Abstract
Azidothymidine (AZT, zidovudine) is used to treat HIV-AIDS and prevent maternal transmission to newborns. Because the azido group replaces the 3′ OH of thymidine, AZT is believed to act as a chain terminator during reverse transcription of viral RNA into DNA, although other mechanisms of viral inhibition have been suggested. There is evidence that AZT is genotoxic, particularly to the mitochondria. In this study, we use the bacterium Escherichia coli to investigate the mechanism of AZT toxicity and the cellular mechanisms that aid survival. We show that that replication arrests quickly after treatment with induction of the SOS DNA damage response. AZT appears to produce single-strand DNA gaps, as evident by RecF-dependent induction of the SOS response and visualization of single-strand DNA binding protein foci within the cell. Some of these gaps must be converted to breaks, since mutants in the RecBCD nuclease, required for recombinational double-strand break repair, are highly sensitive to AZT. Blocks in the late recombination functions, the RuvAB branch migration helicase and RuvC Holliday junction endonuclease, caused extreme AZT sensitivity that could be relieved by mutations in the early recombination functions, such as RecF, suggesting gaps engage in recombination reactions. Finally, our data suggest that the proofreading exonucleases of DNA polymerases play little role in AZT tolerance. Rather, Exonuclease III appears to be the enzyme that removes AZT: xthA mutants are highly AZT-sensitive, with a sustained SOS response, and overproduction of the enzyme protects wild-type cells. Our findings suggest that incorporation of AZT into human nuclear and mitochondrial DNA has the potential to promote genetic instability and toxicity through the production of ssDNA gaps and dsDNA breaks, and predicts that the human Exonuclease III ortholog, APE1, will be important for drug tolerance.
Keywords: gap repair, RecF pathway, recombination, SOS response
1. INTRODUCTION
Azidothymidine (AZT, 3′-azido-3′-deoxythymidine, zidovudine) is a frontline antiviral agent for the treatment of human immunodeficiency virus-1 (HIV-1) and prevention of transmission from mother to offspring. Like many antiviral drugs approved for HIV-AIDS therapy, AZT acts as an inhibitor of HIV reverse transcriptase (RT). AZT is believed to act as a DNA chain terminator because the 3′-hydroxyl group of thymidine is replaced with an azide group, rendering it impossible for the following nucleotide to be incorporated by DNA polymerases. Its therapeutic potential results from the fact that it is readily incorporated by HIV-RT but less well by cellular DNA polymerases (reviewed in [1]). Although AZT is a lifesaving therapy, it has genotoxic effects on cells (reviewed in [1]). Genomic damage is manifest as micronuclei, sister chromatid exchange events and various chromosomal aberrations [2]. AZT treatment has adverse effects on mitochondria with muscle cells being especially vulnerable, leading to severe myopathy (reviewed in [3]). AZT has been demonstrated to be incorporated into nuclear, and particularly, into mitochondrial DNA (reviewed in [1]). Toxicity of various chain terminators appears to correlate with their affinity for mitochondrial DNA polymerase gamma [4]. Although DNA polymerases alpha and beta appear to be resistant to AZT, polymerase gamma was partially inhibited [5].
The particular lesions that AZT can cause in the cell remain poorly understood and mechanisms other than DNA chain termination have been suggested to account for AZT toxicity [6]. Moreover, it is not clear what cellular mechanisms may aid tolerance of the drug. If AZT is incorporated into DNA, it must be removed before DNA synthesis can resume from the blocked primer terminus and should lead to the formation of single-strand DNA gaps in the replication fork. Furthermore, if the AZT is not removed before the next round of replication, a double strand break will result from convergence of the replication fork to the site of the gap. The gaps and breaks that culminate from AZT incorporation may account for the genotoxicity of the compound.
We sought to use the model organism Escherichia coli to identify what types of damage AZT may cause as well as the cellular components that respond to this damage. DNA repair mechanisms are well-established for E. coli, including those that deal with lesions in the replication fork. AZT enters the cell and must then be phosphorylated to the triphosphate form before it can be incorporated into DNA. This is accomplished through the nucleotide salvage pathway, with the first phosphorylation step being catalyzed by thymidine kinase. In E. coli, thymidine kinase readily uses AZT as a substrate with the apparent Km of thymidine kinase for AZT being about 2-fold higher and the rate of phosphorylation being about 6 fold lower than for thymidine [7]. E. coli’s thymidylate kinase (tmk) also readily phosphorylates azidothymidine monophosphate [8]. We therefore expected that E. coli would be a suitable sensitive model system to investigate the genotoxic properties of AZT. Early work demonstrated that E. coli was sensitive to AZT and showed that AZT acted as a chain terminator for E. coli DNA polymerase I (Klenow fragment) in vitro [7]. In this same study, AZT-triphosphate at submicromolar concentrations inhibited replication of permeabilized E. coli cells lacking DNA polymerase I, suggesting that the replicative DNA polymerase III might also inhibited. Our analysis shows that E. coli is indeed quite sensitive to AZT [9-11], with 50% lethality after a hour of treatment with about 100-200 ng/ml (4-8 nM) AZT, although cells appear to tolerate chronic sublethal doses through the activation of stress responses [12]. We and others have shown that AZT induces the SOS response to DNA damage [10, 12, 13]. However, since bacterial antibiotics targeting cell wall synthesis and protein translation, as well as those that directly cause DNA damage, will eventually induce the SOS response [14], this tells us little about the primary mode of action of AZT.
In this report, using genetic analysis, cell visualization and physiological measurements, we demonstrate that AZT very effectively blocks DNA replication of E. coli, leading to the production of single-strand DNA gaps. These gaps are converted at some frequency to double-strand breaks. Tolerance of AZT requires Exonuclease III (ExoIII) and the phenotypes of ExoIII (xthA) mutants suggests that it, rather than the proofreading exonucleases associated with DNA polymerases, is the primary enzyme that removes AZT from DNA. ExoIII is a relatively non-processive 3′ to 5′ nuclease [15, 16] which also possesses endonuclease activity at apurinic/apyrimidinic sites [17]. ExoIII has orthologs in many organisms, including humans, where it is called apurinic/apyrimidinic endonuclease I, or APE1 [18]. One of its preferred substrates is DNA with a recessed 3′ end, similar to that expected to be presented by AZT incorporation into DNA. ExoIII is important in the repair of DNA damaged by oxidation or radiation, including 3′ phosphate or phosphoglycolate termini that, like AZT, must be removed before replication can proceed through the region [19-21].
Our analysis suggests that genetic recombination by the RecFOR pathway, which specifically responds to single-strand gaps (reviewed in [22], is initiated when DNA replication is blocked by AZT incorporation. RecFOR-mediated loading of RecA protein on ssDNA also activates the SOS response to DNA damage. Mutants in the double-strand break recombination pathway, mediated by the RecBCD enzyme, are abnormally sensitive to AZT, implying that some proportion of ssDNA gaps are converted to breaks, which must be repaired by this mechanism. This work therefore confirms the genotoxicity of AZT, through production of ssDNA gaps and DSBs, and defines several important functions for tolerance including homologous recombination, the SOS response and exonuclease III. In addition, this work establishes AZT as a useful agent to study the processing of replication gaps in vivo.
2. MATERIALS AND METHODS
2. 1. Bacterial strains and growth conditions
Escherichia coli K-12 strains in the AB1157 background (Table 1) were grown at 37°C as previously described on Luria-Bertani (LB) medium, consisting of 1% Bacto Tryptone, 0.5% yeast extract, 0.5% sodium chloride and, for plates, 1.5% agar. Antibiotics were used in the following concentrations: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 60 μg/ml; tetracycline (Tc) and chloramphenicol (Cm), 15 μg/ml. New isogenic mutant strains were constructed by P1 transduction [23]. LCG, LB medium supplemented with 1% glucose with an additional 2 mM calcium chloride and 1% agar for plates, was used to make phage lysates and for transductions. Strain constructions were then confirmed by PCR or phenotype. Plasmids were introduced to strains using electroporation [24] or polyethylene glycol/Mg2+ [25] according to standard procedures. 1 mM IPTG was used to induce over-expression of proteins.
Table 1.
AZT survival of nuclease and polymerase deficient strains
| Mutant genotype | Fractional survival | |
|---|---|---|
| 75 ng/ml | 15 ng/ml | |
| (wild-type) | 8.4 × 10−1 | 9.5 × 10−1 |
| xthAΔFRT::cat | 1.1 × 10−4 | 1.6 E-02 |
| xonAΔFRT::kan | 1.0 | nd |
| xseAΔFRT | 9.2 × 10−1 | nd |
| xonAΔFRT::kan xseAΔFRT | 9.8 × 10−1 | nd |
| exoXΔFRT::kan | 1.0 | nd |
|
exoXΔFRT::kan xonAΔFRT xseAΔFRT |
8.1 × 10−1 | nd |
| nfoΔFRT::kan | 8.3 × 10−1 | nd |
| nfiΔFRT::kan | 7.0 × 10−1 | nd |
| dnaQ49 | 9.9 × 10−1 | nd |
| polB ex1 | 8.7 × 10−1 | nd |
| polA480 (5′ to 3′ exo) | 4.1 × 10−1 | nd |
| polA (3′ to 5′ exo) | 5.8 × 10−1 | nd |
| polA Δ kan | 3.7 × 10−1 | nd |
| polB Δ kan | 2.0 × 10−1 | nd |
| polB umuCD dinB | 8.7 × 10−1 | nd |
nd=not determined
All strains are in the AB1157 background. Survival for strains that showed greater than a ten-fold loss of viability at 75 ng/ml AZT is also shown at 15 ng/ml AZT in the split columns. Reported values are averages of at least 8 cultures from at least 2 days. Standard errors are less than 70% or the reported value.
Azidothymidine (AZT) was added to the concentrations indicated in each experiment. For determination of survival, cells were typically grown to an A590 of between 0.3-0.5, serially diluted in 1 × 56/2 salts [26] and plated on LB plates with AZT. Colony formation was determined after incubation at 37° for 24-36 hours. For assays involving AZT in liquid culture, cells were grown at to an A590 between 0.2-0.35 before addition of AZT.
2.2 SOS induction assay
Plasmids carrying vector with Photorhabdus luminescens luxCDABE (pDEW201 ) or luciferase fusions to the E. coli recA (pDEW238) or dinB promoters (pDEW236) [27, 28], were transformed into the appropriate strains and selected for using Ap-resistance. Strains were then grown and treated with either 100 ng/ml AZT or thymidine. At subsequent time points, aliquots of culture were assayed for bioluminescence in a liquid scintillation counter. At least 3 cultures were assayed for each data point. Arbitrary luciferase expression values were then calculated by normalizing the amount of bioluminescence (cpm) to the A590 of the cultures at each time point.
2.3. Measurement of DNA synthesis rate
Standing overnights of wild-type cells were diluted to an A590 of 0.02-0.03 and grown in LB media until the A590 reached approximately 0.2 before induction of protein or treatment with AZT. At the indicated time points, replication was assessed by removing 2 ml aliquots and adding them to 15 ml conical tubes containing 5-ethynyl-2′-deoxuridine (EdU, Invitrogen, ClickIT Edu kit) at a final concentration of 40 μg/ml. After 5 minutes of incubation with EdU, cells were fixed by adding 10 ml 90% methanol. Fixed cells were pelleted at 4° and washed twice with 1.5 ml filtered PBS. Invitrogen’s protocol for Triton X-100 permeabilization and labeling using Alexa-Fluor 488 was followed, except 2-3 additional washes with PBS were added after labeling. Labeled cells were resuspended in 0.5-1.0 ml PBS. For microscopy, cells were spotted onto slides coated with poly-lysine, dried, covered with 2.5 μl Vectashield mounting media, and sealed with a coverslip. Cells were then imaged using an Olympus BX51 microscope equipped with a RGB liquid crystal color filter. Images were acquired and analyzed with a Qimaging Reiga Exi camera using Improvision’s Volocity software. For flow cytometry, cells were diluted further in PBS and counted using an Accuri flow cytometer. For each sample, 50,000 events were collected at a slow flow rate.
2.4 Nucleoid and SSB-foci visualization
Fixed cells were stained with 10 μg/ml DAPI (4′,6′ diamido-2-phenylindole, Sigma-Aldrich) for 15 minutes and then washed thoroughly with PBS. Slides with DAPI labeled cells were prepared and analyzed as outlined above. For visualization of SSB-YPet, a strain with the fusion protein expressed from the ssb natural promoter was used (provided by A. Wright, unpublished). Twenty μl aliquots were spotted onto poly-lysine coated slides at the times indicated. After spotting, cells were incubated at 37° for 8 minutes to allow cells to adhere to the slides. Excess media was then removed and 3 ml Vectashield was added. Slides were then analyzed as above using the U-MYFPHQ filter.
3. RESULTS
3. 1 AZT causes replication inhibition and SOS induction via accumulation of single-strand DNA gaps
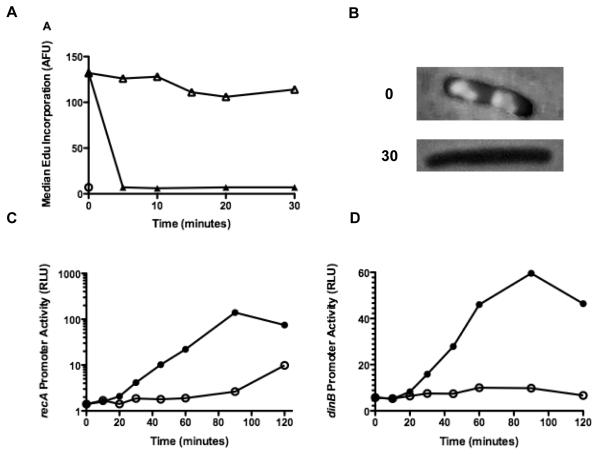
If AZT causes DNA damage by inhibiting replication fork progression, replication should stop or slow when cultures are treated with AZT. The rate of DNA replication in E.coli can be determined by the incorporation of ethenyl deoxyuridine (EdU), a thymidine analog, incorporated into DNA, followed by detection by conjugation with a fluorescent dye [29]. E. coli cultures pulsed with EdU for five minutes incorporate sufficient amounts in newly synthesized DNA to be detectable using fluorescent microscopy (Figure 1B) or flow cytometry (Figure1A). Cells pretreated with thymidine for up to 30 minutes before the pulse of EdU continue to replicate; however, cells pretreated with the same concentration of AZT (100 ng/ml) have diminished EdU incorporation. In fact, replication is reduced to background levels within 5 minutes of pretreatment with 25 ng/ml AZT (Figure 1A and data not shown). We note that this may not represent a full cessation of DNA synthesis and it is possible that slow synthesis continues at low levels during the treatment period.
Figure 1. AZT halts replication and induces SOS.
(A) AB1157 cultures were grown to early log phase and then treated with 100 ng/ml AZT or thymidine for the times indicated. At each time point, 2 ml of culture was removed, EdU was added to 40 μg /ml and cultures were incubated for an additional 5 minutes. The cells were then fixed with 90% methanol, labeled with AlexaFluor488 and analyzed as described in the materials and methods. Median fluorescence for 50,000 cells is shown at each time point (⦿ No Edu, △ Thymidine, ▲AZT). (B) Micrograph showing AB1157 cultures labeled with EdU and visualized with Alexa Fluor 488 at 0 and 30 minutes after the addition of 100 ng/ml AZT. (C) AB1157 cultures harboring a plasmid with the luxCDABE operon under control of the recA promoter were grown as in A. At each time point, 0.1 ml of culture was removed and luminescence was quantified using a liquid scintillation counter (⦿ Thymidine, AZT). Values shown are normalized to the A590 of the culture at each time point. (D) As in C, except the plasmid contains the luxCDABE operon under the dinB promoter (⦿ Thymidine, ● AZT).
DNA chain termination by AZT is predicted to lead to accumulation of single-stranded DNA (ssDNA) gaps in the replication fork and/or double-strand breaks that are processed to reveal ssDNA. In E. coli, production of ssDNA is a signal for a well characterized transcriptional response, the “SOS response” in which over 40 genes are up-regulated [30-32]. Loading of RecA to ssDNA promotes cleavage of the LexA repressor protein thus activating the expression of genes involved in DNA repair, cell cycle arrest and other DNA damage tolerance mechanisms. After treatment with AZT, we measured expression of two SOS-regulated genes, dinB and recA, using luciferase operon fusion constructs. AZT is a potent inducer of both SOS promoters (Figures 1 C and D) after about 20 minutes of treatment of cultures with AZT.
E. coli possesses two DNA substrate-specific systems that load the central RecA recombination protein onto ssDNA (reviewed in [22]). The RecFOR system seems to be designed to load RecA onto ssDNA gaps, such as those we expect to be produced by AZT. In contrast, the RecBCD protein loads RecA on resected DNA it produces from double-strand breaks. Loading of RecA onto ssDNA both initiates the recombination process and potentiates the cleavage of the LexA repressor, leading to induction of the SOS response. We investigated the dependence of AZT induction of the SOS response, as measured with the recA promoter-luciferase fusion construct, on the recF gene (Figure 2). Although expression of the recA promoter was somewhat constitutively elevated in the absence of AZT in recF mutants, induction by AZT was abolished. This supports the notion that the primary lesion caused by AZT is the production of ssDNA gaps.
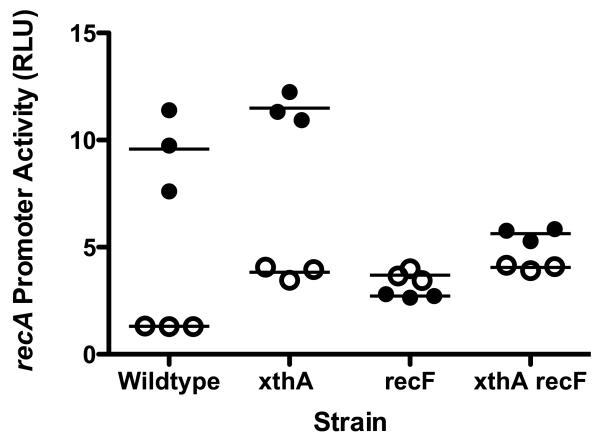
Figure 2. SOS induction in strains treated with AZT.
Strains containing a plasmid with the lux genes under the recA promoter were grown to early log phase and then AZT was added to 150 ng/ml or mock treated. After 40 minutes, culture was removed and luminescence was determined using a liquid scintillation counter. Luminescence was normalized to culture A590 to give relative luciferase units (RLU). (⦿ mock treated cultures, ● treated cultures).
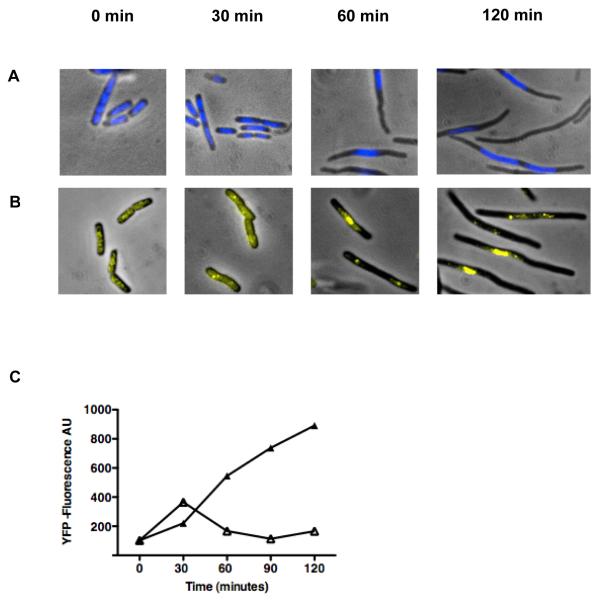
Single-strand DNA binding protein (SSB) protects single-strand DNA in E. coli [33] and can be detected as foci using fluorescence microscopy if there are sufficiently large single-strand DNA regions in the chromosome. The micrograph in Figure 3A shows cells from a strain with SSB-YPet fusion protein expressed from the natural into the chromosomal locus, with and without AZT treatment. Untreated cells have several small diffuse foci (Figure 3A). When these cultures are treated with AZT the SSB-YFP foci become brighter and localize near the middle of the cell suggesting that single-strand DNA is accumulating. Quantification of the SSB-YFP foci by flow cytometry shows that cells accumulate about 7 times the amount of SSB after AZT is added to the growth media indicating that SSB protein is either induced or stabilized under this condition (Figure 3C). The ssb gene has been shown previously to be induced by DNA damage, through a LexA-regulated promoter of the upstream uvrA gene [34].
Figure 3. AB1157 cells elongate and SSB foci accumulate after the addition of AZT.
(A) Wild type cells stained with DAPI at 0, 30, 60, and 120 minutes after addition of AZT to 100 ng/ml. (B) Wild type cells with a SSB-YPet gene under the natural SSB promoter at 0, 30, 60, and 120 minutes after addition of AZT to 100 ng/ml. (C) Quantification of SSB-YPet accumulation (50,000 cell sample) using flow cytometry (△ Thymidine, ▲ AZT).
Microscopic visualization of E. coli cells also confirmed that cell division is inhibited in AZT-treated cells. Cells treated with AZT begin to elongate (Figure 3B) and the nucleoid condenses about 30 minutes after the addition of AZT to the growth media. It is not clear what processes are responsible for the nucleoid compaction, but could result from ongoing repair processes.
3. 2 Exonuclease III facilitates survival in the presence of AZT
AZT-monophosphate (AZT-MP), the incorporated form of AZT, must be removed from the 3′ end of a nascent DNA strand before synthesis can be continued, presumably through direct removal by an exonuclease or endonuclease. This could occur through the activity of a proofreading subunit of a DNA polymerase or by an exogenous exonuclease or endonuclease. We isolated the gene for Exonuclease III (xthA) as a high copy suppressor of an AZT sensitive strain (Cooper, unpublished results), the first indication that this enzyme might play a role in AZT metabolism. Subsequent experiments with an xthA null strain demonstrated a strong requirement for xthA for growth and survival during chronic AZT treatment (Table 1), at AZT doses (15-75 ng/ml) well-tolerated in wild-type strains. We screened several of the other exonucleases and endonucleases in E. coli, including Exonucleases I (xonA), VII (xseA) and X (exoX) and Endonucleases IV (nfo) and V (nfi), that share certain properties with ExoIII, including 3′ to 5′ exonuclease or abasic endonuclease activity . None of these showed appreciable sensitivity, indicating that Exonuclease III is likely to be the primary nuclease that removes AZT-MP from DNA. In addition, the 3′ to 5′ exonucleases associated with DNA Polymerases I (3′exo-), II (polBex1) and III (dnaQ) as well as the 5′ to 3′ exonuclease (polA430) activity of Polymerase I have little effects on the cell’s ability to survive in the presence of AZT.
E. coli encodes five DNA polymerase activities (reviewed in [35, 36]. DNA Polymerase III, a multisubunit holoenzyme, is responsible for the bulk of DNA synthesis during replication; Polymerase I is involved the small amount of synthesis that accompanies Okazaki fragment maturation; Polymerases II, IV and V are normally expressed at low levels but induced during the SOS response and play a role in translesion DNA synthesis during repair. Deletion of all three SOS DNA polymerases, Pol II (polB), IV (dinB) or V (umuCD), did not affect survival to chronic AZT exposure (Table 1). This, and the fact that DNA replication arrests very rapidly upon AZT exposure (Fig. 1) argues that one or both of the replicative DNA polymerases, I and/or III must incorporate AZT into DNA. Since deletion of the gene for Polymerase I (polA) did not enhance resistance to AZT (Table 1), DNA polymerase III must have some ability to incorporate AZT during DNA synthesis.
3.3 The SOS response is amplified in xthA deletion strains, which fail to recover from AZT treatment
Strains deleted for ExoIII (xthA) have normal growth rates and cell morphology (data not shown), although promoter activity of the SOS response gene recA is elevated 1.5-2 fold, indicating a low level of constitutive DNA damage (Figure 2). RecA promoter activity is induced by AZT in the xthA strain to levels higher than wild type strains. This is consistent with the expectation that removal of AZT and subsequent gap-filling is delayed in ExoIII mutants. Most of the SOS response to AZT in xthA mutants was dependent on RecF (Figure 2) although some weak induction was evident in xthA recF double mutants. This may result from chromosomal breakage in the absence of ExoIII and induction via the RecBCD pathway (see below).
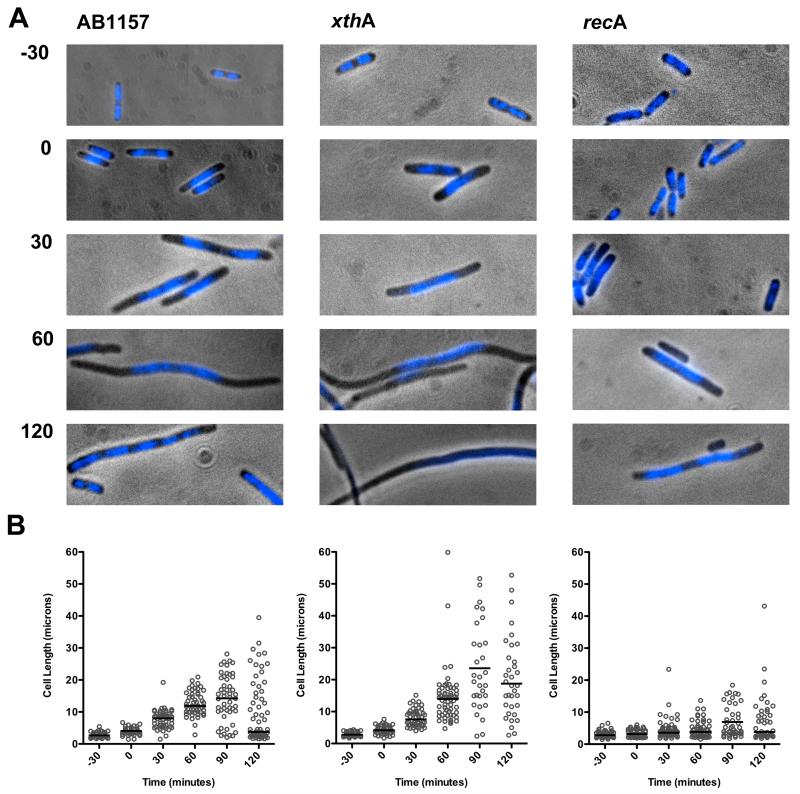
During the SOS response of E. coli, cell division is inhibited by the SOS-inducible SulA protein [37]. Because cell growth continues, this leads to the production of long cell filaments (Figure 4). By microscopy and cell length measurements we measured the ability of cells to recover division capacity after removal of AZT (Figure 4). In addition, DNA was visualized by DAPI fluorescence microscopy to determine any changes in positioning or morphology of the bacterial nucleoid. In wild-type and xthA cells, DNA typically aggregated near midcell within 30 minutes of addition of AZT and the cells continue to elongate even 60 min after AZT has been removed. After 120 minutes following AZT removal, wild-type cells began to segregate chromosomes throughout the length of the cell and divide. Sometimes cell volume in the filamentous cells appeared to be decreased by formation of anucleate cells. Bursting cells also were detected at this time. In xthA cells, nucleoid distribution and cell division was delayed and very few cells assumed normal lengths within 2 hours. This is consistent with the idea that xthA cells accumulate irreversible damage that is repaired in Exonuclease III proficient cells. In recA cells that cannot induce SOS division inhibition via SulA, filamentation was much reduced relative to wild type or xthA cells as expected, although some cells elongated. In addition, the recA nucleoids did not appear to aggregate mid-cell or condense as extensively in the same way as in xthA or wild-type cells (Figure 4). More anucleate cells were observed after AZT treatment and removal in recA cells.
Figure 4. Cellular morphology during recovery from AZT treatment.
Wild-type, xthA, or recA cultures were grown to early log phase. AZT (100 ng/ml) was then added to the media for 30 minutes after which the cells were collected by centrifugation, washed, and then resuspended in LB. Growth was continued in media without AZT and samples were collected at the times indicated. (A) Micrographs showing cultures stained with DAPI before treatment with AZT (−30), immediately after treatment with AZT for 30 minutes (0), or after AZT was removed from the cultures for 30, 60, or 120 minutes. (B) Lengths of at least 50 cells from 2 slides were measured using Volocity Openlab software. Each individual cell length is represented with a ○ and the median cell length for each population is indicated by the horizontal bar.
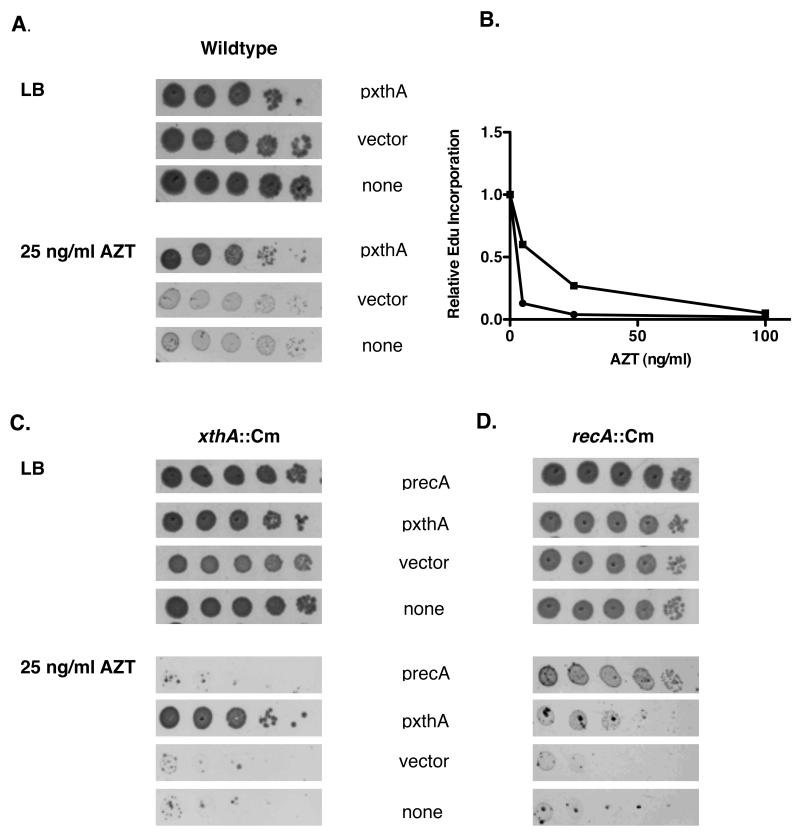
3. 4 Overexpression of xthA mitigates AZT sensitivity
If ExoIII acts immediately to remove AZT monophosphate from the growing fork, it is possible that the SOS response is induced only after the cell has become overwhelmed with damage. In this case, exonuclease III overexpression should decrease the sensitivity of cells to AZT. In wild type cells, overexpression of exonuclease III allowed more robust colony growth in the presence of AZT, as seen in Figure 5A. In addition, higher levels of ExoIII increased the amount of AZT required to halt DNA replication in a wild type strain (Figure 5B). Overexpression of ExoIII also reduced the extreme sensitivity to AZT seen for xthA and recA null strains; however, RecA overexpression did not suppress AZT sensitivity of xthA mutants. The suppression of recA sensitivity to AZT by xthA overexpression was not complete, although it does show that in the absence of homologous recombination, higher levels of ExoIII can compensate somewhat. This indicates that ExoIII acts earlier in the cell’s response to AZT incorporation (Figure 5 B and C), consistent with a role in direct removal.
Figure 5. Overproduction of Exonuclease III but not RecA mitigates AZT toxicity.
(A) Wild type cultures transformed with vector (pBSSK-) or a plasmid over-expressing xthA (in pBSSK-) were grown at 37° to an A590 of approximately 0.4 in LB media with 100 mg/ml Ap. Cultures were then serially diluted with 1X 56/2 media supplemented with 2mM IPTG. An additional 50 μg /ml ampicillin was added to serial dilutions when a plasmid was present. Diluted cultures were plated on either LB plates or LB plates with 25 ng/ml AZT. Plates were then incubated at 37° for approximately 24 hours. (B) 0-100 ng/ml AZT was added to early log phase wild type cultures transformed with either pBSSK- or xthA-pBSSK- and incubated for 5 minutes. Replication after addition of AZT was then assessed by using flow cytometry to measure the amount of EdU incorporated in 5 additional minutes after AZT was washed from the media. (C)xthA::Cm cultures transformed with vector or plasmids over-producing ExoIII (XthA) or RecA were grown to early log phase and then plated on LB or LB with 25 ng/ml AZT. Plasmids were maintained with Ap at 100 μg /ml and ExoIII or RecA was induced with 2 mM IPTG. (D) As in C except the host cultures were recA::Cm.
3.5 Capabilities for recombination and the SOS response are required for survival in the presence of AZT
RecA, orthologous to the Rad51 protein of eukaryotes, is both the primary recombination protein in E. coli and is necessary for induction of the SOS response. We found that mutants in recA were very sensitive to AZT, as shown previously [11] and in Table 2. To understand which function of recA is necessary for survival in the presence of AZT, we examined other mutants associated with the SOS response and recombination.
Table 2.
AZT survival of recombination and SOS deficient strains.
| Mutant genotype | Fractional survival | ||
|---|---|---|---|
| 75 ng/ml AZT | 15 ng/ml AZT | 7.5 ng/ml AZT | |
| (wild-type) | 8.4 × 10−1 | 9.5 × 10−1 | nd |
| recA | < 10−5 | 8.7 × 10−3 | 2.1 × 10 −1 |
| recB | < 10−5 | 4.9 × 10−3 | nd |
| recF | 8.4 × 10−1 | nd | nd |
| recO | 4.0 × 10−1 | nd | nd |
| recR | 9.3 × 10−1 | nd | nd |
| recX | 1.0 | nd | nd |
| recF recB | nd | < 10−4 | nd |
| ruvAB | < 10−5 | 6.5 × 10−3 | 4.4 × 10−2 |
| ruvC | < 10−5 | 2.4 × 10−3 | 1.5 × 10 −2 |
| ruvC recF | nd | 8.2 × 10−1 | nd |
| ruvC xthA | nd | < 10−5 | nd |
| lexA3 | < 10−4 | 1.9 × 10−2 | nd |
| sulA | 3.4 × 10−3 | 1.1 | nd |
nd=not determined
All strains are in the AB1157 background. Survival for strains that showed greater than a ten-fold loss of viability at 75 ng/ml AZT is also shown at reduced AZT concentrations. Reported values are averages of at least 8 cultures from at least 2 days. Standard errors are less than 70% of the reported value.
The lexA3 allele produces a LexA repressor protein that cannot be cleaved after DNA damage. Thus, strains with this mutation cannot induce the SOS repair transcriptional response but are recombination-proficient [38]. The data in Table 2 show that lexA3 mutants are sensitive to AZT, but not as sensitive as recA null mutants, suggesting that both recombination capacity and induction of the SOS response play key roles in survival to AZT. Since transcription of recA itself increases when SOS is induced, it is also possible that there is an optimal level of RecA necessary for AZT tolerance.
SulA inhibits cell division and is highly induced during the SOS response [37]. Null mutants in sulA are sensitive to AZT in high doses (Table 2), indicating that cell division regulation plays a role in promoting survival to AZT. The SOS-induced recX gene, implicated in negative regulation of the RecA filament [39], had no effect on AZT tolerance. The SOS-controlled ruvAB genes, which encode a DNA helicase that processes recombination intermediates such as Holliday junctions (reviewed in [40]), had a strong effect on growth in the presence of AZT; ruvAB mutants were even more sensitive than those in recA (Table 2). Mutants in RuvC, a Holliday junction cleavage protein that interacts with RuvAB, were also extremely AZT-sensitive. (RuvC, unlike RuvAB, is not SOS-regulated).
The RuvABC proteins affect late steps in genetic recombination whereas RecA affects the very earliest step of homologous pairing. The higher sensitivity of mutants in ruvC compared to recA (Table 2) suggests that the inability to complete recombination (ruvC) may trap lethal intermediates; in the absence of initiation of recombination (recA), other mechanisms may contribute to tolerance of AZT damage. A prediction, therefore, is that early blocks in recombination should suppress somewhat the AZT-sensitivity of ruvC mutants. Indeed, AZT sensitivity of ruvC could be completely suppressed by mutations in recF (Table 2). This is consistent with the model that RecFOR loads RecA onto AZT-promoted ssDNA gaps, which are then filled by recombination; in the absence of Holliday junction processing, recombination intermediates cannot be resolved and are lethal to the cell.
3.6 Synergy between SOS or recombination functions and Exonuclease III
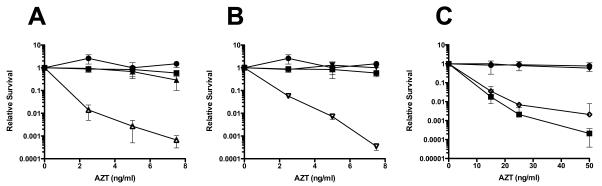
To examine the interaction between xthA and other recombination and repair genes, we constructed xthA recA, xthA lexA, xthA ruvC as well as xthA sulA mutants. The xthA recA and xthA lexA double mutants are extraordinarily sensitive to AZT, at doses as low as 5 ng/ml (Figure 6 A and B, note lower dose scale), levels much greater than the single mutants. The xthA ruvC mutant was one of the most sensitive strains examined in this study, with over 5-orders of magnitude of killing at the low dose of 15 ng/ml (Table 2). This strong genetic synergy is consistent with the idea that the SOS response and genetic recombination mechanisms mediate repair or tolerance of AZT that is distinct from ExoIII-promoted removal of AZT-monophosphate from DNA. In contrast, the combination of xthA and sulA produced a strain more resistant to AZT than the single xthA mutant (Figure 6C). This indicates that the failure of xthA mutants to form colonies in the presence of chronic exposure to AZT is in part due to sustained SOS-induced inhibition of cell division and not cell death. Nevertheless, the xthA sulA strain did retain a considerable degree of sensitivity to AZT, so cell division inhibition is not entirely responsible for the failure of xthA mutants to tolerate AZT.
Figure 6. xthA recA or xthA lexA but not xthA sulA are more sensitive to AZT than xthA single mutants.
Cultures were grown to early to mid log phase and then plated on plates containing the concentrations of AZT as indicated. Survival for wild type (●) and xthA (■) strains are shown in all panels. (A) recA (▲) and xthA recA (△)strains. (B) lexA (▼) and xthA lexA (▽)strains. (C) sulA (◆) and xthA sulA (◇) strains. Note the higher AZT concentrations in panel C.
3.7 RecFOR mutations suppress AZT sensitivity of xthA mutants
The dependence of AZT induction of the SOS response on RecF and suppression of ruvABC sensitivity by mutations in recF demonstrated above suggests that RecFOR loads RecA directly onto the ssDNA gaps caused by replication in the presence of AZT. These gaps would be subsequently filled by RecA-mediated recombination, followed by RuvABC resolution. In the absence of gap-filling recombination, a second round of replication or endonuclease cleavage in the vulnerable ssDNA region would convert gaps to double-strand breaks. RecBCD would be expected to act at these breaks to mediate recombinational double-strand break repair with the sister chromosome. Because RecBCD degrades dsDNA by a helicase/endonuclease mechanism (reviewed in [41]), it should be competent to degrade AZT-containing DNA, promoting its release. Mutants in recB were strongly sensitive to AZT, confirming expectations that some gaps are converted to DSBs. Surprisingly, recF, recO, and recR strains are not appreciably sensitive (Table 2). This might be expected if unfilled gaps could be efficiently converted to breaks and then repaired. Indeed, recB recF strains where both recombination pathways are inactivated are somewhat more sensitive to AZT than recB strains. (Quantification of the viability of the recB recF strain on AZT was difficult to assess because of rapidly arising suppressing mutations.)
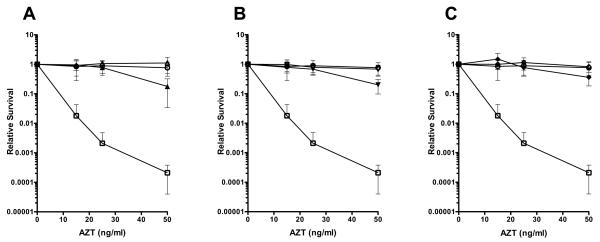
We then tested recF, recO, and recR mutants in combination with xthA and found that, in each case, a mutation in the RecFOR pathway suppressed xthA sensitivity at low to moderate doses of AZT (Figure 7 A-C). Therefore, RecFOR mediated RecA-loading and/or recombination map trap intermediates that require ExoIII for full resolution. In the absence of RecFOR, breakage of the chromosome would allow the RecBCD nuclease to remove the AZT moiety and repair the break, as diagrammed in Figure 8. Exonuclease III could remove AZT before gaps engage with RecFOR and RecA, allowing some gaps to be filled directly by DNA synthesis. Alternatively, AZT-promoted gaps may participate in RecAFOR recombination reactions, with AZT removed by Exonuclease III in the recombination intermediates or products.
Figure 7. Mutations in recFOR suppress the AZT sensitivity of xthA deletion strains.
Cultures were grown to early to mid log phase and then serially diluted and plated on LB plates containing 0-50 ng AZT/ml. Colony forming units were determined after incubation at 37° for approximately 36 hours. Survival for wild type (○) and xthA (□) strains are shown on all panels for comparison. (A) recF (△)and recF xthA (▲)strains. (B) recO (▽) and recO xthA (▼)strains (C) recR (◇) and recR xthA (◆) strains.
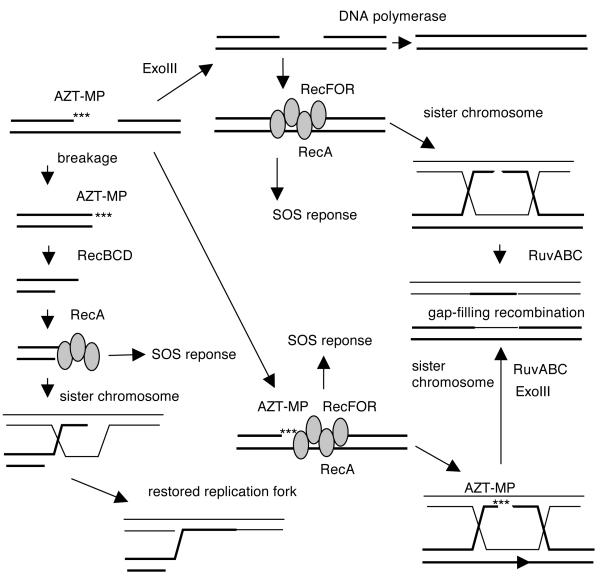
Figure 8. Possible repair pathways for AZT-induced lesions in E. coli.
Incorporation of AZT by DNA polymerases leads to replication gaps. ExoIII can remove the 3′ AZT monophosphate moiety, leaving a clean ssDNA gap. The gap may be filled by DNA polymerase or may engage in recombinational reaction. The RecFOR proteins load RecA onto gaps, leading to induction of the SOS transcriptional response and gap-filling recombination with a sister chromosome. Alternatively, RecAFOR recombination may occur before the 3′ AZT-MP is removed; in such a case, ExoIII is required to remove AZT in the recombination intermediates or products. In the absence of RecFOR, gaps are converted to double-strand breaks by converging replication forks or by endonucleases. RecBCD nuclease processes the break, removing AZT in the process and generating recombinogenic 3′ strands onto which RecA is loaded. This RecA filament may also signal the SOS reponse. Recombination with the sister chromosome restores an intact replication fork.
4. DISCUSSION
4.1 Toxicity of AZT as a DNA replication inhibitor
At concentrations in the nM range, AZT rapidly arrests most replication of E. coli and produces a strong, RecF-dependent induction of the SOS response, consistent with the formation of single-strand gaps. Microscopic examination shows that cells accumulate SSB foci and delay nucleoid segregation and cell division, in a RecA-dependent manner. Therefore, at least in E. coli, AZT appears to act as a chain-terminator, leading to the production of single-strand DNA gaps in replicating DNA. These gaps may occur directly at the fork or be left behind by an advancing fork.
The extremely low doses at which AZT can be toxic (in low nM for mutants deficient in ExoIII and recombination factors), compared to nucleotide triphosphate pools estimated in the 50-100 μM range [42], suggests that there must be minimal discrimination against AZT compared to thymidine; it must be readily transported, phosphorylated and incorporated into DNA. Prolonged treatment with AZT selects variants reduced in thymidine kinase activity [7] and we have repeatedly isolated AZT-resistant variants by transposon insertion in its gene, tdk (Cooper, Mirzai and Lovett, unpublished results), confirming that AZT must be phosphorylated to produce the toxic species. Experiments with permeabilized E. coli cells demonstrated that AZT-triphosphate is more toxic than AZT or its diphosphate, monophosphate derivatives [7], as expected if its incorporation into DNA is source of its lethality. Our genetic analysis and speed at which replication is blocked suggests that AZT is incorporated into DNA by the replicative DNA polymerase III, although we cannot rule out additional incorporation by DNA polymerase I or E. coli’s other DNA repair polymerases II, IV and V.
4. 2 Exonuclease III but not polymerase proofreading exonucleases aid tolerance of AZT
The proofreading activities of E. coli’s DNA polymerases appear to play little role in the tolerance of AZT. Rather, our data are consistent with the idea that Exonuclease III, a 3′ to 5′ exonuclease, 3′ phosphatase and apurinic/apyrimidinic (AP) endonuclease, is the enzyme that removes AZT-monophosphate from DNA. Mutants in ExoIII were strongly sensitive to AZT, exhibited a heightened SOS response and poor recovery of cell division capacity. Overexpression of Exonuclease III alone ameliorated the toxicity of AZT and its ability to block replication, including in wild-type strains. ExoIII has been shown to remove 3′ replication-blocking lesions that are produced by oxidative damage [19], similar to the role proposed here for removal of AZT. Previous biochemical assays with purified yeast and human DNA polymerase gamma have suggested that AZT is poorly proofread by polymerase-associated 3′ exonucleases [43, 44] and we provide biological verification of this property. The human homolog of exonuclease III is APE1 (apurininc/apyrimidinic endonuclease 1), which is found both in the nucleus and in the mitochondria [45]. Our results, and the verification that APE1 can excise AZT from DNA in vitro [46], suggests that the function of APE1 will likely be critical for toleration of AZT therapy in human patients. In addition, because ExoIII plays a critical role in survival both to AZT and to oxidative damage, oxidative damage is likely to predispose cells to toxic effects from AZT.
4. 3 AZT tolerance via induction of stress responses and recombination proteins
The LexA and RecA-regulated SOS DNA damage response allows E. coli cells to tolerate levels of AZT exposure that would otherwise be toxic. Delay in cell division, via the SulA inhibitor of FtsZ, promotes survival to chronic high level AZT exposure. In addition, the SOS-induced RuvAB recombination protein is also a key determinant of toleration of chronic AZT exposure.
Resolution of Holliday junctions or other recombination intermediates by RuvAB helicase and RuvC endonuclease is required to sustain viability in the presence of AZT, suggesting that gap-filling homologous recombination is employed as one of the mechanisms that aids tolerance of AZT exposure. In E. coli, this gap-filling recombination mechanism can be distinguished from that for double-strand break repair by its requirements of RecFOR loading factors for RecA. RecFOR-dependent recombination must also trap AZT-containing DNA in a configuration that (1) requires Holliday junction resolution via RuvABC and (2) requires ExoIII for removal of AZT, since recF, recO or recR mutations have suppressive effects on sensitivity to AZT for mutants deficient for RuvABC or ExoIII. Surprisingly, RecFOR mutants themselves show little sensitivity to AZT. This survival may be a consequence of efficient conversion of gaps to double-strand breaks, followed by DNA digestion and recombination catalyzed by RecBCD (see models in Figure 8). The suppression of ruvABC by mutations in recFOR also must mean that RecFOR recombination has a higher requirement for RuvABC than does the RecBCD pathway. Even in the presence of the RecFOR pathway, AZT appears to promote DSB formation, as evident by the sensitivity of recB (recFOR+) strains.
4.4 Conclusions
Although genetic recombination has long been proposed as a mechanism for replication gap repair, this process has been difficult to study because of the lack of specific gap-producing treatments. We demonstrate here that AZT has properties in E. coli consistent with production of ssDNA gaps in replicating DNA. The study of AZT sensitivity in the model genetic organism Escherichia coli therefore provides an opportunity to define the factors required to survive damage sustained at the active replication fork, an area of intense interest. In addition to Exonuclease III and the SOS response and homologous recombination functions identified here, there are likely several other cellular pathways that play key roles in the tolerance of agents that block replication.
Supplementary Material
5. Acknowledgments
We thank Jawad Mirsai, Katie Pennington, and Houra Merrikh for technical assistance. We thank Cathy Joyce for the polA strains, Myron Goodman for the polB Exo- strain, Andrew Wright for providing the SSB::YPet fusion construct, Ken Knight for the RecA expression plasmid and Tina van Dyk for the luciferase reporter plasmids. The work was supported by a grant from the National Institutes of Health, RO1 GM51753.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. BIBLIOGRAPHY
- [1].Olivero OA. Mechanisms of genotoxicity of nucleoside reverse transcriptase inhibitors. Environ Mol Mutagen. 2007;48:215–223. doi: 10.1002/em.20195. [DOI] [PubMed] [Google Scholar]
- [2].International Agency for Research on Cancer IARC monographs on the evaluation of carcinogenic risks to humans. Some antiviral and antineoplastic drugs and other pharmaceutical agents. 2000. pp. 1–521. (IARC Monographs).
- [3].Kang D, Hamasaki N. Alterations of mitochondrial DNA in common diseases and disease states: aging, neurodegeneration, heart failure, diabetes, and cancer. Curr Med Chem. 2005;12:429–441. doi: 10.2174/0929867053363081. [DOI] [PubMed] [Google Scholar]
- [4].Lee H, Hanes J, Johnson KA. Toxicity of nucleoside analogues used to treat AIDS and the selectivity of the mitochondrial DNA polymerase. Biochem. 2003;42:14711–14719. doi: 10.1021/bi035596s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].König H, Behr E, Löwer J, Kurth R. Azidothymidine triphosphate is an inhibitor of both human immunodeficiency virus type 1 reverse transcriptase and DNA polymerase gamma. Antimicrob Agents Chemother. 1989;33:2109–2114. doi: 10.1128/aac.33.12.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Scruggs ER, Naylor A.J. Dirks. Mechanisms of zidovudine-induced mitochondrial toxicity and myopathy. Pharmacology. 2008;82:83–88. doi: 10.1159/000134943. [DOI] [PubMed] [Google Scholar]
- [7].Elwell L, Ferone R, Freeman G, Fyfe J, Hill J, Ray P, Richards C, Singer S, Knick V, Rideout J, et al. Antibacterial activity and mechanism of action of 3′-azido-3′-deoxythymidine (BW A509U) Antimicrob Agents Chemother. 1987;31:274–280. doi: 10.1128/aac.31.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lavie A, Ostermann N, Brundiers R, Goody RS, Reinstein J, Konrad M, Schlichting I. Structural basis for efficient phosphorylation of 3′-azidothymidine monophosphate by Escherichia coli thymidylate kinase. Proc Natl Acad Sci U S A. 1998;95:14045–14050. doi: 10.1073/pnas.95.24.14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sutera VA, Lovett ST. The role of replication initiation control in promoting survival of replication fork damage. Mol Microbiol. 2006;60:229–239. doi: 10.1111/j.1365-2958.2006.05093.x. [DOI] [PubMed] [Google Scholar]
- [10].Goldfless SJ, Morag AS, Belisle KA, Sutera VA, Lovett ST. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell. 2006;21:595–604. doi: 10.1016/j.molcel.2006.01.025. [DOI] [PubMed] [Google Scholar]
- [11].Foti JJ, Schienda J, Sutera VA, Lovett ST. A bacterial G protein-mediated response to replication arrest. Mol Cell. 2005;17:549–560. doi: 10.1016/j.molcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- [12].Merrikh H, Ferrazzoli AE, Bougdour A, Olivier-Mason A, Lovett ST. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc Natl Acad Sci U S A. 2009;106:611–616. doi: 10.1073/pnas.0803665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mamber SW, Brookshire KW, Forenza S. Induction of the SOS response in Escherichia coli by azidothymidine and dideoxynucleosides. Antimicrob Agents Chemother. 1990;34:1237–1243. doi: 10.1128/aac.34.6.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- [15].Thomas KR, Olivera BM. Processivity of DNA exonucleases. J Biol Chem. 1978;253:424–429. [PubMed] [Google Scholar]
- [16].Richardson CC, Lehman IR, Kornberg A. A Deoxyribonucleic Acid Phosphatase-Exonuclease from Escherichia coli. II. Characterization of the Exonuclease Activity. J Biol Chem. 1964;239:251–258. [PubMed] [Google Scholar]
- [17].Weiss B. Endonuclease II of Escherichia coli is exonuclease III. J Biol Chem. 1976;251:1896–1901. [PubMed] [Google Scholar]
- [18].Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Demple B, Johnson A, Fung D. Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci U S A. 1986;83:7731–7735. doi: 10.1073/pnas.83.20.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Niwa O, Moses RE. Synthesis by DNA polymerase I on bleomycin-treated deoxyribonucleic acid: a requirement for exonuclease III. Biochem. 1981;20:238–244. doi: 10.1021/bi00505a002. [DOI] [PubMed] [Google Scholar]
- [21].Henner WD, Grunberg SM, Haseltine WA. Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J Biol Chem. 1983;258:15198–15205. [PubMed] [Google Scholar]
- [22].Persky NS, Lovett ST. Mechanisms of recombination: lessons from E. coli. Crit Rev Biochem Mol Biol. 2008;43:347–370. doi: 10.1080/10409230802485358. [DOI] [PubMed] [Google Scholar]
- [23].Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Press; New York: 1992. [Google Scholar]
- [24].Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Willetts NS, Clark AJ, Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Van Dyk TK, DeRose EJ, Gonye GE. LuxArray, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J Bacteriol. 2001;183:5496–5505. doi: 10.1128/JB.183.19.5496-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Van Dyk TK, Wei Y, Hanafey MK, Dolan M, Reeve MJ, Rafalski JA, Rothman-Denes LB, LaRossa RA. A genomic approach to gene fusion technology. Proc Natl Acad Sci U S A. 2001;98:2555–2560. doi: 10.1073/pnas.041620498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ferullo DJ, Cooper DL, Moore HR, Lovett ST. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Simmons LA, Foti JJ, Cohen SE, Walker GC. The SOS regulatory network. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, Ussery D, editors. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: Jul 25, 2008. posting date. http://www.ecosal.org. [Google Scholar]
- [31].Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- [32].Lovett ST. The DNA damage response. In: Storz GT, Hengge R, editors. Bacterial Stress Responses. 2nd Edition. American Society for Microbiology; Washington, D. C.: in press. [Google Scholar]
- [33].Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Brandsma JA, Bosch D, de Ruyter M, van de Putte P. Analysis of the regulatory region of the ssb gene of Escherichia coli. Nucleic Acids Res. 1985;13:5095–5109. doi: 10.1093/nar/13.14.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- [36].Kornberg A, Baker TA. DNA replication. second edition W. H. Freeman; 1991. [Google Scholar]
- [37].Huisman O, D’Ari R, Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci U S A. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lovett ST, Clark AJ. Genetic analysis of regulation of the RecF pathway of recombination in Escherichia coli K-12. J Bacteriol. 1983;153:1471–1478. doi: 10.1128/jb.153.3.1471-1478.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cox MM. Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- [40].West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- [41].Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mathews CK, Ji J. DNA precursor asymmetries, replication fidelity, and variable genome evolution. Bioessays. 1992;14:295–301. doi: 10.1002/bies.950140502. [DOI] [PubMed] [Google Scholar]
- [43].Johnson AA, Ray AS, Hanes J, Suo Z, Colacino JM, Anderson KS, Johnson KA. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J Biol Chem. 2001;276:40847–40857. doi: 10.1074/jbc.M106743200. [DOI] [PubMed] [Google Scholar]
- [44].Eriksson S, Xu B, Clayton DA. Efficient incorporation of anti-HIV deoxynucleotides by recombinant yeast mitochondrial DNA polymerase. J Biol Chem. 1995;270:18929–18934. doi: 10.1074/jbc.270.32.18929. [DOI] [PubMed] [Google Scholar]
- [45].Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair (Amst) 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- [46].Chou K-M, Cheng Y-C. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.