Abstract
To broaden the applicability of adoptive T-cell therapy for the treatment of hematologic malignancies, we aim to start a clinical trial using HA-1-TCR transferred virus-specific T cells. TCRs directed against the minor histocompatibility antigen (MiHA) HA-1 are good candidates for TCR gene transfer to treat hematologic malignancies because of the hematopoiesis-restricted expression and favorable frequency of HA-1. For optimal anti-leukemic reactivity, high cell-surface expression of the introduced TCR is important. Previously, however, we have demonstrated that gene transferred HA-1-TCRs are poorly expressed at the cell-surface. In this study several strategies were explored to improve expression of transferred HA-1-TCRs.
Keywords: gene transfer, T-cell receptors, adoptive immunotherapy, TCR optimization
Introduction
Patients with hematologic malignancies can be successfully treated with allogeneic stem cell transplantation (allo-SCT). After allo-SCT, relapse of the hematologic malignancy can occur. This can be successfully treated with donor lymphocyte infusion (DLI) from the original stem cell donor which can result in complete remissions.1,2 This graft-versus-leukemia (GvL) effect of DLI, however, is often accompanied by graft-versus-host disease (GvHD) causing serious mortality and morbidity. It has been demonstrated that donor T cells recognizing minor histocompatibility antigens (MiHA) selectively expressed on hematopoietic cells mediate anti-leukemic reactivity after allo-SCT without causing GvHD.3,4 The MiHA HA-1 is exclusively expressed on hematopoietic cells,5 making it an attractive target antigen to treat hematologic malignancies relapsing after allo-SCT when the patient is HA-1+ and the donor is HA-1-. In addition, HA-1 is presented in the context of HLA-A*02016 and has a favorable population frequency.7,8 In contrast to unmodified DLI, early administration of cytomegalovirus (CMV) or Epstein-Barr virus (EBV)-specific donor T cells does not lead to the development of GvHD. Genetic engineering of donor T cells recognizing viral antigens with HA-1-specific TCRs is, therefore, an attractive strategy to induce anti-leukemic responses without the induction of GvHD. We have previously shown the feasibility of this strategy by the efficient generation of redirected virus-specific T cells transduced with MiHA-specific HA-2-TCRs.9–11 However, transfer of the HA-1-TCR, although of interest for clinical application, did not result in efficient expression at the cell surface of HA-1-TCR engineered T cells.12 In this study, we describe different strategies to improve HA-1-TCR cell-surface expression after TCR gene transfer.
Design and Methods
Construction of HA-1-TCR encoding retroviral vectors
TCRAV and TCRBV gene usage of the HA-1-specific T-cell clone HA1.M7, determined as previously described,13 was demonstrated to be AV32 and BV6S4. Wild-type (WT) and modified TCRα and TCRβ chains were cloned separately into the retroviral vector LZRS, either in combination with eGFP (TCRα) or with NGF-R (TCRβ) marker gene. The single constructs in which the WT and modified HA-1-TCR chains were linked with a picornavirus-derived self-cleaving T2A sequence14 were either expressed in the LZRS combined with the NGF-R marker gene or in the MP71 vector without marker gene. The HA-1-TCRβ chain comprising the CDR1-region of the HA-2.20-TCR15 was obtained by two-step PCR. Primers used are shown in the Online Supplementary Table S1. Codon-optimized TCR genes were designed and produced by GENEART (Regensburg, Germany). In addition, cysteine modified HA-1-TCR chains were constructed as previously described.16 φ-NX-A cells were used to generate viral supernatant as previously described.9,15
HLA class I tetramers and flow cytometric analyses
PE- or APC-conjugated tetramers were constructed as described with minor modifications.17 Tetramers used were composed of HLA-A2 and HA-1 peptide VLHDDLLEA (HA-1), HLA-A1 and CMV-pp50 peptide VTEHDTLLY (pp50 VTE), HLA-B7 and CMV-pp65 TPRVTGGAM (pp65 TPR) or CMV-pp65 RPHERNGFTVL (pp65 RPH) and HLA-B8 and EBV-EBNA3A FLRGRAYGL (EBNA3A FLR). For flow cytometric analyses or cell sorting, cells were labeled with tetramers for 1 h at 4ºC. During the last 30 min, CD4-FITC (Beckton Dickinson [BD], San Diego, CA, USA), CD40-FITC (Bio-connect, Huissen, The Netherlands) or NGF-R-PE [BD] were added. Staining with TCRαβ-PE-Cy5 (Beckman Coulter, Mijdrecht, The Netherlands) or CD3-APC (BD Pharmingen, San Diego, CA, USA) was performed for 30 min at 4ºC.
Isolation of virus-specific T cells and retroviral transduction
All studies were conducted with approval of the institutional review board at Leiden University Medical Center. Virus-specific T cells were isolated from PBMCs of CMV and EBV seropositive individuals. After informed consent, PBMCs were harvested, labeled with the relevant tetramers for 1 h at 4ºC, and sorted at 4ºC using the FACS Vantage (BD) into weak competitor phenotype pp50 VTE or pp65 RPH specific T cells and strong competitor phenotype EBNA3A FLR or pp65 TPR specific T cells with more than 95% purity. Virus-specific T cells were stimulated as described previously18 and transduced two days after stimulation with retroviral supernatant using CH-296 as previously described.18 Marker gene eGFP and NGF-R double positive T cells were subsequently sorted. TCRαβ−/− Jurkat clone 7615 (J76) needed no stimulation prior to transduction.
qRT-PCR
A qRT-PCR was performed to measure the mRNA level of TCRα chain and TCRβ chain. Total RNA was isolated from several T-cell clones, PHA-blasts and as a negative control mesenchymal stem cells (MSCs), using the RNeasy mini kit (Qiagen). First strand cDNA synthesis was performed with oligo dT primers using M-MLV reverse transcriptase (Invitrogen). Samples were run on a 7900HT Fast Real-Time PCR System of Applied Biosystems. Primers used are depicted in the Online Supplementary Table S1. Probes used TET as a dye and TAMRA as a quencher and were chosen over an intron/exon boundary. Each sample was run in duplo with 1 and 10 ng cDNA from 2 μg of total RNA and normalized to the internal Porphobilinogen Deaminase (PBGD) gene. The normalized Ct value of PHA-blasts was set at 1 and expression of other samples was shown referenced to that sample using the following formula [(Cα or CβTCR Ct sample – PBGD Ct sample) / (Cα or CβTCR Ct pha – PBGD Ct pha)].
Cytokine secretion assay and cytotoxicity assay
To test HA-1-specific functionality, 5,000 purified TCR transduced or mock transduced T cells were cocultured with 20,000 different target cells and after overnight incubation specific IFN-γ production was measured by standard ELISA.16 In addition, 50,000 virus-specific T cells were tested one week after transduction with the clinical vector against 20,000 target cells. In the cytotoxicity assay purified TCR transduced or mock transduced T cells were cocultured with different target cells at a 10:1 effector-to-target ratio and cytotoxic reactivity was determined after 4 h.9 The tests were carried out in triplicate. Targets used were HLA-A2+ HA-1+ or HA-1− EBV-transformed lymphoblastoid cell lines (LCLs), and acute myeloid or lymphoblastoid leukemia (AML and ALL, respectively) primary cells.
Results and discussion
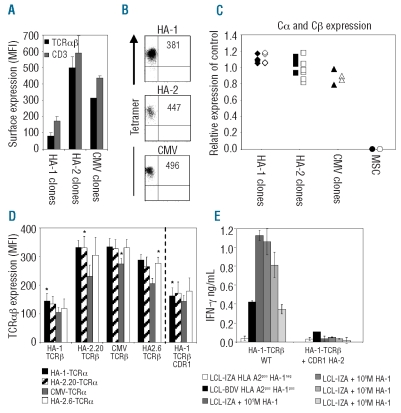
To investigate whether the sub-optimal cell-surface expression of the HA-1-TCR after gene transfer was due to intrinsic properties of the TCR, we determined the HA-1-TCR cell-surface expression of five different parental HA-1-specific T-cell clones derived from different individuals. As demonstrated in Figure 1A, the HA-1-specific T-cell clones expressed significant lower levels of TCR-CD3 complexes at the cell-surface compared to HA-2-15 or CMVA2-specific18 T-cell clones. The HA-1 T-cell clones, however, stained with similar intensity with their respective tetramer to other T-cell clones (Figure 1B), and were fully functional T cells (data not shown). To exclude the possibility that the low TCR expression was due to lower transcriptional activity, TCRα and β mRNA levels were determined, and no significant differences in HA-1-TCRα or β mRNA expression levels compared to other T-cell clones could be detected (Figure 1C ). These results indicate that low HA-1-TCR expression observed in HA-1-TCR transferred T cells is an intrinsic feature of the HA-1-TCR.
Figure 1.
Low HA-1-TCR cell surface expression due to intrinsic properties of the HA-1-TCRβ chain. (A) Several different T-cell clones including 5 HA-1 clones, 6 HA-2 clones, and 2 CMV clones were stained with anti-TCRαβ and anti-CD3 mAbs and analyzed using flow cytometry. Mean fluorescence intensities (MFIs) shown are means of the different clones. (B) The clones were stained with their respective tetramers and MFI is depicted in the dot plots. (C) mRNA levels of TCRα (closed symbols) and TCRβ chains (open symbols) were analyzed for the different clones using q-RT-PCR. As a negative control, cDNA of MSCs was included. (D) J76 cells were transduced with different combinations of TCRα and TCRβ chains of 3 HA-1 clones and 14 other antigen-specific clones.20 TCR cell surface expression was measured by staining with anti-TCRαβ mAbs and analyzing eGFP/NGF-R double positive J76 cells using flow cytometry 5 days after transduc-tion. As a representative example the TCR cell surface expression of 4 TCRα chains derived from the HA-1.M7, HA-2.20, CMV, and HA2.6 clones combined with the TCRβ chains of these clones are depicted in the left panel. In the right panel TCR expression of the 4 TCRα chains combined with the HA-1-TCRβ containing the HA-2-TCRβ CDR1 region are depicted. Non td J76 cells showed little background staining with anti-TCRαβ mAbs (MFI = 16). Parental TCR combinations are indicated with an asterisk. (E) Virus-specific T cells were transduced with the HA-1-TCRα combined with either the unmodified HA-1-TCRβ or exchanged with the HA-2-TCRβ CDR1 region, sorted on bases of eGFP and NGF-R double positivity and subsequently analyzed for HA-1-specific IFN-γ production. Targets used were: HLA-A2+ HA-1− LCL-IZA, HLA-A2+ HA-1+ LCL-BDV and LCL-IZA pulsed with different HA-1-peptide concentrations as indicated in the figure. Data shown are representative for 2 independent experiments.
To investigate which TCR chain was responsible for low TCR expression, TCRαβ-deficient J76 cells15 (J76) were transduced with the individual HA-1-TCRα and β chains derived from 3 different HA-1-specific T-cell clones in combination with 14 different TCRα and β chains derived from different antigen-specific T cells,18 and TCR cell-surface expression was measured. Whereas the HA-1-TCRα chains in combination with any other TCRβ chain resulted in TCR expression comparable to TCR expression of all other parental TCR transduced J76 cells, low TCR expression was observed in all the transductions of the HA-1-TCRβ chains with any of the other TCRα chains, indicating that low HA-1-TCR cell-surface expression was not due to the HA-1-TCRα chain but due to the HA-1-TCR BV6S4 β chain (Figure 1D, left panel). Since there is exclusive TCRBV chain usage of HA-1-specific T cells,19,20 this ruled out the possibility of selecting for other HA-1-TCRs for use in clinical studies.
To improve HA-1-TCR expression after gene transfer, we investigated whether we could determine the specific region of the HA-1-TCR BV6S4 β chain responsible for this low TCR cell-surface expression. For this purpose, the sequences of 4 different TCRβ chains belonging to the BV6 variable domain family and known to exhibit high cell-surface expression after gene transfer were aligned with the sequences of the HA-1-TCR BV6S4 (Online Supplementary Figure S1). Based on these sequence alignments, we hypothesized that primarily the CDR1-region of HA-1-TCR BV6S4 may influence cell-surface expression of the HA-1-TCRβ chain. However, exchange of the HA-1-TCR BV6S4 CDR1-region with the HA-2-TCR BV6S2 CDR1-region did not improve HA-1-TCR cell-surface expression (Figure 1D, right panel), and resulted in an interruption of HA-1-specific reactivity (Figure 1E), indicating that the CDR1-region is modestly involved in low HA-1-TCR cell-surface expression but crucial for HA-1-specificity.
Since apparently HA-1-TCR expression could not be improved by modification of specific sequences of the HA-1-TCRβ chain, other strategies described to improve TCR cell-surface expression of gene transferred TCRs were explored. We studied whether TCR codon optimization21 or cysteine modification22–24 of both the HA-1-TCRα and β chain resulted in potent HA-1-specific T cells after gene transfer into virus-specific T cells known to possess endogenous TCRs which strongly compete for cell-surface expression9 (Figure 2, left panel) and virus-specific T cells known to possess endogenous TCRs which weakly compete for cell-surface expression9 (Figure 2, right panel). Transfer of the unmodified HA-1-TCR into strong competitor EBNA3A FLR T cells (Figure 2A, left panel) resulted in undetectable HA-1-TCR expression, whereas transfer of the unmodified HA-1-TCR into weak competitor pp50 VTE T cells (Figure 2A, right panel) resulted in 40% of HA-1 tetramer positive T cells. Cysteine modification of the HA-1-TCR (combination #6) improved HA-1-TCR expression especially in the strong competitor virus-specific T cells. As expected, cysteine modification of only the HA-1-TCRα (#2) or β chain (#5) significantly diminished HA-1-TCR expression. Codon optimization (#11), in addition, improved HA-1-TCR expression both in strong and weak competitor virus-specific T cells. The combination of codon optimization and cysteine modification (#8) resulted in the most marked improvement in HA-1-TCR expression.
Figure 2.
Analyses of HA-1-TCR cell surface expression and functionality of strong and weak competitor T cells transduced with different modified HA-1-TCRs. Strong competitor phenotype (EBNA3A FLR or pp65 TPR, left panel) or weak competitor phenotype (pp50 VTE or pp65 RPH T-cells, right panel) were transduced with different modified HA-1-TCRs and analyzed for cell surface expression and functionality. (A) T cells were transduced with either unmodified (AV32 WT), cysteine modified (AV32 SS), codon optimized (AV32 opt) or codon optimized and cysteine modified (AV32 opt SS) HA-1-TCRα chains in combination with either unmodified (BV6 WT), cysteine modified (BV6 SS), or codon optimized (BV6 opt) HA-1-TCRβ chains and HA-1 tetramer staining was analyzed. Dot plots depict HA-1 tetramer staining of eGFP and NGF-R double positive virus-specific T cells. (B) The HA-1-TCR td T cells shown in A were tested for HA-1-specific IFN-γ production. Numbers in the figures correspond with the numbers indicated in the dot plots of A. Targets used were HLA-A2+ HA-1- LCL-IZA, HLA-A2+ HA-1+ LCL-BDV and LCL-IZA pulsed with different concentrations of HA-1-peptide. (C) Other strong and weak competitor T cells, as well as total CD8+ T cells were transduced with either a single construct encoding the unmodified (WT td) or the codon optimized and cysteine modified HA-1-TCR chains (opt SS td) and HA-1 tetramer staining was analyzed. Dot plots depict HA-1 tetramer staining of NGF- positive virus-specific T cells. (D–E) T cells transduced with a single construct encoding either HA-1-TCR WT or HA-1-TCR opt SS, or empty vectors were tested against different targets for HA-1-specific cytotoxic reactivity (D) and IFN-γ production (E). Targets used were HLA-A2+ LCLs, AML and ALL primary cells that were either positive or negative for HA-1. (F) T cells transduced with single vectors encoding for unmodified (WT td) HA-1-TCR chains in combination with NGF-R or with the clinical MP71 vector (Clin vect td) HA-1-TCR or empty vector (mock td) were tested one week after transduction for HA-1-specific IFNγ production against HLA-A2+ LCLs, AML and ALL primary cells that were either positive or negative for HA-1. Data shown are representative for 3 independent experiments using T cells of 3 different healthy individuals.
To determine whether improved HA-1-TCR expression resulted in improved HA-1-specific functionality, the transduced T cells were tested against peptide loaded and target cells endogenously expressing the HA-1 antigen (Figure 2B). The combination of codon optimized and cysteine modified HA-1-TCRα chain with the cysteine modified HA-1-TCRβ chain (#8) demonstrated significantly improved HA-1 specific reactivity in strong competitor T cells. In agreement, this TCR combination was able to exert the highest HA-1-specific reactivity also in weak competitor T cells.
To confirm these data, polyclonal peripheral CD8+ T cells, as well as other strong and weak competitor T cells, were transduced with the unmodified or codon optimized and cysteine modified HA-1-TCRs and tested for HA-1-TCR cell-surface expression (Figure 2C). Similarly, transduction with the modified HA-1-TCR resulted in the most efficient cell-surface expression in both strong and weak competitor T cells. The polyclonal CD8+ T cells demonstrated significant HA-1-TCR cell-surface expression after transfer of the modified HA-1-TCR and this was comparable to strong competitor T cells.
To study whether this improved HA-1-TCR cell-surface expression coincided with clinically relevant HA-1-specific functionality, strong and weak competitor phenotype T cells were transduced with single constructs encoding either unmodified HA-1-TCRα and β chains or modified HA-1-TCRα and β chains in combination with NGF-R, and analyzed for HA-1-specific cytotoxic activity (Figure 2D) and IFN-γ production (Figure 2E). Strong competitor T cells transduced with the modified HA-1-TCR demonstrated significant cytotoxic activity and IFN-γ production directed against HA-1+ malignant cells in contrast to T cells transduced with the unmodified HA-1-TCR. In addition, weak competitor T cells transduced with the modified HA-1-TCR demonstrated an increased HA-1-specific reactivity against AML and ALL compared to T cells transduced with the unmodified HA-1-TCR. These results demonstrate that introduction of the modified HA-1-TCR into both strong as well as weak competitor phenotype T cells generated potent redirected HA-1-specific T cells.
For use in clinical therapy, the introduced TCR has to be encoded by a retroviral construct without potentially immunogenic marker genes. Therefore, we constructed a MP71 vector without marker gene encoding the modified HA-1-TCRα and β chain, and analyzed whether strong and weak competitor T cells transduced with this clinical vector demonstrated similarly improved anti-leukemic reactivity (Figure 2F). Both competitor subsets transduced with the clinical vector encoding the modified HA-1-TCR exerted significant anti-leukemic reactivity.
In conclusion, these results illustrate that TCR transfer with a codon optimized and cysteine modified HA-1-TCR resulted in efficient expression of introduced HA-1-TCRs and robust HA-1-specific functionality against clinically relevant target cells, irrespective of the properties of the endogenous TCR. Transduction with this modified HA-1-TCR exhibiting a dominant expression phenotype thus allows for a simple strategy to isolate and transduce CMV-or EBV-specific T-cell populations, without prior selection for weak competitor T cells. Therefore, modified HA-1-TCR transduced virus-specific T cells will be used for a gene therapy trial in patients with leukemia.
Acknowledgments
The authors thank Michel Kester, Dirk van der Steen, Guido de Roo and Menno van der Hoorn for expert technical assistance.
Footnotes
Funding: this work was supported by the Dutch Cancer Society (grant NKB 2005-3251 and NKB 2007-3927) and ZonMw (grant ZonMw 433.00.001).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.de Bueger M, Bakker A, van Rood JJ, Van der Woude F, Goulmy E. Tissue distribution of human minor histocompatibility antigens. Ubiquitous versus restricted tissue distribution indicates heterogeneity among human cytotoxic T lymphocyte-defined non-MHC antigens. J Immunol. 1992;149 (5):1788–94. [PubMed] [Google Scholar]
- 2.Klein CA, Wilke M, Pool J, Vermeulen C, Blokland E, Burghart E, et al. The hematopoietic system-specific minor histocompatibility antigen HA-1 shows aberrant expression in epithelial cancer cells. J Exp Med. 2002;196(3):359–68. doi: 10.1084/jem.20011838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mommaas B, van Halteren AG, Pool J, van der Veken L, Wieles B, Heemskerk MH, et al. Adult and cord blood T cells can acquire HA-1 specificity through HA-1 T-cell receptor gene transfer. Haematologica. 2005;90 (10):1415–21. [PubMed] [Google Scholar]
- 4.Collins RH, Jr, Shpilberg O, Drobyski WR, Porter DL, Giralt S, Champlin R, et al. Donor leukocyte infusions in 140 patients with relapsed malignancy after allogeneic bone marrow transplantation. J Clin Oncol. 1997;15(2):433–44. doi: 10.1200/JCO.1997.15.2.433. [DOI] [PubMed] [Google Scholar]
- 5.Kolb HJ, Mittermuller J, Clemm C, Holler E, Ledderose G, Brehm G, et al. Donor leukocyte transfusions for treatment of recurrent chronic myelogenous leukemia in marrow transplant patients. Blood. 1990;76 (12):2462–5. [PubMed] [Google Scholar]
- 6.Marijt WA, Kernan NA, az-Barrientos T, Veenhof WF, O'Reilly RJ, Willemze R, et al. Multiple minor histocompatibility antigen-specific cytotoxic T lymphocyte clones can be generated during graft rejection after HLA-identical bone marrow transplantation. Bone Marrow Transplant. 1995;16(1):125–32. [PubMed] [Google Scholar]
- 7.Marijt WA, Heemskerk MH, Kloosterboer FM, Goulmy E, Kester MG, van der Hoorn MA, et al. Hematopoiesis-restricted minor histocompatibility antigens HA-1- or HA-2-specific T cells can induce complete remissions of relapsed leukemia. Proc Natl Acad Sci USA. 2003;100(5):2742–7. doi: 10.1073/pnas.0530192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Haan JM, Sherman NE, Blokland E, Huczko E, Koning F, Drijfhout JW, et al. Identification of a graft versus host disease-associated human minor histocompatibility antigen. Science. 1995;268(5216):1476–80. doi: 10.1126/science.7539551. [DOI] [PubMed] [Google Scholar]
- 9.Spierings E, Hendriks M, Absi L, Canossi A, Chhaya S, Crowley J, et al. Phenotype frequencies of autosomal minor histocompatibility antigens display significant differences among populations. PLoS Genet. 2007;3(6):e103. doi: 10.1371/journal.pgen.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffioen M, van Egmond HM, Barnby-Porritt H, van der Hoorn MA, Hagedoorn RS, Kester MG, et al. Genetic engineering of virus-specific T cells with T-cell receptors recognizing minor histocompatibility antigens for clinical application. Haematologica. 2008;93(10):1535–43. doi: 10.3324/haematol.13067. [DOI] [PubMed] [Google Scholar]
- 11.Heemskerk MH, Hoogeboom M, Hagedoorn R, Kester MG, Willemze R, Falkenburg JH. Reprogramming of virus-specific T cells into leukemia-reactive T cells using T cell receptor gene transfer. J Exp Med. 2004;199(7):885–94. doi: 10.1084/jem.20031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Loenen MM, Hagedoorn RS, Kester MG, Hoogeboom M, Willemze R, Falkenburg JH, et al. Kinetic preservation of dual specificity of coprogrammed minor histocompatibility antigen-reactive virus-specific T cells. Cancer Res. 2009;69(5):2034–41. doi: 10.1158/0008-5472.CAN-08-2523. [DOI] [PubMed] [Google Scholar]
- 13.van der Veken LT, Hoogeboom M, de Paus RA, Willemze R, Falkenburg JH, Heemskerk MH. HLA class II restricted T-cell receptor gene transfer generates CD4+ T cells with helper activity as well as cytotoxic capacity. Gene Ther. 2005;12(23):1686–95. doi: 10.1038/sj.gt.3302586. [DOI] [PubMed] [Google Scholar]
- 14.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589–94. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 15.Heemskerk MH, Hoogeboom M, de Paus RA, Kester MG, van der Hoorn MA, Goulmy E, et al. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood. 2003;102(10):3530–40. doi: 10.1182/blood-2003-05-1524. [DOI] [PubMed] [Google Scholar]
- 16.Boulter JM, Glick M, Todorov PT, Baston E, Sami M, Rizkallah P, et al. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 2003;16(9):707–11. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- 17.Cohen CJ, Li YF, El-Gamil M, Robbins PF, Rosenberg SA, Morgan RA. Enhanced anti-tumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 2007;67(8):3898–903. doi: 10.1158/0008-5472.CAN-06-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuball J, Dossett ML, Wolfl M, Ho WY, Voss RH, Fowler C, et al. Facilitating matched pairing and expression of TCR chains introduced into human T cells. Blood. 2007;109 (6):2331–8. doi: 10.1182/blood-2006-05-023069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burrows SR, Kienzle N, Winterhalter A, Bharadwaj M, Altman JD, Brooks A. Peptide-MHC class I tetrameric complexes display exquisite ligand specificity. J Immunol. 2000;165(11):6229–34. doi: 10.4049/jimmunol.165.11.6229. [DOI] [PubMed] [Google Scholar]
- 20.Heemskerk MH, Hagedoorn RS, van der Hoorn MA, van der Veken LT, Hoogeboom M, Kester MG, et al. Efficiency of T-cell receptor expression in dual-specific T cells is controlled by the intrinsic qualities of the TCR chains within the TCR-CD3 complex. Blood. 2007;109(1):235–43. doi: 10.1182/blood-2006-03-013318. [DOI] [PubMed] [Google Scholar]
- 21.van Loenen MM, de Boer R, Amir AL, Hagedoorn RS, Volbeda GL, Willemze R, et al. Mixed T cell receptor dimers harbor potentially harmful neoreactivity. Proc Natl Acad Sci USA. 2010;107(24):10972–7. doi: 10.1073/pnas.1005802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goulmy E, Pool J, van den Elsen PJ. Interindividual conservation of T-cell receptor beta chain variable regions by minor histocompatibility antigen-specific HLA-A*0201-restricted cytotoxic T-cell clones. Blood. 1995;85(9):2478–81. [PubMed] [Google Scholar]
- 23.Verdijk RM, Mutis T, Wilke M, Pool J, Schrama E, Brand A, et al. Exclusive TCRVbeta chain usage of ex vivo generated minor Histocompatibility antigen HA-1 specific cytotoxic T cells: implications for monitoring of immunotherapy of leukemia by TCRBV spectratyping. Hematol J. 2002;3(6):271–5. doi: 10.1038/sj.thj.6200197. [DOI] [PubMed] [Google Scholar]
- 24.Scholten KB, Kramer D, Kueter EW, Graf M, Schoedl T, Meijer CJ, et al. Codon modification of T cell receptors allows enhanced functional expression in transgenic human T cells. Clin Immunol. 2006;119(2):135–45. doi: 10.1016/j.clim.2005.12.009. [DOI] [PubMed] [Google Scholar]