Abstract
The fragile X mental retardation 1 (FMR1) gene contains a CGG repeat within its 5′ untranslated region (5′UTR) that, when expanded to 55–200 CGG repeats (premutation allele), can result in the late-onset neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome. The CGG-repeat is expected to form highly-stable secondary structure capable of inhibiting 5′-cap-dependent translation. Paradoxically, translation in vivo is only mildly impaired within the premutation range, suggesting that other modes of translational initiation may be operating. To address this issue, a set of reporter mRNAs containing between 0 and 99 CGG repeats, in either native (FMR1) or unrelated (heterologous) 5′UTR context, were translated in vitro. The 5′-cap dependence of translation was assessed by inserting a stable hairpin near the 5′ end of the mRNAs. Results of the current studies indicate that translational initiation of the FMR1 mRNA occurs primarily by scanning, with little evidence of internal ribosome entry or shunting. Additionally, the efficiency of translational initiation depends on transcription start site selection, with the shorter 5′UTR (downstream transcription start site I) translating with greater efficiency than the longer mRNA (start site III) for all CGG-repeat elements studied. Lastly, a hairpin previously shown to block translation gave differing results depending on 5′UTR context, in one case initiating translation from within the hairpin.
Keywords: Fragile X syndrome, FXTAS, premutation, autism, neurodegeneration
INTRODUCTION
The 5′ untranslated region (5′UTR) of the human fragile X mental retardation 1 (FMR1) gene (OMIM ID *309550) harbors a CGG repeat that may expand generationally.1; 2 Whereas the general population has fewer than 45 CGG repeats (mode ~30 CGG repeats), full mutation allelic expansions (>200 CGG repeats) are normally accompanied by FMR1 gene silencing3; 4; 5; 6 and loss of FMR1 protein (FMRP).7; 8; 9 This absence of FMRP gives rise to fragile X syndrome, the most common known heritable form of intellectual impairment and leading single-gene form of autism. (Reviews: 8; 10) Smaller “premutation” expansions (55 to 200 CGG repeats) are also associated with increased risk of developmental delay and autism.11; 12 Premutation expansions are additionally linked to two premutation-specific disorders, primary ovarian insufficiency (POI), with loss of ovarian function before the age of 4013; 14; 15; 16; and the neurodegenerative disorder fragile X-associated tremor/ataxia syndrome (FXTAS),17; 18; 19 which involves the core features of intention tremor and gait ataxia and, more variably, cognitive decline, parkinsonism, and peripheral neuropathy. (Reviewed in 20; 21) In contrast to full-mutation alleles, which are generally transcriptionally silent, premutation alleles express up to 8 times more FMR1 mRNA than normal alleles.22; 23; 24; 25; 26 Though the mechanism is not well understood, FXTAS, and perhaps also POI, is thought to be due to a toxic gain-of-function of the expanded-CGG-repeat mRNA.17; 27; 28; 29
The CGG repeat is representative of a growing number of known trinucleotide repeat disorders that include both non-coding repeats (e.g., CTG, myotonic dystrophy; GAA, Friedreich’s ataxia; CGG, fragile X syndrome) and protein-coding repeats (e.g. CAG, Huntington’s disease, spinocerebellar ataxias).30; 31 Because the CGG repeat of the FMR1 mRNA lies outside of the coding region, it does not have a direct effect on the composition of FMRP; however, the location of this structure-forming CG-rich element in the 5′UTR of the mRNA directly affects the efficiency of FMRP production.
Ribo-CGG repeats have been shown experimentally to base-pair intramolecularly and to form hairpin-like secondary structures of C-G and G-G base pairs in vitro.32; 33; 34; 35 Some investigators have also reported tetraplex formation by CGG-repeat RNA.36; 37 This highly-GC-rich secondary structure is thought to inhibit the translation of expanded-repeat FMR1 mRNAs, since strong secondary structure in the 5′UTR can inhibit ribosomal scanning.38; 39; 40 Secondary structures with free energies of stabilization more negative than approximately −50 kcal/mol greatly inhibit translation initiation.39; 41 By comparison, CGG repeats at the lower end of the premutation range have estimated free energies below −100 kcal/mol; a repeat of 55 CGGs has an in silico estimated free energy of stabilization of −117 kcal/mol.42; 43 Therefore, CGG repeats are predicted to pose a substantial energetic barrier for scanning of the FMR1 5′UTR by the 40S ribosome and associated proteins. In addition to the repeat tract, the FMR1 5′UTR otherwise is relatively long (198 bases) and GC-rich (77%).
Paradoxically, patients with mRNAs carrying 50 to 100 CGG repeats have only slightly reduced levels of FMRP.22; 26; 44; 45 Furthermore, polysome-profile analysis of lymphoblastoid cells carrying 97 CGG repeats shows a substantial amount (58%) of FMR1 mRNA in polysomes, suggesting maintenance of at least a moderate rate of protein synthesis.44 In this regard, transient transfections of mammalian cells with plasmids containing 99 CGG repeats resulted in only a ~ 50% loss in reporter translation efficiency relative to 30 CGG-repeat constructs.46; 47 Thus, it remains a fundamental puzzle as to why FMR1 messages in the premutation range can be translated while harboring substantial 5′UTR secondary structures.
It is possible that canonical “ribosomal scanning” (reviewed in 48) is not operational for initiation of translation of the FMR1 mRNA; alternative models of translation initiation have been developed in order to interpret unusual forms of initiation not compatible with scanning. For example, the higher-order RNA structures within internal ribosomal entry sites (IRESs) are thought to have an intrinsic ability to directly bind the 40S ribosome, at or near a transcript’s AUG start codon, without 5′-m7G cap binding or ribosomal scanning.49 Often seen in viral50; 51 as well as cell cycle and apoptotic messages,52; 53 IRESs facilitate protein synthesis in a 5′ cap-independent manner.
Another alternative to ribosomal scanning is ribosomal shunting, a rare mechanism of initiation that is primarily seen in plant viruses.54; 55 Although shunting does require 5′ cap binding by ribosomal initiation factors, the ribosome is able to bypass large, highly-structured RNA domains within the 5′UTR, usually by way of initiation at upstream open reading frames, followed by reinitiation downstream of the structured domain. Since the FMR1 5′UTR is long and highly structured, especially messages with premutation-length CGG repeats, this alternative form of translation initiation in principle could allow for more efficient translation of FMR1.
To address the issue of translational initiation for the FMR1 mRNA, we have examined the above alternative mechanisms. Our results confirm earlier observations22; 44; 47; 56 that CGG repeats inhibit translation initiation in a length-dependent manner. Moreover, when the CGG repeat is placed in an unrelated (heterologous) 5′UTR context, a similar effect is seen. Replacing the CGG repeat with a double-stranded hairpin completely blocks translation in a heterologous 5′UTR, whereas the same hairpin decreases by about 5-fold, but does not abolish translation in the FMR1 context. We provide evidence that translation of FMR1 is 5′cap-dependent, does not substantially involve IRES-mediated initiation, and most likely occurs by ribosomal scanning. We also give possible explanations for hairpin read-through in the FMR1 context.
RESULTS
Plasmid construction
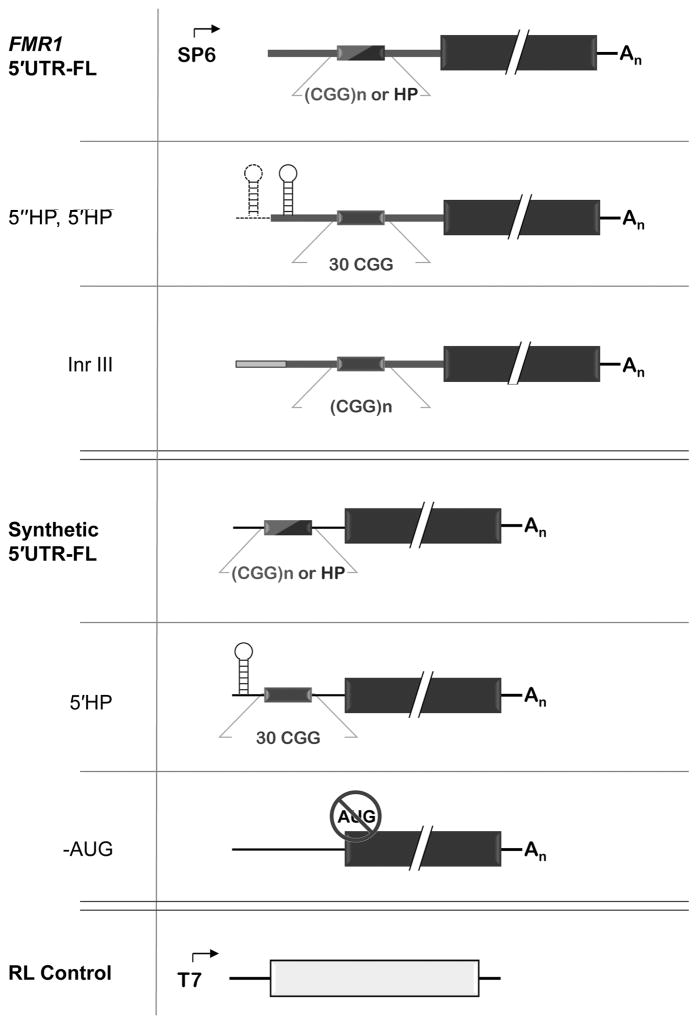
To study the effect of CGG repeats on translation initiation, firefly luciferase (FL) reporter plasmids with varying numbers of CGG repeats within either the FMR1 5′UTR or a heterologous 5′UTR were constructed (Figure 1, Table 1, and Supporting Material). The heterologous (synthetic) 5′UTR was designed as a non-native sequence of 82 bases, created to have minimal secondary structure (−14.5 kcal/mol predicted free energy), moderate length, as well as restriction sites that facilitate insertion of CGG repeats. In addition to synthetic 5′UTR-FL plasmids, we also constructed FL plasmids preceded by the FMR1 5′UTR, starting at FMR1 transcription start site (Initiator region; Inr) I.57 All plasmids were created with the SP6 promoter for in vitro expression and, depending on the 5′UTR employed, are called synthetic 5′UTR(n CGG)-FL or FMR15′UTR(n CGG)-FL.
Figure 1.
FL (dark bars) and RL constructs (light bar). CGG repeats are located within FMR1 and synthetic 5′UTRs and have the following CGG-repeat lengths (n): 0, 16, 30, 62, 99 CGG. FMR15′UTR(HP)-FL and synthetic 5′UTR(HP)-FL have a strong hairpin (HP) replacing the CGG element. 5′HP-FMR15′UTR(30 CGG)-FL, 5″HP-FMR15′UTR(30 CGG)-FL, and 5′HP-synthetic 5′UTR(30 CGG)-FL each have a strong HP at the 5′ ends of the mRNAs. FL and RL messages are transcribed via SP6 and T7 promoters, respectively.
Table 1.
FMR1- and synthetic-5′UTR FL mRNAs used in the current study. All FL mRNAs were generated by SP6-promoted in vitro transcription and are 5′ m7G-capped
| RNA Product | Mid-5′UTR Insert | Additional Modifications |
|---|---|---|
| Inr I; FMR15′UTR(n CGG or HP)-FL | 0, 16, 30, 62, 99 CGGs; HP, HP(GAG) | |
| Inr III; FMR15′UTR-FL(n CGG)-FL | 0, 16, 30, 62, 99 CGGs | |
| Synthetic 5′UTR(n CGG or HP)-FL | 0, 16, 30, 62, 99 CGGs; HP, HP(GUG/no stop) | |
| Inr I; 5′HP-FMR15′UTR(30 CGG)-FL | 30 CGGs | 5′HP, 5″HP |
| 5′HP-Synthetic 5′UTR(30 CGG)-FL | 30 CGGs | 5′HP |
| Synthetic 5′UTR(0 CGG)-FL[-AUG] | 0 CGGs | k.o. AUG start codons |
CGG repeats were inserted into both 5′UTR contexts, either midway through the synthetic 5′UTR or into their native location within the 5′UTR (about two-thirds of the way to the native ATG start codon). The repeats span the normal range (0, 16, 32 CGG) through the lower end of the premutation range (62, 99 CGG). Following in vitro transcription, each message possesses a 5′ cap, synthetic or FMR1 5′UTR, FL reporter, and polyA tail.
CGG-repeat length negatively correlates with translation efficiency in vitro
To assess the effect of CGG repeats outside of their native 5′UTR context, varying CGG-repeat lengths were placed in the synthetic 5′UTR and translation efficiency measured in vitro. Our rationale for using the in vitro (RRL) system for the current studies was twofold. First, in initial studies using the RRL system, we observed that the extent of reduction of reporter (luciferase) protein with increasing CGG repeat was comparable to the reductions observed for either in vivo transfection experiments (SK-N-MC or HEK293 cell lines47) or for native FMRP in cultured premutation cell lines.22; 44 Second, the in vitro system, despite its own caveats, allows for tighter control of RNA levels, for a wide variety of RNA constructs, than would be possible for an in vivo system.
In the current experiments, equimolar amounts (0.2 pmol) of capped synthetic-5′UTR messages with either 0, 16, 30, 62, or 99 CGG repeats were translated in rabbit reticulocyte lysate (RRL), followed by termination of the reactions with passive lysis buffer. Messages were translated within the linear range of the lysate, previously determined to be 20 minutes at this mRNA concentration (data not shown). In addition to FL mRNAs, an internal control Renilla luciferase (RL) mRNA was added to each translation reaction at a 1:40 ratio of FL:RL. Following in vitro translation, FL and RL protein levels were measured in a dual luciferase experiment, which measures the chemiluminescent activities of the FL and RL enzymes produced by each translation reaction. FL values were normalized to RL internal control values, giving relative FL translation efficiencies for each mRNA.
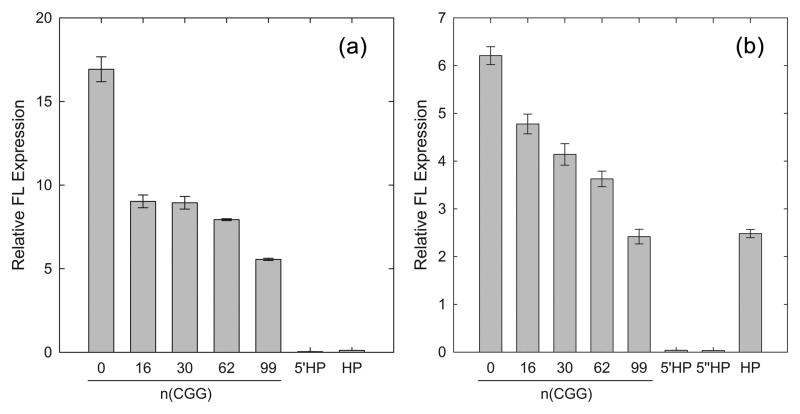
As can be seen in Figure 2A, translation of synthetic-5′UTR mRNAs is reduced for all CGG-repeat lengths. The addition of only 16 CGGs drops the translation efficiency of synthetic 5′UTR(0 CGG)-FL by approximately two-fold, after which the degree of inhibition with increasing numbers of repeats is more gradual. As with the synthetic 5′UTR constructs, translation of FMR1 mRNAs also shows a decrease in translation efficiency with increasing numbers of CGG repeats (Figure 2B), in agreement with previous studies in which increased CGG-repeat length within the FMR1 5′UTR resulted in decreased translation efficiency.22; 44; 47 However, in comparison to translations of synthetic-5′UTR mRNAs, the inhibition of FMR1 mRNAs is more gradual with increasing repeat length. The addition of 16 CGG repeats results in only a 23% decrease in FL expression relative to the FMR1 5′UTR(0 CGG)-FL construct, followed by an incremental decrease in translation efficiency with additional CGG repeats. Note, however, that translation of each synthetic-5′UTR mRNA species is more efficient by 2- to 3-fold than the corresponding CGG repeat in the FMR1 5′UTR, in accord with more favorable translation of an otherwise unstructured synthetic 5′UTR.
Figure 2.
In vitro translation of capped synthetic and FMR1 5′UTR messages. (A) Synthetic 5′UTR(n CGG or HP)-FL and 5′HP mRNAs. (B) FMR15′UTR(n CGG or HP)-FL, 5′HP and 5″HP mRNAs. All experiments were performed at least in triplicate. Relative FL expression levels were obtained by dividing each FL measurement by the corresponding RL measurement within the same in vitro translation reaction. Using t-tests, all expression levels (within either the FMR1 or synthetic 5′UTR) were significantly different from each other with p<.001, except for the following: FMR1 99 CGG vs. HP p=.414. Synthetic 16 CGG vs. 30 CGG p=.709.
In contrast to the expectation based on the computed free energies (Table 2), the reduction in translational efficiency of FMR1 mRNAs is quite gradual within the premutation range, with the 99-CGG-repeat construct translating at a rate reduced by only 60% relative to 0 repeats despite a high predicted free energy of stabilization (−227 kcal/mol; Table 2). The FMR1 5′UTR, at 198 nt excluding the CGG repeats, is unusually long, GC-rich, and highly structured, with a predicted free energy estimated to be around −100 kcal/mol. Thus, compared to shorter, less structured 5′UTRs, the 43S ribosome pre-initiation complex is predicted to experience greater impairment of scanning through the FMR1 5′UTR than the synthetic 5′UTR, both with and without CGG repeats.
Table 2.
Predicteda free energies of CGG- and HP-RNA secondary structures
| 5′UTR Insert | kcal/mol |
|---|---|
| 0 CGG | 0 |
| 16 CGG | −29.9 |
| 30 CGG | −63.5 |
| 62 CGG | −140.3 |
| 99 CGG | −226.7 |
| HP | −45.3 |
| Synthetic UTR | −14.5 |
| FMR1 UTR | −105.0 |
Free energies were estimated using the Zucker algorithm (http:://mfold.bioinfo.rpi.edu).42; 43
Initiation of FMR1 translation is predominantly 5′ end-dependent
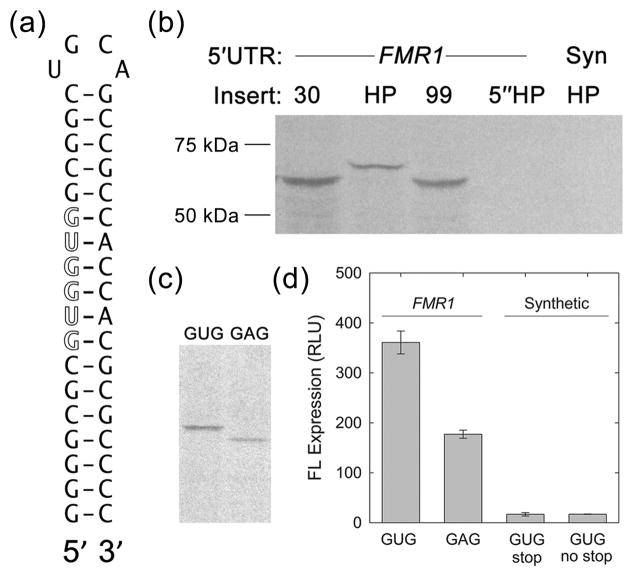
To determine whether the seemingly paradoxical experimental results, given the predicted structural stability of the CGG-repeat element, might be due to 5′-end-independent translational initiation, we investigated the 5′ end-dependence of FMR1 translation. Initiation-factor interactions with the 5′ end of the mRNA are required for translation initiation in both scanning and shunting mechanisms, but not for mRNAs that utilize an internal ribosome entry site (IRES).48 Structures located within 15 nt of the 5′-terminal m7G cap are expected to be especially inhibitory, as they prevent cap recognition by, and initial binding of, the initiation factor eIF4F.39; 58; 59 Accordingly, a fully base-paired hairpin (HP) was inserted 13 nt from the 5′ ends of both synthetic and FMR1 5′UTRs containing 30 CGG repeats [5′HP-synthetic 5′UTR(30 CGG)-FL and 5′HP-FMR15′UTR(30 CGG)-FL]. The 40-nt hairpin sequence, originally described by Kozak as being capable of blocking translational initiation,39 possesses a stem of ~18–20 bp (unknown loop size) and a predicted −45 kcal/mol free energy of stabilization (Table 2; hairpin depicted in Figure 3A). Translation of 5′-HP-containing mRNAs was completely blocked for both synthetic and FMR1 5′UTRs (Figures 2A and B, 5′HP), thus ruling out an IRES-based mechanism as a major mode of initiation, in accord with earlier studies.60; 61
Figure 3.
Upstream initiation from GUG codon(s) in the HP FMR1 context. (A) Sequence and presumed structure of HP. GUG sequence is indicated in outlined font. (B) Autoradiograph of [35S] methionine-labeled, in vitro-translated FL protein products separated by SDS-PAGE. FMR15′UTR(HP)-FL initiates translation early and runs high, while there is no product from either 5″HP-FMR15′UTR(30 CGG)-FL or 5′HP-synthetic 5′UTR(30 CGG)-FL constructs. Abbreviation: Syn, Synthetic. (C) Autoradiograph of [35S]-labeled proteins produced from FMR15′UTR(HP)-FL mRNAs with and without GUG codons. U-to-A changes in the GUG sequences within the hairpin results in normal-length protein product. (D) In vitro translation of FMR1- and synthetic-5′UTR mRNAs with internal HPs. Results are given in RLUs of FL expression and are not normalized to RL levels. GUG: FMR15′UTR(HP)-FL; GAG: FMR15′UTR(HP/GAG)-FL; GUG (stop): synthetic 5′UTR(HP)-FL; GUG (no stop): synthetic 5′UTR(HP/GUG)[no stop]-FL. All messages were translated at least in triplicate.
To account for the possibility that inserting the hairpin into the FMR1 5′UTR disrupted sequence and/or secondary structure necessary for alternative forms of translation, an additional FMR1 5′UTR construct (5″HP-FMR1 5′UTR(30 CGG)-FL) was generated in which the hairpin was added as an extension onto the 5′ end of the message (5″HP). The HP in this extension begins 12 bases from the new 5′ end of the mRNA. Translation of this 5′-extended message was also blocked (Figure 2B, 5″HP).
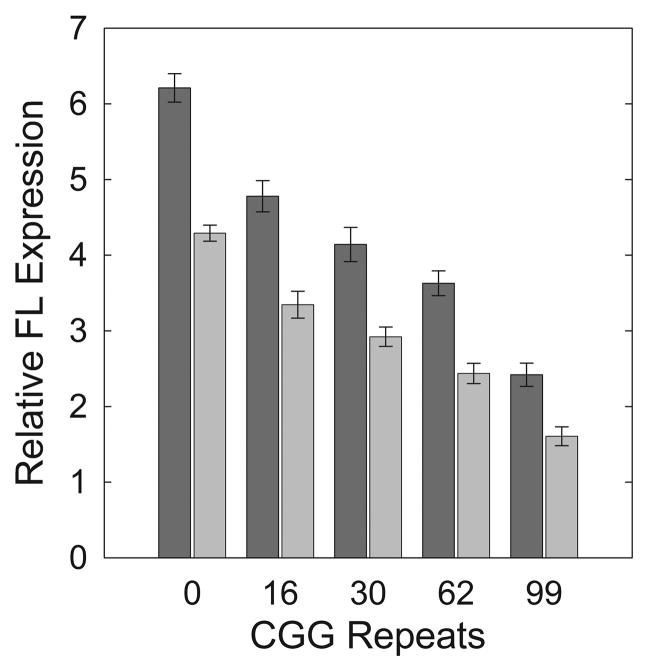
Figure 5.
Effect of transcription start site (length of 5′UTR 5′ to the CGG repeat) on translation efficiency of FMR1 message. In vitro translations of FMR15′UTR(n CGG)-FL (Inr I) and FMR1(InrIII)5′UTR(n CGG)-FL mRNAs with either 0, 16, 30, 62, or 99 CGG repeats. mRNAs were transcribed from start site I (dark bars) or start site III (light bars). T-tests of site I vs. site III for each CGG-repeat length were significant, with all P<0.001. Each translation was performed in quadruplicate.
A stable hairpin within the 5′UTR of FMR1 does not block translation
Highly-stable secondary/tertiary structure within the 5′UTR region is expected to block translation initiation that occurs via scanning.48 In the current instance, whereas the CGG repeats are predicted to form strong hairpin-like secondary structures, such hairpins would necessarily possess non-canonical (e.g., GG) base pairs at every third stem position. While such non-canonical pairings can and do occur,33 they may lower the overall stabilization free energy density (i.e., kcal/mol/bp), thereby still permitting scanning/initiation.
To test whether a lower-than-predicted relative stability of the CGG repeat might be permitting at least some scanning, we first replaced the CGG repeat sequence, which lies approximately two-thirds of the way toward the 3′ end of the FMR1 5′UTR, with the HP used above for 5′-dependency experiments (Figure 2B, HP). We also replaced the CGG repeats in the synthetic 5′UTR with the same HP element. In vitro (RRL) translation of FMR15′UTR(HP)-FL and synthetic 5′UTR(HP)-FL mRNAs demonstrate that the mid-5′UTR HP blocked translation of the synthetic-5′UTR mRNA, as expected. However, the HP only partially blocked translation within the FMR1 5′UTR context (Figure 2B) - to a degree comparable to the 99-CGG-repeat FMR1 5′UTR, or a 60% reduction vs. no insert. These results further support our conclusions that initiation occurs via scanning, a 5′-dependent mechanism that is sensitive to the stabilities of elements of RNA structure within the 5′UTR (e.g., HP; CGG-repeat length). This latter observation is not consistent with a pure shunting mechanism (see Discussion).
In the FMR1 5′UTR, the central HP lies 130 nt from the 5′ end of the message, whereas this distance is 40 nt in the synthetic 5′UTR (synthetic 5′UTR(HP)-FL). As mentioned, hairpins very near to the 5′ ends of messages have been shown to inhibit translation initiation.39 However, the mid-UTR hairpins in both FMR1 or synthetic 5′UTRs are believed to be at sufficient distances from the 5′ cap as to not inhibit loading of the 40S ribosome, since a hairpin 23 bases from the 5′ end was previously shown to not inhibit translation initiation.62
To better understand the difference in behavior between HP and the 99-CGG repeat in the FMR1 context, we sought to visualize the FL protein product produced in RRL translations. [35S]methionine was added to in vitro translation reactions, which were analyzed by SDS-PAGE/autoradiography (Figure 3B). Surprisingly, translation of FMR1 5′UTR(HP)-FL mRNA resulted in a larger protein product than other FL translation reactions, indicating upstream initiation from this mRNA. By contrast, the FMR1 constructs with both 30 and 99 CGGs initiate from the canonical AUG; whereas neither the 5″HP-FMR1 5′UTR(30 CGG)-FL nor the synthetic 5′UTR(HP)-FL mRNAs yielded any protein product, consistent with a block in 5′-end-dependent translation.
Closer examination of the HP sequence revealed two GUGs in the 5′ strand of the HP stem (Figure 3A), which could theoretically be the site of upstream initiation seen in the FMR1 context. Translation initiation primarily occurs from the most 5′ AUG, but can also (less commonly) initiate at CUG or GUG sites.63; 64 Indeed, the two upstream GUG codons are both in frame with FL, without any intervening stop codons. When the GUG sequences in the hairpin were modified to GAGs (and the opposite stem modified to CUC to preserve the hairpin’s structure and stability), upstream initiation ceased, as can be seen by normal-length FL product (FMR15′UTR(HP/GAG)-FL) (Figure 3C). Evidence that aberrant initiation occurred at a GUG within the HP are as follows: 1) Other than changing the GUG sequence itself (U-to-A), there were only 2 other nt changes to the RNA (A-to-U on the 3′ side of the HP) which led to elimination of the early-initiation product, suggesting that GUGs are required for upstream initiation; 2) Sequence changes did not result in loss of other putative start sites in the 5′UTR; 3) The size of the upstream-initiation protein product corresponds to size of predicted GUG-initiated product (64.1 kDa vs. original 60.6 kDa). Thus, the larger translation product was the result of initiation at GUG codon(s) within the HP, establishing that unwinding of the HP does occur, again consistent with scanning. The lack of a normal-length FL protein from FMR1 5′UTR(HP)-FL mRNA, in either Figure 3B or 3C, indicates that all of the scanning ribosomes initiate within the HP, with no leaky scanning occurring. Once the sequence of the hairpin was altered to prevent upstream initiation, the translation efficiency of the FMR15′UTR(HP/GAG)-FL mRNA decreased by an additional ~50%, to about 20% of the efficiency observed for the FMR15′UTR(0 CGG)-FL message (Figure 3D, FMR1 GAG).
We had originally designed the HP mRNAs to block translation initiation, based on the early work of Kozak.39 Since the HP does not block translation in the FMR1 5′UTR context, instead initiating translation within the 5′ stem of the HP, it was important to determine whether continued translation was due to a feature intrinsic to the HP itself (i.e., cryptic start codon), or whether it was specifically a result of the FMR1 5′UTR context. Indeed, in the synthetic 5′UTR context, the GUG codons within the mid-UTR HP are in frame with the FL reporter; however, there is a stop codon between the two GUGs and the FL coding region. Thus, if initiation had occurred at these uGUG sites in the synthetic 5′UTR, an 18-amino acid peptide would have been produced and would not have resulted in either visible full-length FL or FL activity.
To resolve this last ambiguity, synthetic 5′UTR(HP)-FL was modified to detect possible upstream initiation at the HP by removing the intervening stop codon (changing the TAG/stop to TTA/Leu) between the two GUGs and FL [synthetic 5′UTR(HP/GUG)[no stop]-FL]. Attempted translation of this construct resulted in no FL production, either in luciferase activity per se or [35S]protein product (Figure 3D, synthetic GUG no stop; and data not shown). We thus conclude that the ability to translate through the HP depends not only on the sequence intrinsic to the HP itself, but also on its RNA context (synthetic vs. FMR1).
Translational initiation is occurring at the first AUG, located at the start of the FL reporter
As an additional control to establish that translation is initiating at the expected 5′ AUG codon of the FL-reporter coding sequence (i.e., not directed at downstream AUG codons), we knocked out the first two AUG start codons in synthetic 5′UTR(0 CGG)-FL by site-directed mutagenesis targeted to the plasmid, thereby forcing the ribosome to initiate at the next in-frame AUG (87 bases/29 amino acids downstream). The second knocked-out AUG was out of frame, and would therefore not produce active FL.
Following in vitro translation, the original synthetic 5′UTR(0 CGG)-FL message resulted in an average of 603 relative light units (RLUs) per FL-activity measurement, while the same [-AUG] message had only 0.6 RLUs, indicating that active FL in our translations must be the result of initiation at the standard AUG start codon. If initiation were to occur at a downstream AUG, the FL produced would be inactive, and not measured in our dual luciferase experiments. [35S]methionine incorporation confirmed lack of full-length FL from [-AUG] mRNAs (Figure 4). The ribosome does additionally initiate at several locations further downstream, which can be seen as lower bands in the autoradiograph.
Figure 4.
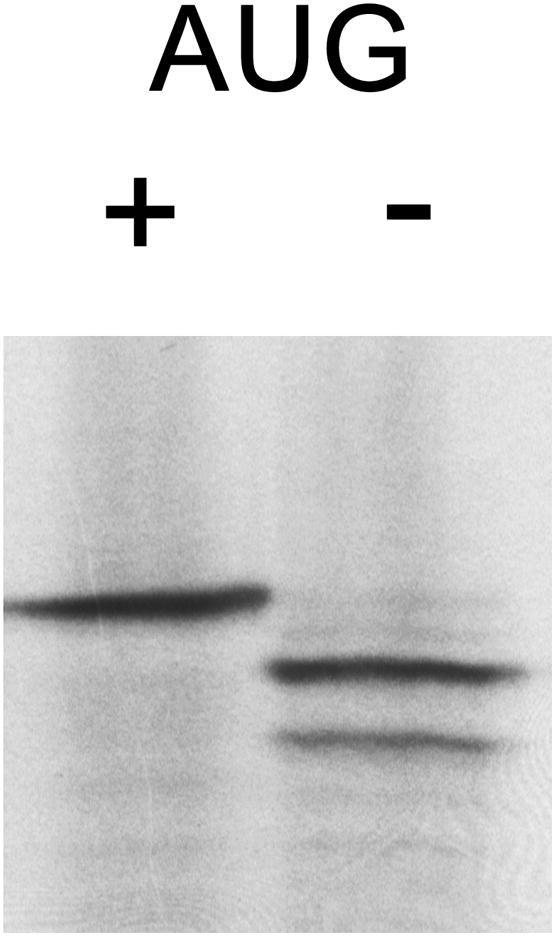
FL[-AUG] translation products. Autoradiograph of [35S]-labeled, in vitro-transcribed, synthetic 5′UTR(0 CGG)-FL and synthetic 5′UTR(0 CGG)-FL[-AUG], with and without an intact AUG start codon, respectively.
Effect of transcription start site on translation efficiency
Transcription of FMR1 has been demonstrated to begin at each of three transcription initiator regions (Inr).57 The most 3′ start site, Inr site I (Inr I), is the major start site for FMR1 alleles with CGG-repeat elements in the normal range (<45 CGG repeats) and also results in mRNAs with the shortest 5′UTRs (not including the CGG repeat). However, for expanded repeats there is increased utilization of Inrs II and III, which lie upstream of Inr I. Site III is the furthest upstream, 52 nt from site I, and becomes the dominant start site for alleles in the premutation range.57 Although we do not understand the mechanism for increased usage of Inrs II and III when the repeat is expanded, it could either be due to cofactors interacting with CGG repeat or that the CGG repeats themselves modulate nucleosome structure/positioning around the 5′UTR, in either instance allowing access to previously masked transcription sites 57; 65; 66. All FMR1-FL mRNAs described thus far have utilized the SP6 promoter directed to Inr I, which results in a 198-nt 5′UTR, excluding the length of the CGG repeats.
To account for any influence of the translation mechanism/efficiency on the length of the 5′UTR extension (Inr III vs. Inr I), we quantified the relative translation efficiencies of Inr III and Inr I mRNAs. Of particular interest were the premutation-length messages (62 and 99 CGG), as they favor initiation from Inr III in humans. To this end, FMR1 5′UTR-FL messages with either 0, 16, 30, 62, or 99 CGG repeats were extended 5′, to Inr III, and in vitro transcription and translation reactions performed as before. Results of these measurements (Figure 5) demonstrate that the expansion of the 5′UTR from Inr I to Inr III significantly and consistently (across CGG-repeat lengths) decreased the protein translation from each CGG-repeat mRNA. These results negate the possibility that an alternative form of translation initiation occurs in messages containing 5′ extensions. Instead, the added length – and perhaps added structure – of the 5′UTR within Inr III messages may decrease the amount of protein that can be made in 20 minutes.
DISCUSSION
Our current results demonstrate that translational initiation of the FMR1 mRNA occurs predominantly, if not exclusively, by scanning. In particular, translational initiation at GUG codon(s) within a canonical hairpin (HP) demonstrates that the ribosome scans the 5′UTR and does not use an alternate mechanism (i.e., IRES- or shunt-mediated initiation) to circumvent secondary or tertiary structure within the 5′UTR. Not only does the canonical hairpin not block translation initiation when in the FMR1 5′UTR context, our results provide direct evidence that the hairpin is disrupted enough for initiation to occur within the stem of the hairpin structure, concurrent with scanning through the 5′UTR.
Previous studies have shown that secondary structure downstream of a non-canonical start codon increases initiation from that codon by causing pausing of the ribosome.67; 68 In principle, it would not be necessary for the entire hairpin stem to be disrupted for initiation to occur at the GUG codons, since the first GUG begins 8 nt into the 5′ end of the stem, after which the rest of the hairpin may stall the ribosome long enough to facilitate initiation at a GUG in a weak Kozak consensus sequence (CGCGTGG or GTGGTGG [HP] vs. A/GNNAUGG [Kozak consensus]). However, scanning through the hairpin appears to be contextual, as synthetic 5′UTR(HP)-FL does not translate. Thus, additional structure within the FMR1 5′UTR appears to modulate the structure/accessibility of the hairpin sequence itself, an observation that underscores a caveat associated with any in silico-based analysis of stability/structure of either DNA or RNA sequence. Alternatively, the FMR1 5′UTR may recruit additional translation factors (e.g., eIF4A and other DEAD-box proteins) that aid in disrupting the hairpin and other local structure. Two such proteins found to aid in the translation of mRNAs with highly structured 5′UTRs are the DExH-Box protein DHX29 and the helicase DDX3.69; 70
The observation of aberrant, upstream initiation from FMR1 5′UTR(HP)-FL alters our initial interpretation that translation of the HP-substituted FMR1 5′UTR occurred at about the same rate as 99-CGG mRNA. Specifically, the HP mRNA (with adenines in place of uracils in GUG sequences) actually translated at a rate about half that of the 99-CGG mRNA. In other words, a hairpin with a predicted free energy of −45 kcal/mol inhibited FMR1 translation initiation more severely than a −227 kcal/mol CGG repeat. The disparity in the abilities of these secondary structures to inhibit translation might be due to the lower density of stable (canonical) base pairs within the CGG-repeat region, although G-G mismatches do form base pairs in the CGG-repeat hairpin context.33 Alternatively, the structure of the putative hairpin (HP) could be altered by base-pairing between the HP (85% GC rich) and the surrounding FMR1 5′UTR (77% GC).
Instead of suggesting that the FMR1 5′UTR employs an alternative mechanism of translation initiation in order to bypass the hairpin, our results indicate that the HP sequence inhibits translation to a greater degree than does the 99-CGG-repeat element. These results are concordant with a model in which the CGG repeats form a moderate-energy secondary structure that is scanned by the ribosome, albeit with some inhibition with longer repeat lengths. Blocked translation of 5′HP messages additionally indicates that FMR1 translation initiation is a 5′-dependent process (i.e. not an IRES).
Ribosomal shunting is employed, albeit rarely, in the translation of messages with long, highly structured leader sequences, particularly those with multiple uAUG initiation start sites.55; 71 Discovered in plant pararetroviruses, few shunting mammalian transcripts have been described.72; 73; 74; 75 In order to bypass the CGG repeats, one would expect an initiation event to occur upstream of the repeats, following by termination and then reinitiation at the FL AUG start codon. Such a mechanism should also operate with the mid-5′UTR HP constructs. Since the HP in the FMR1 5′UTR context does not generate a normal-size protein product, one can conclude that a shunting mechanism is not operating to bypass the HP, which is further illustrated by the internal initiation seen within the FMR1 5′UTR(HP)-FL mRNA.
In silico analyses of CGG repeat secondary structures yields substantial computed free energies of stabilization of CGG-repeat structures (Table 2). Considering the free energies of secondary structures previously shown to block translation in the literature (Table 3), one would predict that CGG-repeat elements even in the high-normal range (30–45 CGG repeats) would have a strong inhibitory effect on translation, as there is a trend in which translation inhibition is greater with increasing predicted secondary structure, which may be sequence- and context-dependent. However, the addition of 30 CGGs in the FMR1 5′UTR context, with a computed −63 kcal/mol free energy of stabilization, reduces the translation efficiency in vitro by only 34% (Figure 2B). Weak translation inhibition is consistent with a low-to-intermediate stability of the CGG repeat, which is surprising in view of the known stability of the G-G mismatches in the context of the CGG-repeat stem.33 However, Zumwalt et al.33 also demonstrated that the G-G base pair was in a dynamic conformational equilibrium, in which guanine bases flip between syn and anti conformations. This dynamic may render the CGG-repeat stem more amenable to local disruption, as for the case of scanning, than would be expected based on free energy computations alone.
Table 3.
Literature values for hairpin stability and degree of inhibition of translation.38; 39; 40; 76; 77; 78; 79; 80
| ΔG (kcal/mol) | % Inhibition | System | Measured by |
|---|---|---|---|
| −11.0 | 17 | Drosophila S2 cells | [35S] Hsp70/mRNA 79 |
| −13.4 | none | NTera2D1 cells | chemilum/cell extracts 76 |
| −14.2 | 74 | RRL | CAT activity 78 |
| −18.0 | 83–90 | S. cerevisiae | CAT or FL activity 77 |
| −22.0 | 92 | Drosophila S2 cells | [35S] Hsp70/mRNA 79 |
| −30.0 | 0–20 | Cos cells | [35S] proinsulin 40 |
| −30.0 | none | RRL | [35S] CAT 39 |
| −49.4 | 75 | RRL | CAT activity 78 |
| −50.0 | 85–95 | Cos cells | [35S] proinsulin 40 |
| −50.0 | 90–95 | Xenopus embryos | CAT activity 80 |
| −61.0 | 100 | RRL | [35S] CAT 39 |
| −61.1 | 61 | RRL, wheat germ | TK activity 38 |
| −61.1 | 82 | Cos cells | TK activity 38 |
| −74.8 | 79 | NTera2D1 cells | chemilum/cell extracts 76 |
| −195.3 | 93 | Cos cells | TK activity 38 |
| −195.3 | >99 | RRL, wheat germ | TK activity 38 |
Abbreviations: CAT, chloramphenicol acetyl transferase; Hsp70, 70 kDa heat shock protein; TK, thymidine kinase; chemilum, chemiluminescence.
The CGG repeats may well form the type of secondary structure predicted by folding RNA in silico. Similar hairpin structures were found by both NMR spectroscopy and limited enzyme digestion of CGG-repeat mRNA.33; 34 However, computed free energies of stabilization are clearly inadequate to understand the functional consequences of the expanded CGG repeats on translation initiation. That is, except for specific, well-characterized structures (specific hairpins, for example), secondary structure with large, computed free energies of stabilization in the 5′UTR does not necessarily imply blocked translation. The effect of an inhibitory structure is both sequence- and context-specific, and, therefore, further testing to confirm secondary structure, as well as functional studies, is necessary.
An additional factor adding to the uncertainty associated with free energy estimates is the gradual CGG-dependent repression on translational initiation seen in FMR1 and FMRP measurements from human samples,22; 26; 44; 47 in which only a reduction of approximately 50% in FMRP levels is seen as the CGG-repeat length increases from 30 to ~100 repeats. Even taking into consideration the two- to four-fold increase in mRNA levels over this same size range, there is less than a five-fold reduction in protein/mRNA ratios over a range of CGG-repeat lengths where the stabilization free energy is computed to go from approximately −64 kcal/mol to lower than −220 kcal/mol.
Interestingly, in a mouse model of the CGG-repeat expansion, which, unlike human cells, does not undergo methylation-coupled transcription silencing,81 the translation efficiency of FMR1 alleles with greater than 200 repeats is reported to be about 25% of wildtype (38% FMRP divided by 150% FMR1). Extrapolating these results to humans, FMRP is predicted to be translated from all repeat lengths that are transcriptionally expressed in humans (normal and premutation alleles), albeit inefficiently.
In addition to a translational deficit due to CGG repeats, we also found that transcription initiation from the upstream start site (Inr III) resulted in less-efficiently translated messages. While this may simply be due to an increase in the time it takes to scan a 52-base extended 5′UTR and therefore be predicted to have little effect on steady-state FMRP levels in vivo, there may be implications for individuals with premutation-length alleles. FMRP has been shown to be locally translated within post-synaptic spines in response to glutaminergic stimulation.82 If the requirement for FMRP at these loci is time-dependent, the delay in FMRP translation (in addition to the decreased translational efficiency of premutation-length CGG repeats) may have negative connotations for synaptogenesis and synaptic plasticity in terms of delayed timing of the post-synaptic response rather than simply altered levels of induced protein.
Finally, as a cautionary note, we found that a hairpin previously employed as a translational block, and as a tool for providing initial support for the scanning model of translation initiation, not only did not completely inhibit translation, but also displayed a level of inhibition that was context-dependent. Thus, even well-defined helix elements are not guaranteed to block translation in all 5′UTR contexts. In the FMR1 5′UTR, in vitro translation was inhibited by about 80% compared to no insert. If the hairpin/GUGs had caused upstream initiation in the original study39 as it did in ours, the product would not have been in frame with the CAT reporter downstream, resulting in the production of an 18-amino acid peptide.
MATERIALS AND METHODS
Construction of FMR1 and synthetic reporter plasmids
A series of firefly luciferase (FL) reporter plasmids containing specified numbers of CGG repeats was constructed in which the CGG repeat was placed either in its native FMR1 5′UTR context or in an unrelated (heterologous) 5′UTR. The heterologous 5′UTR was designed as a multiple cloning site of 82 bases having minimal secondary structure, with restriction sites that facilitate insertion of CGG-repeat cassettes (thus designated “synthetic”). In addition to CGG-repeat constructs, we also inserted a stable hairpin into both FMR1 and synthetic 5′UTRs, either near the 5′ cap or in place of the CGG repeats. A final set of plasmids involved alteration of the FMR1 5′UTR to start from either FMR1 transcription initiator region (Inr) I or III,57 with varying numbers of CGG repeats. The 5′UTR, CGG-repeat length, hairpin insertion, and transcription start site of each construct are described in Figure 1 and Table 1. Plasmid cloning for each construct is included as Supporting Material. All FL plasmids have an SP6 promoter and PolyA tail.
Bacterial maintenance
Top10 E. coli cells (Invitrogen Corp, Carlsbad, CA) were maintained in LB containing 50 μg/ml ampicillin. Miniprep cultures for plasmid constructs containing 62 or 99 CGG repeats were grown in 4 ml LB overnight at 33°C; the reduced temperature was found to improve stability of the CGG repeat during cloning. All other cultures were grown at 37°C.
In vitro transcription
FL and RL luciferase plasmids were linearized with EcoRI and BamHI (all restriction enzymes from New England Biolabs, Inc, Ipswich, MA; NEB), respectively, after which the DNAs were purified using the Qiagen MinElute reaction cleanup kit (Qiagen Inc, Valencia, CA). In vitro transcriptions of 5′-capped messages were performed using mMachine SP6 (FL) and T7 (RL) transcription kits (Ambion Inc, Foster City, CA) according to the manufacturer’s protocol (2 hours 37°C), followed by RQ1 DNase digestion (Promega Corp; Madison, WI). The manufacturer’s protocol for in vitro transcription utilizes a ratio of cap analog to GTP of 4:1, which predicts that ~80% of the 5′ ends will be capped (Ambion mMessage mMachine kit), although this ratio was not separately determined. Following the capping reaction, transcripts were isolated using RNeasy reaction cleanup kit (Qiagen); mRNA concentrations were measured on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc, Waltham, MA).
In vitro translation and dual luciferase experiments
We chose to study the translation initiation of FMR1 in rabbit reticulocyte lysate (RRL), which represents a standard in vitro model for the study of translation. Each FL mRNA (0.2 pmol) was translated with 5.0 fmol of RL mRNA in 50 μl of nuclease-treated RRL (Promega) for 20 min at 30°C, followed by termination of the reactions with 12.5 μl passive lysis buffer (Promega). FL and RL levels were measured using the Dual-Luciferase Reporter Assay System (Promega), in which FL and RL activities are obtained by quantifying the chemiluminescent output (given in relative light units; RLUs) upon luciferase substrate addition. Briefly, 10 μl of the in vitro translation reaction was pipetted into each well in a 96-well format, followed by the addition of 100 μl of luciferase assay reagent. After a 2-second delay, FL luminescence was read for 5 seconds, followed by the addition of 100 μl Stop-and-Glo and a further 2-second delay, and finally a 5-second RL luminescence reading. An Lmax luminometer (Molecular Devices Corp, Sunnyvale, CA) running SOFTmax PRO software automated the procedure. Relative FL luminescence was achieved by dividing the measured FL luminescence by its corresponding RL value for each in vitro translation reaction. Three aliquots of each in vitro reaction were used for each set of conditions. Translations of FMR1 and synthetic messages were performed simultaneously, and in the same RRL lots.
Translation/[35S] labeling and SDS-PAGE autoradiography
Each mRNA (0.1 pmol) was added to RRL supplemented with ~10 μCi [35S]-methionine (1,000 Ci/mmol) (MP Biomedicals, LLC, Solon, OH) and translated in a 25-μl reaction for 20 min at 30°C, followed by termination with 6.25 μl passive lysis buffer. Three μl of each reaction were electrophoresed on a 15% SDS Tris-HCl polyacrylamide gel.83 Gels were placed on Filter Paper Backing (Bio-Rad Laboratories; Hercules, CA) and allowed to dry overnight, then exposed to Blue Basic Autorad double-emission film (ISC BioExpress, Kaysville, UT) for periods ranging from 4 hours to overnight.
In silico RNA folding
FMR1 and synthetic 5′UTR RNA secondary structures and free energies of stabilization were obtained using the mFold web server http://mfold.bioinfo.rpi.edu.42; 43
Supplementary Material
Acknowledgments
FUNDING
This work was funded by the National Institutes of Health through a research award, [R01 HD040661], and Roadmap Interdisciplinary Research Consortium (IRC) grants [UL1 DE019583] and [RL1 AG032119]; and through generous donations from families in support of fragile X research.
The authors wish to thank Greg Mayeur for his assistance in the methionine labeling experiments, and the patients and families that participated in our research who made these studies possible.
Abbreviations
- FL
firefly luciferase
- FMR1
fragile X mental retardation 1
- FMRP
fragile X mental retardation protein
- FXTAS
fragile X-associated tremor/ataxia syndrome
- IRES
internal ribosome entry site
- HP
hairpin
- Inr
initiator region
- POI
primary ovarian insufficiency
- RL
Renilla luciferase
- RRL
rabbit reticulocyte lysate
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Anna L Ludwig, Email: alludwig@ucdavis.edu.
John WB Hershey, Email: jwhershey@ucdavis.edu.
Paul J Hagerman, Email: pjhagerman@ucdavis.edu.
References
- 1.Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Jr, Warren ST, Oostra BA, Nelson DL, Caskey CT. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 2.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR- 1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–14. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 3.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas M, Mandel J. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 4.Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 5.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 6.Garber K, Smith KT, Reines D, Warren ST. Transcription, translation and fragile X syndrome. Curr Opin Genet Dev. 2006;16:270–275. doi: 10.1016/j.gde.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Pfeiffer BE, Huber KM. The state of synapses in fragile X syndrome. Neuroscientist. 2009;15:549–567. doi: 10.1177/1073858409333075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 10.Hagerman RJ, Berry-Kravis E, Kaufmann WE, Ono MY, Tartaglia N, Lachiewicz A, Kronk R, Delahunty C, Hessl D, Visootsak J, Picker J, Gane L, Tranfaglia M. Advances in the treatment of fragile X syndrome. Pediatrics. 2009;123:378–390. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey DB, Jr, Raspa M, Olmsted M, Holiday DB. Co-occurring conditions associated with FMR1 gene variations: findings from a national parent survey. Am J Med Genet A. 2008;146A:2060–2069. doi: 10.1002/ajmg.a.32439. [DOI] [PubMed] [Google Scholar]
- 12.Farzin F, Perry H, Hessl D, Loesch D, Cohen J, Bacalman S, Gane L, Tassone F, Hagerman P, Hagerman R. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J Dev Behav Pediatr. 2006;27:S137–144. doi: 10.1097/00004703-200604002-00012. [DOI] [PubMed] [Google Scholar]
- 13.Uzielli ML, Guarducci S, Lapi E, Cecconi A, Ricci U, Ricotti G, Biondi C, Scarselli B, Vieri F, Scarnato P, Gori F, Sereni A. Premature ovarian failure (POF) and fragile X premutation females: from POF to to fragile X carrier identification, from fragile X carrier diagnosis to POF association data. Am J Med Genet. 1999;84:300–303. [PubMed] [Google Scholar]
- 14.Marozzi A, Vegetti W, Manfredini E, Tibiletti MG, Testa G, Crosignani PG, Ginelli E, Meneveri R, Dalpra L. Association between idiopathic premature ovarian failure and fragile X premutation. Hum Reprod. 2000;15:197–202. doi: 10.1093/humrep/15.1.197. [DOI] [PubMed] [Google Scholar]
- 15.Gersak K, Meden-Vrtovec H, Peterlin B. Fragile X premutation in women with sporadic premature ovarian failure in Slovenia. Hum Reprod. 2003;18:1637–40. doi: 10.1093/humrep/deg327. [DOI] [PubMed] [Google Scholar]
- 16.Bodega B, Bione S, Dalpra L, Toniolo D, Ornaghi F, Vegetti W, Ginelli E, Marozzi A. Influence of intermediate and uninterrupted FMR1 CGG expansions in premature ovarian failure manifestation. Hum Reprod. 2006;21:952–957. doi: 10.1093/humrep/dei432. [DOI] [PubMed] [Google Scholar]
- 17.Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 18.Jacquemont S, Hagerman RJ, Leehey M, Grigsby J, Zhang L, Brunberg JA, Greco C, Des Portes V, Jardini T, Levine R, Berry-Kravis E, Brown WT, Schaeffer S, Kissel J, Tassone F, Hagerman PJ. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am J Hum Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, Hessl D, Becker EJ, Papazian J, Leehey MA, Hagerman RJ, Hagerman PJ. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 20.Berry-Kravis E, Abrams L, Coffey SM, Hall DA, Greco C, Gane LW, Grigsby J, Bourgeois JA, Finucane B, Jacquemont S, Brunberg JA, Zhang L, Lin J, Tassone F, Hagerman PJ, Hagerman RJ, Leehey MA. Fragile X-associated tremor/ataxia syndrome: clinical features, genetics, and testing guidelines. Mov Disord. 2007;22:2018–2030. doi: 10.1002/mds.21493. quiz 2140. [DOI] [PubMed] [Google Scholar]
- 21.Leehey MA. Fragile X-associated tremor/ataxia syndrome: clinical phenotype, diagnosis, and treatment. J Investig Med. 2009;57:830–836. doi: 10.231/JIM.0b013e3181af59c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 23.Allen EG, He W, Yadav-Shah M, Sherman SL. A study of the distributional characteristics of FMR1 transcript levels in 238 individuals. Hum Genet. 2004;114:439–447. doi: 10.1007/s00439-004-1086-x. [DOI] [PubMed] [Google Scholar]
- 24.Tassone F, Beilina A, Carosi C, Albertosi S, Bagni C, Li L, Glover K, Bentley D, Hagerman PJ. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007;13:555–562. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tassone F, Hagerman RJ, Loesch DZ, Lachiewicz A, Taylor AK, Hagerman PJ. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am J Med Genet. 2000;94:232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 26.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J Biol Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amiri K, Hagerman RJ, Hagerman PJ. Fragile X-associated tremor/ataxia syndrome: an aging face of the fragile X gene. Arch Neurol. 2008;65:19–25. doi: 10.1001/archneurol.2007.30. [DOI] [PubMed] [Google Scholar]
- 29.Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am J Hum Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brouwer JR, Willemsen R, Oostra BA. Microsatellite repeat instability and neurological disease. Bioessays. 2009;31:71–83. doi: 10.1002/bies.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dick KA, Margolis JM, Day JW, Ranum LP. Dominant non-coding repeat expansions in human disease. Genome Dyn. 2006;1:67–83. doi: 10.1159/000092501. [DOI] [PubMed] [Google Scholar]
- 32.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zumwalt M, Ludwig A, Hagerman PJ, Dieckmann T. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 2007;4:93–100. doi: 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]
- 34.Napierala M, Michalowski D, de Mezer M, Krzyzosiak WJ. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handa V, Saha T, Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisman-Shomer P, Cohen E, Fry M. Interruption of the fragile X syndrome expanded sequence d(CGG)(n) by interspersed d(AGG) trinucleotides diminishes the formation and stability of d(CGG)(n) tetrahelical structures. Nucleic Acids Res. 2000;28:1535–1541. doi: 10.1093/nar/28.7.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)(n) is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 39.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 42.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 44.Primerano B, Tassone F, Hagerman RJ, Hagerman P, Amaldi F, Bagni C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002;8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 45.Hessl D, Tassone F, Loesch DZ, Berry-Kravis E, Leehey MA, Gane LW, Barbato I, Rice C, Gould E, Hall DA, Grigsby J, Wegelin JA, Harris S, Lewin F, Weinberg D, Hagerman PJ, Hagerman RJ. Abnormal elevation of FMR1 mRNA is associated with psychological symptoms in individuals with the fragile X premutation. Am J Med Genet B Neuropsychiatr Genet. 2005;139:115–121. doi: 10.1002/ajmg.b.30241. [DOI] [PubMed] [Google Scholar]
- 46.Khateb S, Weisman-Shomer P, Hershco-Shani I, Ludwig AL, Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–5788. doi: 10.1093/nar/gkm636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen LS, Tassone F, Sahota P, Hagerman PJ. The (CGG)n repeat element within the 5′ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum Mol Genet. 2003;12:3067–3074. doi: 10.1093/hmg/ddg331. [DOI] [PubMed] [Google Scholar]
- 48.Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Hershey NSJWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- 49.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 51.Doudna JA, Sarnow P. Translation Initiation by Viral Internal Ribosome Entry Sites. In: Mathews MB, Sonenberg N, Hershey JW, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2006. pp. 129–153. [Google Scholar]
- 52.Graber TE, Holcik M. Cap-independent regulation of gene expression in apoptosis. Mol Biosyst. 2007;3:825–834. doi: 10.1039/b708867a. [DOI] [PubMed] [Google Scholar]
- 53.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 54.Mauro VP, Chappell SA, Dresios J. Analysis of ribosomal shunting during translation initiation in eukaryotic mRNAs. Methods Enzymol. 2007;429:323–354. doi: 10.1016/S0076-6879(07)29015-9. [DOI] [PubMed] [Google Scholar]
- 55.Ryabova LA, Pooggin MM, Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006;119:52–62. doi: 10.1016/j.virusres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 56.Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, Warren ST. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 57.Beilina A, Tassone F, Schwartz PH, Sahota P, Hagerman PJ. Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum Mol Genet. 2004;13:543–549. doi: 10.1093/hmg/ddh053. [DOI] [PubMed] [Google Scholar]
- 58.Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawson TG, Cladaras MH, Ray BK, Lee KA, Abramson RD, Merrick WC, Thach RE. Discriminatory interaction of purified eukaryotic initiation factors 4F plus 4A with the 5′ ends of reovirus messenger RNAs. J Biol Chem. 1988;263:7266–7276. [PubMed] [Google Scholar]
- 60.Dobson T, Kube E, Timmerman S, Krushel LA. Identifying intrinsic and extrinsic determinants that regulate internal initiation of translation mediated by the FMR1 5′ leader. BMC Mol Biol. 2008;9:89. doi: 10.1186/1471-2199-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang PW, Carpenter LE, Hagerman PJ. The 5′-untranslated region of the FMR1 message facilitates translation by internal ribosome entry. J Biol Chem. 2001;276:37916–37921. doi: 10.1074/jbc.M105584200. [DOI] [PubMed] [Google Scholar]
- 62.Kozak M. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res. 1990;18:2828. doi: 10.1093/nar/18.9.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 64.Algire MA, Lorsch JR. Where to begin? The mechanism of translation initiation codon selection in eukaryotes. Curr Opin Chem Biol. 2006;10:480–486. doi: 10.1016/j.cbpa.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Godde JS, Kass SU, Hirst MC, Wolffe AP. Nucleosome assembly on methylated CGG triplet repeats in the fragile X mental retardation gene 1 promoter. J Biol Chem. 1996;271:24325–24328. doi: 10.1074/jbc.271.40.24325. [DOI] [PubMed] [Google Scholar]
- 66.Wang YH, Gellibolian R, Shimizu M, Wells RD, Griffith J. Long CCG Triplet Repeat Blocks Exclude Nucleosomes: A Possible Mechanism for the Nature of Fragile Sites in Chromosomes. J Mol Bio. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 67.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kochetov AV, Palyanov A, Titov II, Grigorovich D, Sarai A, Kolchanov NA. AUG_hairpin: prediction of a downstream secondary structure influencing the recognition of a translation start site. BMC Bioinformatics. 2007;8:318. doi: 10.1186/1471-2105-8-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hemmings-Mieszczak M, Hohn T, Preiss T. Termination and peptide release at the upstream open reading frame are required for downstream translation on synthetic shunt-competent mRNA leaders. Mol Cell Biol. 2000;20:6212–6223. doi: 10.1128/mcb.20.17.6212-6223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishimura K, Okudaira H, Ochiai E, Higashi K, Kaneko M, Ishii I, Nishimura T, Dohmae N, Kashiwagi K, Igarashi K. Identification of proteins whose synthesis is preferentially enhanced by polyamines at the level of translation in mammalian cells. Int J Biochem Cell Biol. 2009;41:2251–2261. doi: 10.1016/j.biocel.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 73.Koh DC, Mauro VP. Reconciling contradictory reports regarding translation of BACE1 mRNA: initiation mechanism is altered by different expression systems. RNA Biol. 2009;6:54–58. doi: 10.4161/rna.6.1.7567. [DOI] [PubMed] [Google Scholar]
- 74.Morley SJ, Coldwell MJ. A cunning stunt: an alternative mechanism of eukaryotic translation initiation. Sci Signal. 2008;1:pe32. doi: 10.1126/scisignal.125pe32. [DOI] [PubMed] [Google Scholar]
- 75.Sherrill KW, Lloyd RE. Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol Cell Biol. 2008;28:2011–2022. doi: 10.1128/MCB.01446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McMillan JP, Singer MF. Translation of the human LINE-1 element, L1Hs. Proc Natl Acad Sci U S A. 1993;90:11533–11537. doi: 10.1073/pnas.90.24.11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oliveira CC, van den Heuvel JJ, McCarthy JE. Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: the roles of the stability and position of stem-loops in the mRNA leader. Mol Microbiol. 1993;9:521–532. doi: 10.1111/j.1365-2958.1993.tb01713.x. [DOI] [PubMed] [Google Scholar]
- 78.Vega Laso MR, Zhu D, Sagliocco F, Brown AJ, Tuite MF, McCarthy JE. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J Biol Chem. 1993;268:6453–6462. [PubMed] [Google Scholar]
- 79.Hess MA, Duncan RF. Sequence and structure determinants of Drosophila Hsp70 mRNA translation: 5′UTR secondary structure specifically inhibits heat shock protein mRNA translation. Nucleic Acids Res. 1996;24:2441–2449. doi: 10.1093/nar/24.12.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bouvet P, Paris J, Phillippe M, Osborne HB. Degradation of a developmentally regulated mRNA in Xenopus embryos is controlled by the 3′ region and requires the translation of another maternal mRNA. Mol Cell Biol. 1991;11:3115–3124. doi: 10.1128/mcb.11.6.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brouwer JR, Huizer K, Severijnen LA, Hukema RK, Berman RF, Oostra BA, Willemsen R. CGG-repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J Neurochem. 2008;107:1671–1682. doi: 10.1111/j.1471-4159.2008.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grossman AW, Aldridge GM, Weiler IJ, Greenough WT. Local protein synthesis and spine morphogenesis: Fragile X syndrome and beyond. J Neurosci. 2006;26:7151–7155. doi: 10.1523/JNEUROSCI.1790-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.