 50% acetonitrile/0.1% TFA, equilibrated five times with 10 μl of 0.1% TFA and then the sample was bound to the tip. Prior to elution of the digest, the tip was again washed five times with 10 μl of 0.1% TFA (to remove salts) and the sample was eluted in three μl of 0.1% formic acid/50% acetonitrile/50% water. The eluted sample was subjected to nanospray MS on a Micromass Q-Tof Ultima mass spectrometer. Micromass type B nanovials (#M956230AD1) were used to spray the sample into the mass spectrometer with a voltage of 1.4 kV applied. Masses were chosen manually for MS/MS fragmentation based on the predicted tryptic masses from the Edman sequencing results and the MALDI-MS spectra.
A final intact mass of non-alkylated callinectin was acquired using LCMS on a LTQ Orbitrap mass spectrometer. This instrument was equipped with a Waters nanoACQUITY UPLC system, and used a Waters Symmetry® C18 180 μm × 20 mm trap column and a 1.7 μm, 75 μm × 250 mm nanoAcquity™ UPLC™ column at 35°C for peptide separation. Trapping was done at 15 μl/min, 99% Buffer A (100% water, 0.1% formic acid) for one min. Separation was performed at 300 nl/min with Buffer A (0.1% formic acid in water) and Buffer B (0.075% formic acid in acetonitrile). A linear gradient (51 min) was run with 5% Buffer B at initial conditions, 50% Buffer B at 50 min, and 85% Buffer B at 51 min. MS was acquired in the Orbitrap using one microscan, and a maximum inject time of 900 milliseconds.
2.2 Bioinformatics
Homology searches were performed against the SwissProt, NCBInr, PAT and Month databases with the Basic Local Alignment Search Tool (BLAST) provided by the NCBI server (http://www.ncbi.nlm.nih.gov/BLAST). Homology searches of full and partial sequences were also performed against the Antimicrobial Peptide Database (http://aps.unmc.edu/AP/main.html) (Wang and Wang, 2004) and against the ANTIMIC (http://research.i2r.a-star.edu.sg/Templar/DB/ANTIMIC/) and CAMP (http://www.bicnirrh.res.in/antimicrobial) databases of AMP sequences (Brahmachary et al., 2004; Thomas et al., 2010). Isoelectric point was predicted using the Skoog and Wichmann (1986) calculation (in GPMAW Version 8.2, Lighthouse Data).
2.3 Synthesis of Callinectin-Like Peptide (CLP)
While studies were ongoing to identify the complete sequence of native callinectin, we synthesized a peptide that was based upon the sequence data we had available at the time. This callinectin-like peptide (CLP) was eventually found to vary from native callinectin in having an unmodified tryptophan residue at position 1, a glycine at position 8, and a free C-terminus. CLP was synthesized via Fmoc chemistry on a Rainin Symphony instrument that provides on instrument cleavage of the peptide from the resin. After synthesis, CLP was purified via analytical reverse phase HPLC using a YMC C18 column (4 mm × 50 mm, 3 micron particle size, 120 angstrom pore size support) and an acetonitrile gradient that was eluted at one ml/min where Buffer A was 0.05% TFA in water and Buffer B was 80% acetonitrile/0.05% TFA. CLP was detected by its absorbance at 210 nm. Mass spectrometry of an aliquot of purified CLP was carried out on a Micromass TofSpec SE mass spectrometer that was operated in positive ion mode and equipped with a nitrogen laser (337 nm), a reflectron, delayed extraction and a post acceleration detector. The purified peptide was lyophilized from 0.05% TFA/acetonitrile solution and stored dessicated under argon gas until reconstitution in solvent.
2.4 Anti-CLP Antibody
Two milligrams of CLP was conjugated to keyhole limpet hemocyanin (KLH) using carbodiimide chemistry, which linked the peptide to KLH via both the N-terminal and the C-terminal. The conjugation via the terminal amino acid allows tertiary conformation of the peptide that may be expected to mimic that in the native peptide, thus eliciting anti-conformational antibodies important for recognizing the native peptide. Immunogen was mixed with Complete Freund’s Adjuvant (1:1) and KLH-conjugated peptide was injected into two New Zealand white rabbits at 5 subcutaneous sites (0.2 ml per site, 100 μg/injection) on days 0, 14, 28 and 42. Thirty ml of antiserum was collected from each rabbit on days 35 and 45. The antiserum was pooled and was then affinity-purified using CLP conjugated to cyanogen bromide-activated agarose as an immunosorbent (10.5 mg of CLP was reacted with 15 g of agarose). One hundred and twenty ml of antiserum (two 30 ml bleeds from two rabbits) was loaded onto the column (Uniflow 4, Sterogene, Carlsbad, CA). After washing, the affinity-purified antibody was eluted and concentrated.
This method produces greater than 0.1 mg of peptide-specific antibody per mL of antiserum, as determined by recovered affinity-purified antibody. Antibody was greater than 95% IgG, as determined by immunoelectrophoresis using antibodies specific for rabbit IgG, IgM and serum proteins. The titer of the antibody was determined via ELISA, using CLP as the antigen coated onto a microtiter plate. Pure CLP (10 μg/mL in PBS, pH 7.2-7.5) was coated onto a microtiter plate at room temperature for 1 h. The plate was then washed and post-coated with 1% BSA in PBS for 30 min. The plate was washed and then dilutions of antibody in 1% BSA/PBS/0.01% Tween 20 were added, beginning at 1 μg antibody/mL. After incubation for 1 h, the plate was washed, followed by addition of peroxidase-conjugated goat anti-rabbit IgG (h&l) in 1% BSA/PBS/0.01% Tween 20. After incubation for 1 h, the plate was washed and peroxidase substrate was added, incubated for 15 min, and then stopped with 1 N HCl (1:1). The absorbance was then read at 450 nm. The titer was read as the reciprocal of the antibody dilution (dilution of a 1 mg/mL solution) that produced a net optical density of 1.0, compared to a blank (diluent-only well), which had an OD < 0.1. The titer of the antibody used in all assays was approximately 1:159,000. The peptide-specific antibody would have less than 1% cross-reactivity by ELISA, where 1% cross-reactivity is 100 times more antibody than would be required to produce the same optical density with either free KLH or free peptide that shared less than 3 amino acids in the sequence. Western blotting also showed that this antibody strongly reacted with native callinectin (data not shown).
2.5 Immuno-electron microscopy
A 0.5 ml sample of blue crab hemolymph was aspirated into 2.5 ml of 10% NBF. The cells were centrifuged at 2390 × g and then washed twice with 0.1 M Sorensen’s phosphate buffer (pH 7.2-7.4), and vortexed to re-suspend the cells between washes. The cells were then embedded in molten 3% agar (Fisher Scientific, Fair Lawn, NJ), cooled until the agar hardened, and cut into cubes less than 1 mm thick. The agar-embedded cells were dehydrated in a graded ethanol series culminating in 100% ethanol. They were then placed into 100% LR White resin (Electron Microscopy Sciences, Fort Washington, PA) overnight at room temperature. The next day, they were incubated in three changes of fresh LR White resin (1 hr, 1 hr, and 3 hr). The samples were then placed in capped gelatin capsules with fresh LR White resin and polymerized overnight at 55°C. Semi-thin sections (0.5 μm) were cut with a glass knife and stained with toluidine blue O for light microscopic evaluation. The blocks were then trimmed to include as many cells as possible before ultrathin (90 nm) sections were cut with a diamond knife. Sections were placed on carbon-formvar-coated stainless steel grids.
Immunolabeling of grids was accomplished by floating the grids face down on single droplets of the various reagents on Parafilm pushed into a well of a 24-well tissue culture plate. All incubations were done at room temperature while gently rocking the 24-well plate on an Adams nutator (Clay Adams, Parsnippany, NJ) for the duration of the incubation period. Grids were first washed for 10 min in washing buffer (PBS with 1% BSA and 3% normal goat serum), followed by incubation for 1 hr in primary antibody (anti-CLP, 1000 μg/ml, titer 1:159,000 diluted with washing buffer to produce 1:50, 1:100, and 1:200 dilutions). A negative control was incubated in washing buffer alone. Grids were then washed four times (5 min per wash) in washing buffer, followed by incubation for 30 min in secondary antibody (gold-anti-rabbit Ig, [#G7402, Sigma Chemical, St. Louis, MO] diluted in washing buffer at a concentration of 1:40). Grids were then washed five times (4 min per wash) in PBS, followed by a 5 min wash in distilled water. These grids were viewed and photographed with a Philips EM-208S transmission electron microscope (FEI Company, Hillsboro, Oregon).
For routine transmission electron microscopy (TEM), another 0.5 ml sample of blue crab hemolymph was aspirated into 2.5 ml of 4F:1G fixative (McDowell and Trump, 1976). This sample was agar embedded using the same procedure above. The samples were then place into 1% osmium tetroxide for one hour. The osmicated cells were dehydrated in an ascending series of ethanol, ending with 100% ethanol. After the ethanol series, the sample was rinsed 2 times with 100% acetone for 10 minutes each. It was then infiltrated with 1:1 acetone/Spurr’s epoxy resin (Spurr, 1969) for 30 min, followed by two 1 hr changes of 100% resin. The samples were then placed into flat embedding molds filled with fresh resin. Semithin sections (0.5 μm) were prepared with a glass knife and stained with toluidine blue O. The blocks were then trimmed to include as many cells as possible as determined by light microscopic evaluation of the semithin sections. Ultrathin sections (90 nm) were cut with a diamond knife, transferred to 200 mesh copper grids, and post-stained with uranyl acetate and Reynold’s lead citrate. Grids were viewed and photographed with a Philips EM-208S transmission electron microscope.
3. RESULTS
3.1 Primary Structure of Callinectin
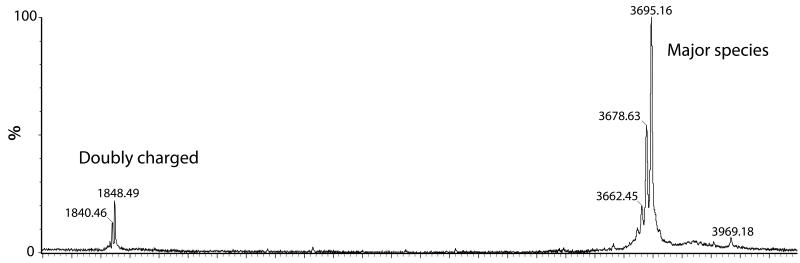
The partial sequence for callinectin that we had previously identified (Khoo et al., 1999) was for a 26-residue peptide. However, the mass of the protein identified via MALDI-MS in our current callinectin study was determined to be MH 3647.0 Da (average mass) with 3 additional masses at +16 Da from this parent mass (suggesting sites of oxidation) with the most intense mass at MH 3695.16 (Fig. 1). This indicated that either the peptide had a large posttranslational modification or that the protein was longer than 26 residues. In order to prepare for an Edman chemical degradation analysis, callinectin was alkylated to modify cysteines/cystines to carboxamidomethylated cysteine (CAM cys). The latter is a cysteine modification that is readily identified during Edman sequencing (Hunkapiller, 1988). Both the alkylated and non-alkylated proteins were HPLC-purified. Edman sequencing of the alklyated callinectin provided 30 amino acids of sequence, with a missing residue at position 8, four carboxamidomethylated cysteines (residues 20, 22, 27 and 29) and an unknown C-terminus termination.
Figure 1.
MALDI-MS of callinectin. The intact mass of callinectin was initially determined using MALDI-MS (Micromass TofSPec SE) in linear mode. The major species was observed at a singly charged M+H of 3695.16 Da with a series of masses at minus 16 Da, 2 × 16 Da and 3 × 16 Da. This suggested that the major state of callinectin is a trioxidized form. The doubly charged M+2 ion was also observed at 1848.49 Da, with a doubly charged series of masses at minus 8 Da.
MALDI-MS peptide mass fingerprinting as well as nanospray MS/MS and LC-MS/MS analysis (see Supplementary Tables 1 and 2; Supplementary Figures 1-7) of a tryptic digestion of callinectin confirmed the Edman sequence and identified the C-terminal peptide. Nanospray MS/MS spectra of the C-terminal peptide revealed the C-terminus of the protein to be the tyrosine at residue 32, with C-terminal amidation (Supplementary Fig. 8).
In order to determine residue 8, another intact mass was obtained on a LTQ Orbitrap XLwith 60,000 resolution. The LC-MS spectra showed a major observed protonated mass (739.55M+5) of 3693.75 Da (Supplementary Fig. 9). Again, masses corresponding to the major mass minus a series of 16 Da were observed, indicating that callinectin has multiple oxidation states. The amino terminal tryptophan (which is the only possible amino acid that could oxidize in callinectin) must therefore oxidize to hydroxy-N-formylkynurenine (+48 amu), N-formylkynurenine (+32 amu), and hydroxy-tryptophan (+16 amu), with the hydroxyl-N-formylkyurenine modification as the major mass. Assuming two disulfide bonds, the predicted monoisotopic mass without residue 8 is 3537.36 Da which is 156.39 Da different from the Orbitrap protonated mass of 3693.75 Da. The only residue mass that fits this mass difference is arginine (156 amu). Hence, based on this mass difference, residue 8 was assigned to be arginine. Unfortunately, the predicted tryptic peptide FRVGR was not observed in either the MALDI-MS digest analysis or the MS/MS analysis (predicted protonated mass 634.37 Da).
Figure 2 shows the Edman sequence that was determined, along with the mass spectrometrically-identified peptides (partially proteolysed peptides that were identified are not shown). A Protein BLAST search of the complete callinectin sequence against the NCBInr database showed homology to arasins, AMPs recently isolated from the small spider crab (Hyas araneus) (Stensvåg et al., 2008). Callinectin is highly basic, with a calculated theoretical PI of 12.8 with disulfide bonds, with a net positive charge of +6.
Figure 2.
Primary structure of callinectin.
a). Amino acid sequence of callinectin. The amino acid sequence of callinectin was determined using multiple approaches of Edman chemical degradation and tryptic digestion followed by LC-MS/MS analysis (Micromass Q-Tof Ultima and Thermo Scientific LTQ Orbitrap XL mass spectrometers) and an accurate intact mass (LTQ Orbitrap). Each residue was identified using Edman sequencing, except for three residues: the proline at position 28 was verified by MS/MS of a tryptic digest of callinectin; the arginine at position 6 was confirmed with a tryptic peptide; the arginine at position 8 was determined based on the accurate intact mass with tryptophan modification. The observed intact M+H mass with the tryptophan modification (+48 Da, called hydroxy-N-formylkynurenine) is 3693.75 Da. The calculated intact M+H mass with hydroxy-N-formylkynurenine and 2 disulfide bonds is 3693.73 Da. The last portion of the sequence was determined by LC-MS/MS of the C-terminal tryptic peptide, which showed that the C-terminus is amidated.
b) Comparison of the amino acid sequences of callinectin, arasin 1 and arasin 2. Residues are numbered in accordance with the callinectin sequence. Gaps are situated to optimize similarities. The cysteines are in bold. Shaded amino acids are identical to arasin 1 and/or arasin 2. The O (°) indicates oxidization, as either hydroxy-N-formylkynurenine, N-formylkynurenine or hydroxy-tryptophan. The asterisk (*) indicates C-terminal amidation.
3.2 Intracellular Localization of Callinectin
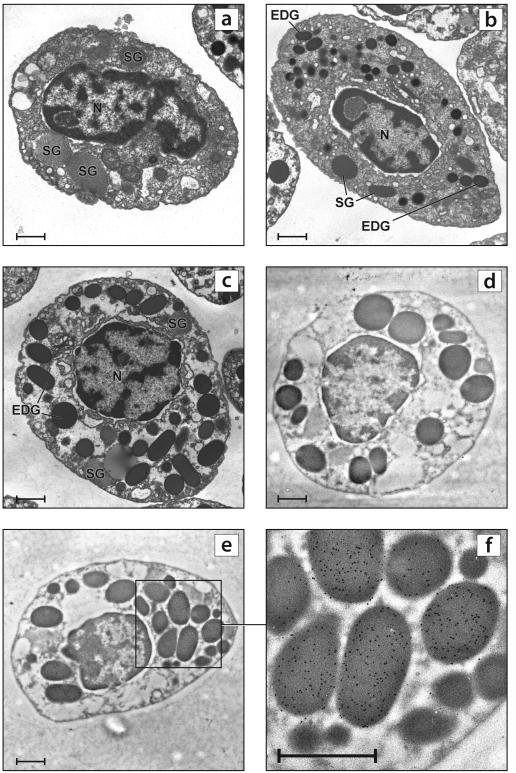
Our definition of hemocyte types was primarily based upon the morphological descriptions of (Clare and Lumb, 1994), who followed the classification scheme of Hose et al. (1990). At the ultrastructural level, hemocytes were typically spherical or elliptical, and granules, when present, were either striated, reticulated, or electron dense. There were three hemocyte types (cell sizes were measured using the longest or widest axis of each cell): hyaline cells, small granule hemocytes, and large granule hemocytes. Hyaline cells ranged from 4.5-8.0 μm; they had a central nucleus and a high nucleocytoplasmic ratio. They typically had between 0-10 reticulated and/or striated granules per cell. These cells appeared agranular via light microscopy, so it would seem that the granules that are apparent correspond to the electron dense granules found in the small and large granule hemocytes. Small granule hemocytes (semi-granulocytes) ranged from 6-11 μm. They had a central nucleus and a low nucleocytoplasmic ratio. They had mostly small (0.5 μm or slightly smaller), electron dense granules. In one particular cell, larger (0.75 μm) electron dense granules were found. The other granule types were rare but present. Large granule hemocytes (granulocytes) also ranged from 6-11 μm in size. They had an eccentric nucleus and a low nucleocytoplasmic ratio. They had primarily large electron dense granules (approximately 1.0 μm) that filled most of the cytoplasm, but an occasional reticulated or striated granule was present (Fig. 3).
Figure 3.
Transmission electron micrographs of blue crab hemocytes. SG = striated granules, EDG = electron-dense granules, N = nucleus. Bar = 1 μm.
a) Hyaline cell
b) Small granule hemocyte
c) Large granule hemocyte
d) Large granule hemocyte reacted only with gold-labeled goat-anti-rabbit antibody.
e) Large granule hemocyte reacted with rabbit anti-CLP antibody and goat anti-rabbit gold-labeled antibody. Note gold particles in the EDG.
f) Closer view of Fig. 3e, showing immunopositive granules
Via electron microscopy, hemocytes prominently reacted with anti-CLP antibody. Immunoreactivity was consistently present in the large, electron-dense granules of the large granule hemocytes (all 10 large granule hemocytes were positive). Immunostaining of the granules was variable, with granules having few to many gold particles. None of the 7 hyaline or 6 small granule hemocytes was positive for callinectin. One grid was not treated with primary antibody and all cells in the section were negative (Fig. 3).
4. DISCUSSION
4.1 Primary Structure of Callinectin
For callinectin, high resolution LC-MS analysis (LTQ Orbitrap) revealed the mass of the trioxidized tryptophan form (oxidation was based on the series of minus 16 Da masses called hydroxy-N-formylkynurenine) to be 3693.75 Da and based on this accurate mass, residue 8 was concluded to be arginine. Edman chemical degradation combined with MALDI-MS, nanospray MS/MS and intact mass (high resolution) spectra determined the sequence of callinectin to be WNSNRRFRVGRPPVVGRPGCVCFRAPCPCSNY, where the C-terminus is amidated. Potential disulfide bridges were not explored but if the three-dimensional structure of callinectin is similar to other cysteine-rich antimicrobial peptides, the four cysteines probably form two disulfide bridges (which we have assumed), giving the peptide a great degree of structural rigidity. Callinectin’s oxidatively decarboxylated aromatic C-terminal residue may also confer proteolytic resistance, as in halocyamines and plicatamide, which are AMPs from tunicates (Galinier et al., 2009). This might also at least partly explain the highly stable nature of this peptide and our ability to reproducibly and accurately measure antibacterial activity in blue crab serum, even after very long periods of storage (years) (Noga et al., 1994). This C-terminal amide also increases its net cationic charge and consequently the electrostatic attraction to targets like negatively charged bacterial membranes (Shai, 2002).
Callinectin exists as three isomers that vary in the functional group on the tryptophan residue. The most prevalent isomer had a hydroxy-N-formylkynurenine group, with the other two modifications (N-formylkynurenine and hydroxytryptophan) being less abundant. Tryptophan modifications have been documented in AMPs of a number of aquatic animals. The most common modification is bromination (bromotryptophan). This modification occurs in styelin D from the tunicate Styela clava (Lehrer et al., 2001), as well as hedistin, a novel antimicrobial peptide in NK cells of the marine polychaete worm, Nereis diversicolor (Tasiemski et al., 2007). Other AMPs with bromotryptophan include strongylocin 2 and centrocins from the green sea urchin (Strongylocentrotus droebachiensis) (Li et al., 2010), and hagfish cathelicidin isolated from the intestine of Atlantic hagfish (Myxine glutinosa) (Shinnar et al., 2003). The presence of bromotryptophan is associated with increased resistance to proteolysis (Shinnar et al., 2003). Hydroxylated tryptophan occurs in MGD-2, an arthropod defensin from the Mediterranean mussel (Mytilus galloprovincialis) (Yang et al., 2000) and in piscidin 4, a member of the piscidin family from hybrid striped bass (Morone saxatilis × M. chrysops) (Noga et al., 2009; Salger et al., 2010).
In some cases, the modified tryptophan is needed for full expression of antimicrobial activity. Synthesis of MGD-2 without hydroxylation of its tryptophan residue results in much lower activity against certain Gram-negative bacteria (Yang et al., 2000). However, in other cases, potent antimicrobial activity is still expressed after deletion of the modified amino acid (Shinnar et al., 2003) and does not appear to affect this function (Tasiemski et al., 2007; Noga et al., 2009). The functional significance of the tryptophan modifications in callinectin are unknown, but it is possible that they might result in diminished activity. Tryptophan oxidation is often an indicator of oxidative stress and frequently accompanies loss of function of a protein (Fedorova et al., 2010). Cobratoxin, a 6.2 kDa peptide that binds to plasma membrane receptors, is inactivated when its single tryptophan residue is oxidized to either N’-formylkynurenine or bromotryptophan (Chang and Hayashi, 1969). Oxidation of the tryptophan in lysozyme abrogates its enzymatic activity (Kuroda et al., 1975). It has been suggested that the most susceptible tryptophan residues in certain proteins are “hot spots” for oxidation in close proximity to a source of reactive oxygen species (Taylor et al., 2003). Thus, it is interesting that the antioxidant enzyme, superoxide dismutase, has been localized to the plasma membrane of some crustacean granulocytes (Johansson et al., 2000), which is the cell type in blue crabs that stores callinectin (see below).
Callinectin has significant amino acid sequence similarity to arasins, AMPs recently isolated from the hemocytes of the small spider crab (Hyas araneus). However, arasin 1, the only arasin yet purified, has a free C-terminus (Stensvåg et al., 2008) and no modifications on its tryptophan residue. Callinectin and arasins both have a chimeric structure, with a proline-, arginine- rich N-terminus (comprising about 16% and 19%, respectively, of callinectin’s amino acids) and a similar cysteine motif in the C-terminus (Fig. 2). This same cysteine array is also expressed in other AMPs including mammalian protegrins as well as tachyplesins, polyphemeusins, androctonin and gomesin from arthropods (Stensvåg et al., 2008). It is likely that if disulfide bonds are present in callinectin, they are most probably C22-C27 and C20-C29, which is the type of motif found in the aforementioned AMPs.
4.2 Immunolocalization of Callinectin
Our identification of the three major hemocyte types in blue crabs agreed well with those previously published by Clare and Lumb (1994), as well as the generally recognized classification scheme for crustaceans (Matozzo and Marin, 2010). The only cell structures that we did not observe were stellate forms of electron-dense granules. The appearance of these granules in Clare and Lumb (1994) is somewhat similar to the “activated” granules of Pacific white shrimp after exposure to pathogenic bacteria (Muñoz et al., 2002). Thus, our failure to observe this granule type in our cell preparation might have been because our cell sample was more quickly preserved. Cell architecture was also well-preserved with use of the 4F:1G, a relatively low osmolality fixative.
Hemocytes that reacted with anti-CLP antibodies were exclusively large granulocytes. The cell we have termed an intermediate cell might be between developmental stages of the small and large granule hemocytes because the granules are in between the sizes of the granules for the respective cells. Variability in the concentrations of antibodies and techniques used could account for the variable labeling results between studies.
While a number of AMPs have been identified from solid tissues of a number of animals (e.g., epithelial defensins), all AMP that have been identified in crustaceans have been isolated from and/or localized to hemocytes, including penaedins (Muñoz et al., 2002), the 6.5 kDa shore crab peptide (Schnapp et al., 1996) and armadillidin (Herbiniére et al., 2005). The 11.5 kDa shore crab protein (crustin) as well as black tiger shrimp ALF are also present in hemocytes (Relf et al., 1999, Somboonwiwat et al., 2005).
Our findings are somewhat similar to those of Muñoz et al (2002), who immunochemically identified penaeidin in the granules of both small and large granule hemocytes, while hyaline cells were negative. While some of the small granules in small granule hemocytes were also significantly positive, they were less so than the large granule hemocytes (Destoumieux et al., 2000). Penaeidins are also synthesized in hemocytes (Destoumieux et al., 2000). Horseshoe crabs (Tachypleus), a chelicerate arthropod, have a single predominant hemocyte type, the amoebocyte, which has both large and small granules (Iwanaga et al., 1998).
Localization of callinectin in granulocytes suggests that they would not be directly secreted into the plasma after synthesis, but rather would be stored as a mature peptide in the granules. How callinectin is released from the granulocytes is unknown. In horseshoe crabs (chelicerates), microbial products such as β-glucan or lipopolysaccharide stimulate granulocytes to degranulate, releasing the AMP tachyplesin and other antimicrobial factors into the plasma (Iwanaga et al., 1998). However, penaedins appear to be released from the granules directly into the hemocyte cytoplam after stimulation by bacteria. This then apparently is followed by cell lysis and the release of the AMPs into the extracellular space (Muñoz et al., 2002).
Supplementary Material
Supplementary Table 1. Predicted b and y ions for the amidated C-terminal tryptic peptide spanning residues 25-32 of callinectin. The observed intact protonated monoisotopic peptide mass (M+H) is 967.30 with a predicted M+H of 967.37. This mass difference of 0.07 Da, along with the observed b and y ion series sequence fragments, indicate that the C-terminus of callinectin is amidated.
Supplementary Table 2. Edman sequencing pmol yields of amino acids. Amino acids identified by Edman sequencing are in bold in the right hand column. % CO is the % carryover as calculated by subtracting the pmol background prior to the amino acid increase and then dividing the following residue pmol yield over the increased pmol yield.
Supplementary Figure 1. MALDI-MS chromatogram of callinectin tryptic digestion. The callinectin tryptic digestion was analyzed using MALDI-MS (Micromass TofSpec SE) in both linear and reflectron modes, with the linear spectra shown. Using peptide mass fingerprinting and comparing the observed masses to the edman sequence of callinectin, the indicated residues were identified. Mass accuracy with internal calibration is better than 70 ppm. Internal calibrants/ standards (INT STD) were 100 fmol of bradykinin (M+H mono-isotopic mass: 1060.57), and 250 fmol of adrenocorticotropic hormone human fragment 18-39 (ACTH Clip) (M+H monoisotopic mass: 2465.2).
Supplementary Figure 2. De-convoluted MS spectra of the tryptic digestion of callinectin. MS and MS/MS analysis was performed on a Micromass Q-Tof Ultima using nanospray. MAXENT 3 (Micromass Masslynx) deconvoluted the MS spectra into singly charged m/z. Callinectin peptide residues identified by MS/MS are indicated.
Supplementary Figure 3. Nanospray MS/MS of the singly charged peptide 798.3 MH; peptide spans residues 19-24 of callinectin.
Supplementary Figure 4. Nanospray MS/MS of singly charged mass 895.3 MH; peptide spans residues 18-24 of callinectin. Internal fragment sequences were observed as marked.
Supplementary Figure 5 Nanospray MS/MS of doubly charged mass 468.7MH; peptide spans residues 9-17 of callinectin. Internal fragment sequences were observed as marked.
Supplementary Figure 6. Nanospray MS/MS of singly charged mass MH 741.3; peptide spans residues 20-24 of callinectin.
Supplementary Figure 7. LTQ Orbitrap MS/MS of doubly charged mass MH 424.71. The peptide spans residues 1-6 of callinectin and verifies residue 6 as arginine.
Supplementary Figure 8. MS/MS spectra of the C-terminal tryptic peptide of callinectin. The singly charged peptide (967.45 Da) was subjected to nanospray MS/MS analysis and was found to span residues 25-32 of callinectin, with amidation of the C-terminus. Internal fragment sequence ions were observed as marked (e.g., PCSN = Pro-CAM-Ser-Asn). Predicted b and y series fragment ions are shown in Supplementary Table 1.
Supplementary Figure 9. Accurate intact mass of callinectin. An accurate intact mass (M+H) was determined using LCMS on a Waters nanoACQUITY UPLC system with detection in a LTQ Orbitrap mass spectrometer.
a) The total ion current intensity (y axis) of the LCMS related to the elution time (x axis). The intact callinectin eluted at ~43 min while degradation products (or proteolytic fragments) of callinectin eluted from 26 to 33 min as shown.
b) Combined mass scans across the intact callinectin peak. The charge state (z) is as indicated for each mass observed. Singly charged (z=1) masses were not of interest due to their low mass.
c) The expanded +5 (739.55 Da) mass which deconvolutes to a singly charged mass of M+H 3693.75 Da. The +6 (616.46 Da) charged mass corresponds to a singly charged mass of 3693.76 Da and is in good agreement with the +5 deconvoluted mass. Mass accuracy at this mass in this mass spectrometer is routinely 0-3 ppm.
Acknowledgements
This research was supported by Fishery Research Grant Project #99-AM-01, Blue Crab Research Project #03-BIOL-01, and Blue Crab Research Project #06-Biol-01, administered by the North Carolina Sea Grant College Program (to EJN), U.S.D.A. Agriculture and Food Research Initiative Grant #2009-03571 (to EJN and JD Oliver), and Yale School of Medicine’s Clinical and Translational Science (CTSA) Award UL1 R024139, National Center for Research Resources, National Institutes of Health. We thank S McKenna for providing blue crabs, and J Stephenson (NCSU), M Mattmuller (NCSU), J Johansen (Kendrick Laboratories) and L Khoo (Mississippi State University) for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anonymous . The Ecological Condition of Estuaries in the Gulf of Mexico. U.S.E.P.A. Office of Research and Development; 1999. EPA 620-R-98-004. [Google Scholar]
- Brahmachary M, Krishnan S, Koh J, Khan A, Seah S, Tan T, Brusic V, Bajic VB. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004;32:586–589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Hayashi K. Chemical modification of the tryptophan residue in cobratoxin. Biochem. Biophys. Res. Commun. 1969;37:841–846. doi: 10.1016/0006-291x(69)90968-1. [DOI] [PubMed] [Google Scholar]
- Clare AS, Lumb G. Identification of haemocytes and their role in clotting in the blue crab, Callinectes sapidus. Mar. Biol. 1994;118:601–610. [Google Scholar]
- Destoumieux D, Munoz M, Cosseau C, Rodriguez J, Bulet P, Comps M, Bachere E. Penaedins, antimicrobial peptides with chitin-binding activity, are produced and stored in shrimp granulocytes and released after microbial challenge. J. Cell Sci. 2000;113:461–469. doi: 10.1242/jcs.113.3.461. [DOI] [PubMed] [Google Scholar]
- Fedorova M, Todorovsky T, Kuleva N, Hoffmann R. Quantitative evaluation of tryptophan oxidation in actin and troponin I from skeletal muscles using a rat model of acute oxidative stress. Proteomics. 2010;10:2692–2700. doi: 10.1002/pmic.201000147. [DOI] [PubMed] [Google Scholar]
- Galinier R, Roger E, Sautiere P-E, Aumelas A, Banaigs B, Mitta G. Halocyntin and papillosin, two new antimicrobial peptides isolated from hemocytes of the solitary tunicate, Halocynthia papillosa. J. Pept. Sci. 2009;15:48–55. doi: 10.1002/psc.1101. [DOI] [PubMed] [Google Scholar]
- Herbiniére J, Braqart-Varnier C, Gréve P, Strub J-M, Frére J, Van Dorsselaer A, Martin G. Armadillidin: a novel glycine-rich antibacterial peptide directed against gram-positive bacteria in the woodlouse Armadillium vulgare (Terrestrial Isopod, Crustacean) Dev. & Comp. Immunol. 2005;29:489–499. doi: 10.1016/j.dci.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Hines AH. Chapter 14: Ecology of juvenile and adult blue crabs. In: Kennedy VS, Cronin LE, editors. The Blue Crab: Callinectes sapidus. University of Maryland Sea Grant; College Park, MD: 2007. pp. 565–664. [Google Scholar]
- Hose JE, Martin GG, Gerard AS. A decapod classification scheme integrating morphology, cytochemistry, and function. Biol. Bull. 1990;178:33–45. doi: 10.2307/1541535. [DOI] [PubMed] [Google Scholar]
- Hunkapiller MW. Gas phase sequence analysis of proteins/peptides. In: Bhown AS, editor. Protein/Peptide Sequence Analysis: Current Methodologies. CRC Press, Inc.; Boca Raton FL: 1988. pp. 87–117. [Google Scholar]
- Imjongjirak C, Amparyup P, Tassanakajon A, Sittipraneed S. Antilipopolysaccharide factor (ALF) of mud crab Scylla paramamosain: molecular cloning, genomic organization and the antimicrobial activity of its synthetic LPS binding domain. Mol. Immunol. 2007;44:3195–3203. doi: 10.1016/j.molimm.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Imjongjirak C, Amparyup P, Tassanakajon A, Sittipraneed S. Molecular cloning and characterization of crustin from mud crab Scylla paramamosain. Mol. Biol. Rep. 2009;36:841–850. doi: 10.1007/s11033-008-9253-0. [DOI] [PubMed] [Google Scholar]
- Iwanaga S, Kawabata S-I, Muta T. New types of clotting factors and defense molecules found in horseshow crab hemolymph: Their structures and functions. J. Biochem. 1998;123:1–15. doi: 10.1093/oxfordjournals.jbchem.a021894. [DOI] [PubMed] [Google Scholar]
- Johansson MW, Keyser P, Sritunyalucksana K, Söderhäll K. Crustacean haemocytes and haematopoesis. Aquaculture. 2000;191:45–52. [Google Scholar]
- Khoo L, Robinette D, Noga EJ. Isolation of callinectin, an antibacterial peptide from blue crab (Callinectes sapidus) hemocytes. Mar. Biotechnol. 1999;1:44–51. doi: 10.1007/pl00011750. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Sakiyama F, Narita K. Oxidation of tryptophan in lysozyme by ozone in aqueous solution. J. Biochem. (Tokyo) 1975;78:641–651. doi: 10.1093/oxfordjournals.jbchem.a130951. [DOI] [PubMed] [Google Scholar]
- Li C, Blencke H-M, Smith LC, Karp MT, Stensvåg K. Two recombinant peptides, SpStrongylocins 1 and 2, from Strongylocentrotus purpuratus, show antimicrobial activity against Gram-positive and Gram-negative bacteria. Dev. & Comp. Immunol. 2010;34:286–292. doi: 10.1016/j.dci.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Li C, Zhao J, Song L, Mu C, Zhang H, Gai Y, Qiu L, Yu Y, Ni D, Xing K. Molecular cloning, genomic organization and functional analysis of an anti–lipopolysaccharide factor from Chinese mitten crab Eriocheir sinensis. Dev. & Comp. Immunol. 2008;32:784–794. doi: 10.1016/j.dci.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Lee RF, Frischer ME. The decline of the blue crab. Am. Scientist. 2004;92:548–553. [Google Scholar]
- Lehrer RI, Lee IH, Menzel L, Waring A, Zhao C. Clavanins and styelins, α-helical antimicrobial peptides from the hemocytes of Styela clava. In: Beck G, Sugumaran M, Cooper EL, editors. Phylogenetic Perspectives on the Vertebrate Immune System. Kluwer Academic; New York: 2001. pp. 71–76. [Google Scholar]
- Matozzo V, Marin MJ. First cytochemical study of haemocytes from the crab Carcinus aestuarii (Crustacea, Decapoda) Eur. J. Histochem. 2010;54:44–49. doi: 10.4081/ejh.2010.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell EM, Trump BF. Histologic fixatives suitable for diagnostic light and electron microscopy. Arch. Pathol. Lab. Med. 1976;100:405–414. [PubMed] [Google Scholar]
- Muñoz M, Vandenbulcke F, Saulnier D, Bachere E. Expression and distribution of penaeidin antimicrobial peptides are regulated by haemocyte reactions in microbial challenged shrimp. Eur. J. Biochem. 2002;269:2678–2689. doi: 10.1046/j.1432-1033.2002.02934.x. [DOI] [PubMed] [Google Scholar]
- Noga EJ. Hemolymph biomarkers of crustacean health. In: Fingerman M, Nagabhushanam R, editors. Recent Advances in Marine Biotechnology. Vol. 5 Immunobiology and Pathology. Science Publishers, Inc.; Enfield, New Hampshire: 2000. pp. 125–163. [Google Scholar]
- Noga EJ, Engel DW, Arroll TW, McKenna S, Davidian M. Low serum antibacterial activity coincides with increased prevalence of shell disease in blue crabs Callinectes sapidus. Dis. Aquat. Org. 1994;19:121–128. [Google Scholar]
- Noga EJ, Sawyer TK, Rodón-Naveira M. Disease processes and health assessment in blue crab fishery management. J. Shellfish Res. 1998;17:567–577. [Google Scholar]
- Noga EJ, Silphaduang U, Park NG, Seo J-K, Stephenson J, Kozlowicz S. Piscidin 4, a novel member of the piscidin family of antimicrobial peptides. Comp. Biochem. Physiol. B. 2009;152:299–305. doi: 10.1016/j.cbpb.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Relf JM, Chisholm JRS, Kemp GD, Smith VJ. Purification and characterization of a cysteine-rich 11.5-kDa antibacterial protein from the granular haemocytes of the shore crab, Carcinus maenas. Eur. J. Biochem. 1999;264:350–357. doi: 10.1046/j.1432-1327.1999.00607.x. [DOI] [PubMed] [Google Scholar]
- Salger SA, Reading BJ, Baltzegar DA, Sullivan CV, Noga EJ. Molecular characterization of two isoforms of piscidin 4 from the hybrid striped bass (Morone chrysops × M. saxatilis) Fish & Shellfish Immunol. 2010 doi: 10.1016/j.fsi.2010.10.009. DOI 10.1016/j.fsi.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Schnapp D, Kemp GD, Smith VS. Purification and characterization of a proline-rich antibacterial peptide, with sequence similarity to bactenecin-7, from the haemocytes of the shore crab, Carcinus maenas. Eur. J. Biochem. 1996;240:532–539. doi: 10.1111/j.1432-1033.1996.0532h.x. [DOI] [PubMed] [Google Scholar]
- Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- Shields JD, Overstreet RM. Chapter 8: Diseases, parasites and other symbionts. In: Kennedy VS, Cronin LE, editors. The Blue Crab: Callinectes sapidus. University of Maryland Sea Grant; College Park, MD: 2007. pp. 223–339. [Google Scholar]
- Shinnar AE, Butler KL, Park HJ. Cathelicidin family of antimicrobial peptides: proteolytic processing and protease resistance. Bioorg. Chem. 2003;31:425–436. doi: 10.1016/s0045-2068(03)00080-4. [DOI] [PubMed] [Google Scholar]
- Skoog B, Wichmann A. Calculation of the isoelectric points of polypeptides from the amino acid composition. Trends Anal. Chem. 1986;5:82–83. [Google Scholar]
- Somboonwiwat K, Marcos M, Tassanakajon A, Klinbunga S, Aumelas A, Romestand B, Gueguen Y, Boze H, Moulin G, Bachére E. Recombinant expression and anti-microbial activity of anti-lipopolysacchride factor (ALF) from the black tiger shrimp Penaeus monodon. Dev. & Comp. Immunol. 2005;29:841–851. doi: 10.1016/j.dci.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Sperstad SV, Haug T, Vasskog T, Stensvåg K. Hyastatin, a glycine-rich multi-domain antimicrobial peptide isolated from the spider crab (Hyas araneus) hemocytes. Mol. Immunol. 2009;46:2604–2612. doi: 10.1016/j.molimm.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Stensvåg K, Haug T, Sperstad SV, Rekdal O, Indrevoll B, Styrvold OB. Arasin 1, a proline–arginine–rich antimicrobial peptide isolated from the spider crab, Hyas araneus. Dev. & Comp. Immunol. 2008;32:275–285. doi: 10.1016/j.dci.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Sung HH, Sun R. Use of monoclonal antibodies to classify hemocyte subpopulations of tiger shrimp (Penaeus monodon) J. Crustacean Biol. 2002;22:337–344. [Google Scholar]
- Tasiemski A, Schikorski D, Le Marrec-Croq F, Pontoire-Van Camp C, Boidin-Wichlacz C, Sautiére P-E. Hedistin: A novel antimicrobial peptide containing bromotryptophan constitutively expressed in the NK cells-like of the marine annelid, Nereis diversicolor. Dev. & Comp. Immunol. 2007;31:749–762. doi: 10.1016/j.dci.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Murray J, Capaldi RA, Ghosh SS. Oxidative post-translational modification of tryptophan residues in cardiac mitochondrial proteins. J. Biol. Chem. 2003;278:19587–19590. doi: 10.1074/jbc.C300135200. [DOI] [PubMed] [Google Scholar]
- Thomas S, Karnik S, Barai R. Shanker, Jayaraman VK, Idicula-Thomas S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010;38:D774–D780. doi: 10.1093/nar/gkp1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Mitta G, Chavanieu A, Calas B, Sanchez JF, Roch P, Aumelas A. Solution stricture and activity of the synthetic four-disulfide bond Mediterranean mussel defensin (MGD-1) Biochemistry. 2000;39:14436–14447. doi: 10.1021/bi0011835. [DOI] [PubMed] [Google Scholar]
- Yedery RD, Reddy KV. Identification, cloning, characterization and recombinant expression of an anti-lipopolysaccharide factor from the hemocytes of Indian mud crab, Scylla serrata. Fish & Shell. Immunol. 2009;27:275–284. doi: 10.1016/j.fsi.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Yue F, Pan L, Miao J, Zhang L, Li J. Molecular cloning, characterization and mRNA expression of two antibacterial peptides: crustin and anti-lipopolysaccharide factor in swimming crab Portunus trituberculatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010;156:77–85. doi: 10.1016/j.cbpb.2010.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Predicted b and y ions for the amidated C-terminal tryptic peptide spanning residues 25-32 of callinectin. The observed intact protonated monoisotopic peptide mass (M+H) is 967.30 with a predicted M+H of 967.37. This mass difference of 0.07 Da, along with the observed b and y ion series sequence fragments, indicate that the C-terminus of callinectin is amidated.
Supplementary Table 2. Edman sequencing pmol yields of amino acids. Amino acids identified by Edman sequencing are in bold in the right hand column. % CO is the % carryover as calculated by subtracting the pmol background prior to the amino acid increase and then dividing the following residue pmol yield over the increased pmol yield.
Supplementary Figure 1. MALDI-MS chromatogram of callinectin tryptic digestion. The callinectin tryptic digestion was analyzed using MALDI-MS (Micromass TofSpec SE) in both linear and reflectron modes, with the linear spectra shown. Using peptide mass fingerprinting and comparing the observed masses to the edman sequence of callinectin, the indicated residues were identified. Mass accuracy with internal calibration is better than 70 ppm. Internal calibrants/ standards (INT STD) were 100 fmol of bradykinin (M+H mono-isotopic mass: 1060.57), and 250 fmol of adrenocorticotropic hormone human fragment 18-39 (ACTH Clip) (M+H monoisotopic mass: 2465.2).
Supplementary Figure 2. De-convoluted MS spectra of the tryptic digestion of callinectin. MS and MS/MS analysis was performed on a Micromass Q-Tof Ultima using nanospray. MAXENT 3 (Micromass Masslynx) deconvoluted the MS spectra into singly charged m/z. Callinectin peptide residues identified by MS/MS are indicated.
Supplementary Figure 3. Nanospray MS/MS of the singly charged peptide 798.3 MH; peptide spans residues 19-24 of callinectin.
Supplementary Figure 4. Nanospray MS/MS of singly charged mass 895.3 MH; peptide spans residues 18-24 of callinectin. Internal fragment sequences were observed as marked.
Supplementary Figure 5 Nanospray MS/MS of doubly charged mass 468.7MH; peptide spans residues 9-17 of callinectin. Internal fragment sequences were observed as marked.
Supplementary Figure 6. Nanospray MS/MS of singly charged mass MH 741.3; peptide spans residues 20-24 of callinectin.
Supplementary Figure 7. LTQ Orbitrap MS/MS of doubly charged mass MH 424.71. The peptide spans residues 1-6 of callinectin and verifies residue 6 as arginine.
Supplementary Figure 8. MS/MS spectra of the C-terminal tryptic peptide of callinectin. The singly charged peptide (967.45 Da) was subjected to nanospray MS/MS analysis and was found to span residues 25-32 of callinectin, with amidation of the C-terminus. Internal fragment sequence ions were observed as marked (e.g., PCSN = Pro-CAM-Ser-Asn). Predicted b and y series fragment ions are shown in Supplementary Table 1.
Supplementary Figure 9. Accurate intact mass of callinectin. An accurate intact mass (M+H) was determined using LCMS on a Waters nanoACQUITY UPLC system with detection in a LTQ Orbitrap mass spectrometer.
a) The total ion current intensity (y axis) of the LCMS related to the elution time (x axis). The intact callinectin eluted at ~43 min while degradation products (or proteolytic fragments) of callinectin eluted from 26 to 33 min as shown.
b) Combined mass scans across the intact callinectin peak. The charge state (z) is as indicated for each mass observed. Singly charged (z=1) masses were not of interest due to their low mass.
c) The expanded +5 (739.55 Da) mass which deconvolutes to a singly charged mass of M+H 3693.75 Da. The +6 (616.46 Da) charged mass corresponds to a singly charged mass of 3693.76 Da and is in good agreement with the +5 deconvoluted mass. Mass accuracy at this mass in this mass spectrometer is routinely 0-3 ppm.