Abstract
Liquid chromatography-selected reaction monitoring (LC-SRM) is a highly specific and sensitive mass spectrometry (MS) technique that is widely being applied to selectively qualify and validate candidate markers within complex biological samples. However, in order for LC-SRM methods to take on these attributes, target-specific optimization of sample processing is required, in order to reduce analyte complexity, prior to LC-SRM. In this study, we have developed a targeted platform consisting of protein immunoaffinity enrichment on magnetic beads and LC-SRM for measuring carbonic anhydrase 12 (CA12) protein in a renal cell carcinoma (RCC) cell line (PRC3), a candidate biomarker for RCC whose expression at the protein level has not been previously reported. Sample processing and LC-SRM assay were optimized for signature peptides selected as surrogate markers of CA12 protein. Using LC-SRM coupled with stable isotope dilution, we achieved limits of quantitation in the low fmol range sufficient for measuring clinically relevant biomarkers with good intra- and inter-assay accuracy and precision (≤17%). Our results show that using a quantitative immunoaffinity capture approach provides specific, accurate, and robust assays amenable to high-throughput verification of potential biomarkers.
Keywords: immunoaffinity enrichment, selected reaction monitoring, enhanced signature peptide predictor, carbonic anhydrase 12, renal cell carcinoma, biomarker, Protein G magnetic beads
Introduction
An important step in the discovery of new clinical blood tests for early diagnosis of disease is the ability to detect protein biomarkers in complex biological samples present in the low fmol range. Secreted proteins, cleaved receptors from the surface of the cell, and proteins that can “leak” from the diseased tissue into the bloodstream have been regarded as attractive candidate biomarkers; however, these are expected to be at low concentrations. Moreover, the high degree of protein structural complexity and wide dynamic range of proteins expressed in the biological matrices that must be detected and measured1, 2 impose a great challenge on proteomic-based studies. Often the dynamic range of analytes exceeds six orders of magnitude in cells and at least ten orders of magnitude in biofluids.3 The principal enabling technology for proteomic discovery is mass spectrometry and although current mass spectrometers can achieve atto-molar sensitivities for detection of single compounds, their working dynamic range typically spans only three to four orders of magnitude for complex mixtures within a single mass spectrum.4, 5 In order to be able to detect biomarkers present at low concentrations, the current trend in protein biomarker discovery or verification studies with or without prior information has been focused on targeted proteomics.6–8 For instance, specific protein(s) of known identity discovered as biomarker candidates at the gene level are targeted in the mass spectrometer by LC-SRM.
Targeted LC-SRM methods coupled with stable isotope dilution are becoming popular strategies for quantitation, validation and therefore early selection, during the biomarker discovery phase, of candidate biomarkers.9 LC-SRM experiments are traditionally performed using a triple quadrupole mass spectrometer (QQQ). The mass filters imposed on the first (Q1) and third (Q3) quadrupoles enable LC-SRM methods to be highly specific for monitoring proteotypic peptides selected as quantitative surrogates for a candidate protein. In addition, the non-scanning nature of this technique (high dwell times) increases its sensitivity by several orders of magnitude compared to the limit of detection achieved in typical LC-MS experiment or even a product ion scan.10 Although quantitative LC-SRM is becoming an indispensable method for detection and quantitation of proteins that may serve as candidate biomarkers, the use of this technique is not yet routine. This can be, at least in part, attributed to the significant time required for biomarker specific optimization and validation of sample processing. This is a required step in order to ensure accurate and reproducible quantitation of protein marker(s). For example, it is necessary to develop and optimize enrichment strategies for the detection of very low abundance species. Several groups have demonstrated that coupling LC-SRM methodology with affinity-based enrichment methods enhances the sensitivity, selectivity, and dynamic range of the proteomic technology.11–14
Over the past decade, significant amounts of information relating to the diagnosis and classification of many diseases have been generated using high-throughput mRNA (cDNA) microarrays. However, this information alone falls short of providing a complete solution to the rather challenging field of biomarker discovery and diagnosis of disease. The expression of mRNA does not necessarily correlate with the expression of the corresponding proteins.15, 16 Additionally, many promising, genomic based leads may not be measurable outside of the cell, as the corresponding protein product could be of very low-abundance. Thus, the process of detecting a biomarker that was previously shown to be up- or down- regulated at the gene level may be difficult at the protein level, either in cell lysates or biological fluids. Furthermore, an impediment to the application of mRNA/cDNA techniques to the discovery and use of clinically usable biomarkers is their limited utility for the analysis of biological fluids such as plasma, urine, cerebrospinal fluid or saliva.
A translational approach to the discovery of disease biomarkers in clinical proteomics can be based on detecting gene expression profile differences in cell lines that harbor the molecular characteristics of the human disease. This information can serve as a valuable first step in discovering candidate biomarkers that can be detected in relevant biological fluids. Despite reasonable concerns about comparing cells cultured in vitro to their corresponding actual tumor, proteomic studies of cancer cell lines have already led to the discovery of differentially expressed proteins of clinical interest (e.g., cathepsin D and cytokeratins for breast cancer).17–20 Benefits associated with the use of cell lines for protein biomarker discovery include their renewable nature and the ability to introduce controlled, tumor-associated, genetic changes.
Several human cell lines are available for study of clear cell renal cell carcinoma (RCC).21 Many of these cell lines display inactivation of von Hippel-Lindau (VHL) tumor suppressor gene, which is deleted in the majority of sporadic and inheritable forms of clear cell RCC. As a background work, we compared gene expression profiles of isogenic VHL +/+ with VHL −/− cell lines and identified proteins that could serve as candidate molecular markers (unpublished data). We and others have demonstrated that carbonic anhydrases 9 (CA9) and 12 (CA12)22–24 are candidate biomarkers for RCC. Carbonic anhydrase 9 has been confirmed as a candidate biomarker in a subset of RCC patients, using ELISA as a detection method.25 The goal of our current study was to develop a quantitative immunoaffinity-liquid chromatography-selected reaction monitoring mass spectrometry (IP-LC-SRM) assay that uses antibodies to enrich these protein targets from cell lines followed by mass spectrometry-based quantitation. In this paper we describe the optimization of the assay, including antibody capture, elution, digestion, and LC-SRM protocols, all which are critical parameters that could impact the overall sensitivity and specificity of the IP-LC-SRM method and therefore affect the limit of detection and quantitation. Using this approach we were able for the first time to observe and measure CA12 protein in the VHL deficient human RCC cell line (PRC3). Additionally, we show the validation of this optimized platform demonstrating the ability to quantitate biomarkers in cell lysates in the low fmol range.
Materials and methods
Materials
Cell lines used in this work were obtained from American Type Culture Collection (Manassas, VA). Dulbecco’s modified eagle medium (DMEM) was from Media Tech (Manassas, VA). FetalClone and 100X penicillin-streptomycin-glutamine were purchased from HyClone (Logan, UT) and Invitrogen (Carlsbad, CA), respectively. Dynabeads Protein G beads were from Invitrogen Dynal AS (Oslo, Norway). For immunoprecipitations (IPs), a polyclonal antibody (rabbit serum) against human carbonic anhydrase 12 was developed by Cell Signaling Technologies (Danvers, MA). Bovine sequencing grade modified trypsin was purchased from Promega (Madison, WI). LC/MS grade formic acid, acetonitrile, water, and other buffer reagents were from Thermo Fisher Scientific (Waltham, MA). Two unlabeled (UNL) peptides of CA12 (LNLPSDMHIQGLQSR and WTYFGPDGENSWSK) were synthesized by GenScript Corp. (Piscataway, NJ) and quantitated by amino acid analysis at Dana Farber Cancer Institute Core Facilities (Boston, MA). The stable isotope labeled versions of these peptides, with 13C- and 15N-labeled proline, were synthesized and quantitated by amino acid analysis by AnaSpec (San Jose, CA). The labeled version of these peptides resulted in a mass shift of +6 Da.
Methods
Cell culture and protein extraction
The human RCC cells 786-O lack wild type VHL. The PRC3 cell line was derived from 786-O cells by stable transfection with empty vector.26 All methods in this study were developed using the PRC3 cell line, which is expected to over-express CA12 due to lack of VHL gene.23 Cells were cultured in DMEM with 10% FetalClone supplemented with penicillin-streptomycin-glutamine solution. The cells were grown in p100 plates in an incubator maintained at 37°C and 5% CO2. The cells were harvested at 80% confluency and prior to lysis, the medium was removed and cells were washed twice with cold phosphate buffered saline (PBS). Two different cell lysis buffers were employed for extraction of proteins from PRC3 cells: 1) urea-based buffer (50 mM Trisbase pH 7.4, 1.0 mM EDTA, 150 mM NaCl, 7 M urea), 2) detergent-based buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% Nonidet P-40, 20 µM iodoacetate, 580 µM AEBSF, 140 µM leupeptin, 1.4 mM Na-Orthovanadate, 50 µM pepstatin A, 20 µM aprotinin, 700 mM NaFl). To each plate, 1 mL of the lysis buffer was added and rocked for 30 min at 4°C. The cells were scraped and centrifuged for 15 min at 13000 × g and 4°C. The supernatants containing the soluble extracted proteins were collected and the total protein content was measured using the Bradford protein assay (Thermo Fisher Scientific, Waltham, MA) as per the manufacturer’s instructions.
Preparation of CA12-FLAG standard
CA12 was amplified with oligonucleotides 5’GCGCGGATCCGCCACCATGCCCCGGCGCAGCCTG-3’ (forward) and 5’-GCGC GAATTCAGCGTGGGCCTCAGTCTCCATCTTG-3’ (reverse), restricted with BamHI and EcoRI and ligated into the pcDNA3.1-C-FLAG vector. The latter vector was created as follows: oligos 5’-AATTCGATTACAAAGATGATGATGATAAATAG GTT-3’ (forward) and 5’-AACCTATTTATCATCATCATCTTTGTAATCG-3’ (reverse) were annealed, kinased and ligated into vectror pcDNA3.1 digested with EcoRI and EcoRV.
For in vitro transcription and translation (IVTT) of the FLAG-tagged CA12 protein standard we used the Promega IVTT (Promega, Madison, WI) according to the manufacturer’s instructions. The CA12-FLAG protein standard was purified with ANTI-FLAG M2 affinity gel and eluted with 1X FLAG peptide (Sigma Aldrich, St. Louis, MO) according to the manufacture’s recommended procedure for purification of FLAG fusion proteins via immunoprecipitation. After purification, the eluate containing CA12-FLAG was frozen at −80 °C before lyophilization using Labconco FreeZone 2.5 Plus Freeze Dry System (Labconco, Kansas, MO). Lyophilized protein standard was reconstituted in 100 µL of water. To determine the concentration of CA12-FLAG, 3 µL of the standard was digested with trypsin and quantitated using the calibration curve as described below.
Non-covalent binding of rabbit CA12 antibody to Protein G beads
An aliquot of Dynabeads Protein G coated magnetic beads was washed twice with citrate-phosphate buffer, pH 5.0. The beads were re-suspended in three times the original volume with 1X PBS containing anti-CA12 polyclonal serum antibody (pAb) at a ratio of 2 µL of serum, equivalent to ~ 20 µg of total IgG, per 50 µL of beads (the amount of total IgG used was calculated assuming that the concentration of total IgG in serum is approximately 10 mg/mL)27. The solution was allowed to rock at room temperature for 1 hour. The supernatant was removed and the beads were washed twice with 0.5 mL of citrate phosphate buffer, pH 5.0 and three times with 0.5 mL of 1X PBS to remove any unbound protein. The beads were resuspended in the original volume with 1X PBS.
Optimization of immunoprecipitation conditions
We optimized the IP conditions for capture of the target antigen on antibody-coated magnetic beads and elution using an affinity purified FLAG-tagged CA12 protein standard and quantitative LC-SRM. In these experiments, 652 fmol of CA12-FLAG was spiked into 1 mL of PRC3 NP-40 cell lysate diluted 100-fold with 1X PBS and 50 µL of beads containing the bound rabbit anti-CA12 pAb were added to the solution. The antigen was captured at 4 °C for 5 hours. The beads were washed four times with 25 mM Tris, pH 7.5, 250 mM NaCl. A series of elution buffers (buffer 1: 50 mM hydrochloric acid; buffer 2: 5% acetic acid; buffer 3: 0.2 M sodium carbonate, pH 11; and buffer 4: 4 M urea/50 mM hydrochloric acid) were tested for their ability to efficiently elute the captured CA12-FLAG. Two aliquots (40 µL) of each of the elution buffers were added to antigen-bound antibody-coated Protein G beads at room temperature in 2-min intervals for a total of 4 min. Each elution was collected and the low pH IP eluates were neutralized, adjusted to pH ~ 7.5, immediately after elution by addition of 100 µL of 40 mM sodium carbonate pH 11.0. The immunoprecipitated proteins were digested and the antigen capture efficiency and protein recoveries were determined by LC-SRM. All spike-and-recovery samples were prepared in duplicate.
Immunoprecipitation of endogenous CA12 from PRC3 cell lysate and trypsin digestion
A 50 µL aliquot of anti-CA12-antibody-coated magnetic beads suspension with approximately 20 µg of the captured IgG was added to an aliquot of PRC3 NP-40 cell lysate containing ~ 8.4 mg of total extracted proteins. The tubes were allowed to rock at 4°C for 5 hours; the beads were extracted with a magnet and washed four times with 0.5 mL of 25 mM Tris, pH 7.5, 250 mM NaCl. The antigen was released by incubating the beads in 40 µL of 4 M urea/50 mM HCl for 2 min in a thermomixer (Eppendorf North America, Westbury, NY) set to 25°C and operating at 1000 rpm. A tryptic digest of the immunoaffinity-enriched sample was prepared as follows: proteins were reduced by addition of 0.5 M TCEP to a final concentration of 10 mM. The mixture was incubated at room temperature for 15 min; 0.5 M iodoacetamide was added to a final concentration of 20 mM and the sample was incubated at room temperature for 30 min in the dark. The reaction was quenched with 10 mM DTT for 5 min at room temperature. Samples were cleaned up of urea and excess reagents with Poros 50 R1 perfusion-reversed-phase packing (R1-RP) (Life Technologies, Carlsbad, CA) spin columns made in house. Briefly, 50 µL of R1-RP media was first washed with 3 × 200 µL 80% acetonitrile in 0.1% formic acid; the beads were equilibrated with 3 × 200 µL 0.1% formic acid. The IP eluate sample was added to the R1-RP spin column, excess reagents were washed 3 × 200 µL 0.1% formic acid; washing and elutions were carried out using a benchtop microcentrifuge. Bound protein was eluted with 2 × 50 µL 80% acetonitrile in 0.1% formic acid. Eluates were frozen at −80°C and the volume was evaporated to dryness via vacuum centrifugation and reconstituted in 10 µL of trypsin digestion buffer (5% acetonitrile in 50 mM ammonium bicarbonate, pH 8.0). Finally, 0.5 µL of 0.5 µg/µL trypsin was added to the sample and digestion was allowed to proceed for 17 hours at 37 °C. The resulting sample was acidified with 1.5 µL of 1% formic acid to achieve a final concentration of 0.1%.
LC-SRM quantitative analysis
Preparation of PRC3 blank matrix
An aliquot of PRC3 cell lysate containing 2.0 mg of 7 M urea extracted proteins was reduced, alkylated, and quenched as described above. Urea concentration was diluted 6.5 fold with water prior to digestion with trypsin using a 1:50 w/w enzyme to substrate ratio. Digestion was allowed to proceed at room temperature for 17 hours and it was terminated with formic acid to a final concentration of 1%. Peptides were extracted from the PRC3 cell lysate digest using R1-RP chromatography column packed in house (4.6 × 100-mm) on a Shimadzu HPLC system. The composition of solvent A was 0.1% (v/v) trifluoroacetic acid in HPLC grade water and that of solvent B, 0.1% (v/v) trifluoroacetic acid in acetonitrile. Digested proteins were loaded onto R1 column at 2 mL/min of 2% solvent B and then washed with 2% solvent B for 10 min to remove salts, urea, and trypsin digestion reagents. The bound peptides were eluted at 4 mL/min by stepping to 35% solvent B for 5 min; partially digested and/or non-digested proteins or very hydrophobic peptides were eluted with 95% solvent B for 3 min. The separation was monitored at both 214 and 280 nm. Peptides that eluted with 35% of solvent B were collected and lyophilized as described above. Lyophilized peptides were reconstituted with 5% acetonitrile/0.1% formic acid at a concentration 100-fold lower than the original.
Chromatographic and mass spectrometric conditions
Nano-flow liquid chromatography and peptide detection was performed using the Agilent 1200 Series LC modules connected to the Agilent HPLC-Chip Cube interfaced to the high-performance Agilent 6410 Triple Quadrupole (QQQ) LC-MS/MS (Agilent Technologies, Santa Clara, CA). Agilent Mass Hunter software (version B.02.01) was used for data acquisition and processing. The HPLC-Chip (Protein ID chip) integrates sample concentration on a small volume (40 nL) enrichment column (11 × 0.075 × 0.05 mm3), and peptide separation on an analytical nano-column (43 × 0.075 × 0.05 mm3) packed with Zorbax 300SB-C18, 5 µm. Direct spraying of eluting compounds into the MS instrument was accomplished with a nano-electrospray-tip. Processed samples or peptide standards were loaded onto the enrichment column using an autosampler. Enrichment of the analytes prior to gradient separation was performed by the capillary LC pump delivering 97:5 water/0.1% formic acid (mobile phase A):acetonitrile/0.1% formic acid (mobile phase B) at 4 µL/min. The sample was eluted in backward flush from the enrichment column and transferred to the analytical column by automatic switching of the HPLC-Chip nano-rotary valve. Peptides were separated at a flow rate of 600 nL/min by a nano-pump delivering a linear gradient of 5–70% mobile phase B in 7 min followed by a post-time of 2 min for column re-equilibration. The analyses were performed in positive ionization mode with a capillary voltage set at 1750 V and a Delta EMV of 200 V. The drying gas flow rate was 5 L nitrogen/min and an interface heater temperature of 325 °C. Using MassHunter Optimizer software for peptides, collision energy (CE) voltage was optimized for each SRM transition via the HPLC with column workflow. The MS fragmentor voltage was fixed at 130 V. During an SRM experiment, cycle time did not exceed 1 s; SRM transition dwell times were 100 ms, with Q1 set to “wide” and Q3 to “unit”.
Preparation of standard samples
The internal standard was prepared by adding 64 pmol of each of the SIL peptides to 2 mL of PRC3 blank matrix. The concentration of SIL in the internal standard was 32 pmol/mL. During each analysis, solutions containing 200, 80, 32, 13, 5.1, 2.1 and 0.8 pmol/mL of the UNL peptides were prepared in the presence of a fixed amount, 32 pmol/mL, of the SIL peptides. Four replicates of 1 µL aliquot of each working standard solution were used for LC-SRM analysis. Blank matrix and internal standard samples were prepared to determine the lack of potentially interfering substances present in the cell lysate and internal standard. Using MassHunter quantitative analysis software, calibration curve was obtained by plotting the peak area ratios of UNL versus SIL peptides against the nominal amount of the UNL peptide. A linear curve fitting with 1/× weighting (where × is the concentration of a given standard) was used. The regression equation for the calibration curve was used to back-calculate the measured concentration at each standard level and the result was compared with the theoretical concentration to obtain the accuracy, expressed as a percentage of the theoretical value, for each standard level measured. This calibration curve was also used to back-calculate concentrations of CA12-FLAG standard samples and endogenous CA12 present in PRC3 cell lysate following enrichment on antibody-coated magnetic beads.
To determine the sensitivity of the IP-LC-SRM platform, protein samples were prepared in PRC3 cell lysate diluted 100-fold with 1X PBS at 15, 7.5, 3.8, and 1.9 fmol of CA12 peptide on-column equivalent to 652, 326, 163, and 82 fmol of CA12 protein, respectively. After IP, the samples were digested with trypsin as described above and protein recoveries were quantitated by LC-SRM. Three replicates were prepared for each concentration level.
Results and discussion
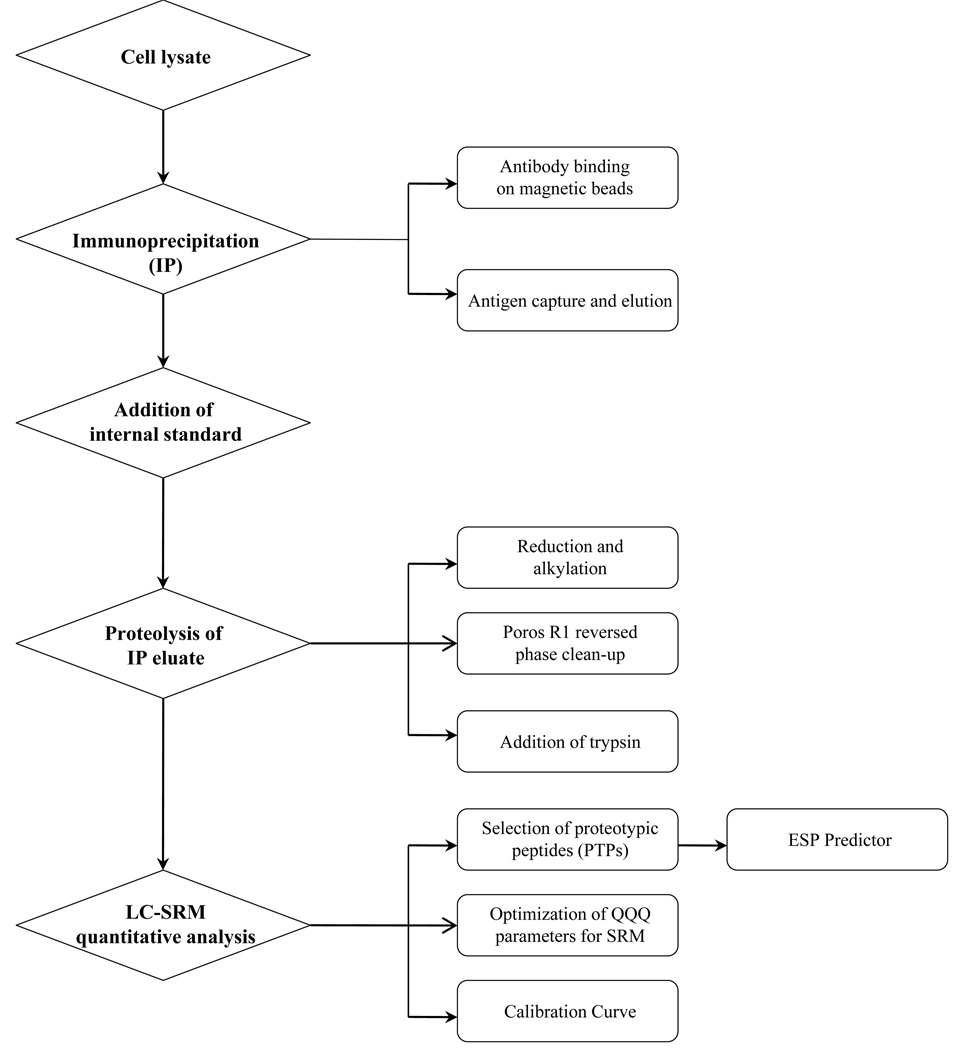
We first explored the detection of the CA12 protein in PRC3 cell lysate using standard proteomic protocols.28 The whole cell lysate was separated by gel electrophoresis on a 1D SDS-PAGE. A band corresponding to the expected migration position of CA12 was digested with trypsin and analyzed by LC-MS/MS in a data-dependent mode or SRM (data not shown). Although 1D gel electrophoresis is a useful means of reducing sample complexity and/or removing common sample contaminants such as detergents and salts, we were not able to detect CA12 using this technique. The limitations of direct LC-MS/MS analysis and 1D SDS-PAGE include potential ion suppression of lower abundance analytes, limited sample loading capacity onto capillary columns for LC-MS/MS and anomalous migration in the gel due to possible post-translational modifications. To overcome these limitations, we developed a quantitative, sensitive, and specific proteomic platform consisting of magnetic bead-based antibody enrichment step and selected reaction monitoring (IP-LC-SRM). A schematic flowchart of the method is presented in Figure 1. The major steps of the method include: (1) immunoprecipitation of target protein from a cell lysate, (2) spiking IP eluate with a stable isotope labeled internal standard, (3) trypsin digestion of IP eluate, and (4) LC-SRM quantitative analysis. The aim of this work was to define each step of the sample preparation process as well as quantitative mass spectrometry that are essential to improve sensitivity of this platform. We also demonstrate the method’s analytical performance and its effectiveness in detecting endogenous CA12 present in RCC cell lysate at low fmol level.
Figure 1.
Workflow for preparation of cell lysate samples and quantitative analysis by IP-LC-SRM.
Selection of proteotypic peptides for LC-SRM quantitation of CA12
Quantitative measurement of disease related changes in protein abundances by LC-SRM uses one or more peptides as surrogate markers, rather than the whole protein as the analyte. To select the proteotypic peptides, we used the enhanced signature peptide (ESP) predictor, a computational method to predict high-responding peptides in an ESI-MS experiment that has been described elsewhere.29 Briefly, the ESP predictor is a classification algorithm that evaluates 550 physicochemical properties for a peptide and predicts the likelihood that the peptide will generate a high response in the mass spectrometer. Some of the top ranked features from the ESP predictor that are used to represent specific characteristics of the peptide include, mass, length, positive charge, hydrophobicity, and gas-phase basicity. CA12 was computationally digested, in silico (no missed cleavages, 600 – 2,800 Da), to produce a set of predicted tryptic peptides. Peptide sequences were input into the ESP predictor and five peptides, which showed the highest probabilities of response (Table 1) were selected. Peptide 1 with a sequence of LNLPSDMHIQGLQSR and the highest probability of response value of 0.56 and peptide 5 with a sequence of WTYFGPDGENSWSK and the least probability ranking of 0.28 were selected for optimization and configuration of the LC-SRM assay. Furthermore, peptide selection was confirmed experimentally by analysis of trypsin digested CA12-FLAG standard by nano-LC-MS/MS on an LTQ linear ion trap mass spectrometer (Thermo, San Jose, CA) with data-dependent acquisition and database matching of collected MS/MS spectra using Bioworks software (Thermo Fisher Scientific) (see supporting information).
Table 1.
Enhance signature peptide (ESP) predictions of the top five SRM peptides for CA12.
| CA12 | |||
|---|---|---|---|
| Peptide Sequence | m/z | ESP prediction |
m/z of transition ions |
| 1. LNLPSDMHIQGLQSR | 570.3 | 0.56 | 684.8, 741.4, 535.3, 798.4, 801.5 |
| 2. GVIYKPATK | 488.8 | 0.53 | 416.3, 707.4, 410.8, 544.4, 820.5 |
| 3. QFLLTNNGHSVK | 679.4 | 0.42 | 856.4, 969.5, 1082.6, 755.4, 541.8 |
| 4. SLHAAAVLLLVILK | 731.0 | 0.31 | 630.9, 811.6, 698.5, 585.4, 910.7 |
| 5. WTYFGPDGENSWSK | 837.4 | 0.28 | 1076.5, 1019.4, 1223.5, 807.4, 1386.6 |
Note. Peptides 1 and 5 were selected as signature peptides of CA12; three transitions per peptide were monitored by SRM (in bold) and the underlined transition was used for quantitation.
Signature peptide characterization and optimization of LC-SRM assay
In order to maximize sensitivity of the SRM assay it is critical to optimize collision energy voltages to give the most abundant fragmentation; hence, the collision energy voltages were optimized for each of the five transitions predicted by the ESP predictor. The collision energy was automatically determined using the QQQ operating in a targeted fashion whereby only m/z corresponding to the doubly, [M + 2H]2+, and triply, [M + 3H]3+, charged form of each peptide were transmitted through Q1 during run. All product ions for each of the candidate peptides and each of their charge states were monitored in Q3. Both Q1 and Q3 resolution was set to “unit” and the dwell time was 5 ms. If more than one charge state was detected the predominant charge state and sequence specific product ions were used to setup the SRM acquisition method for subsequent quantitative analyses of the CA12 protein. Transitions were chosen based upon relative abundance and m/z closest to the precursor m/z on the high m/z side. Both of the signature CA12 peptides are dominated by the y ion formed by gas phase cleavage N-terminal to proline (see supporting information for the list of predicted transition ions).30 Consequently the m/z 570.3 → 684.8 (peptide 1) and m/z 837.4 → 1019.4 (peptide 2) pairs were chosen for quantitation by LC-SRM. To provide qualitative information, we also monitored two additional transitions for each of the peptides, m/z 570.3 → 741.4, m/z 570.3 → 801.5, and m/z 837.4 → 1076.5, m/z 837.4 → 1386.6. The results presented in this paper were derived using the response of the most abundant transition for precursor ion m/z 570.3 → 684.8. However, similar behavior was observed for the most abundant transition for precursor ion m/z 837.4 → 1019.4 (data not shown).
Linearity, reproducibility, and limit of quantitation of peptides
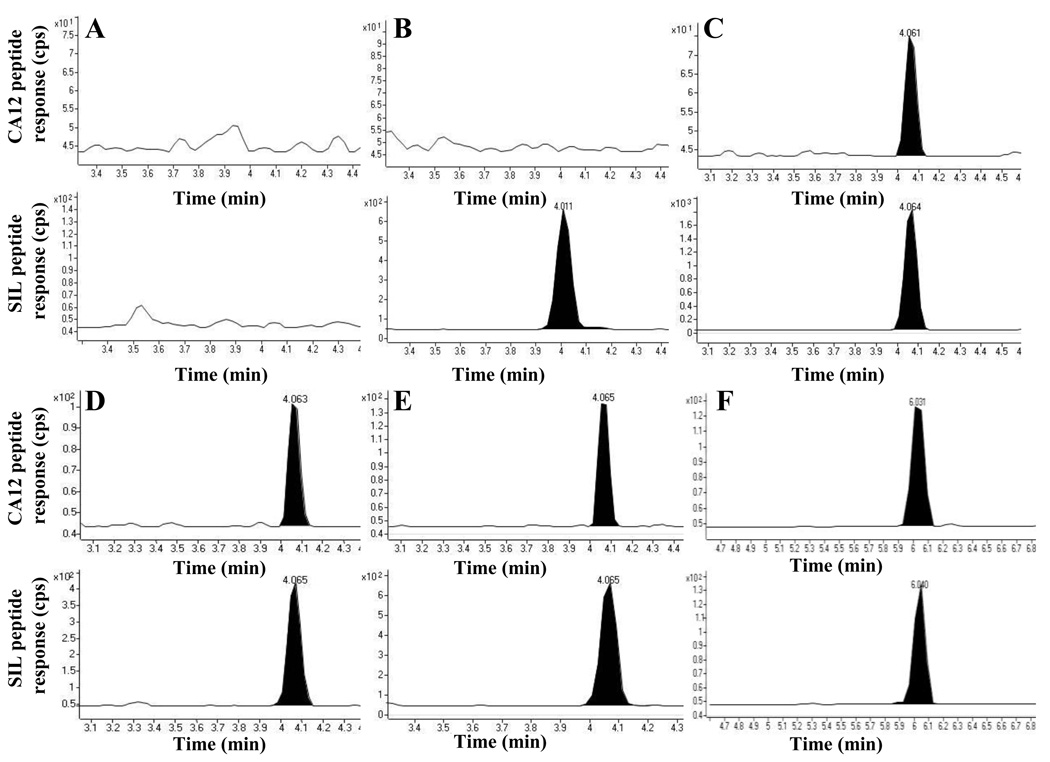
The major factors affecting the reproducibility and quantitative capability of IP-LC-SRM assays are antigen capture/elution efficiency and/or ion suppression in the eluate. Therefore, when analyzing complex samples such as cell lysate or plasma, matrix interference must be taken into consideration in order to define a quantitation limit of the analyte of interest. Hence, linearity, accuracy and precision, limit of quantitation were determined for CA12 peptides using the sample matrix. A blank matrix, produced as described in materials and methods, was selected as a model matrix to define these values and to mimic matrix effects of IP eluate during sample analysis. Peptide standard solutions containing a fixed amount of the SIL peptides and different amounts of the UNL peptides from CA12 were prepared in the blank matrix. A 1 µL injection of each standard solution containing 32 fmol of SIL peptides and seven different levels of the UNL peptides ranging from 0.8 to 200 fmol/µL were analyzed by LC-SRM. Each calibration point was determined in quadruplicate. The calibration curve is shown in Figure 2 (see supporting information); the peak area ratios of UNL versus SIL peptide are plotted against the amounts of the UNL peptide. The linearity was evaluated by comparing the correlation coefficient (r2), accuracy and precision of back-calculated and theoretical amounts of calibration standard samples. A linear correlation was obtained between the peak area ratios and the amounts of the UNL peptide (r2 = 0.9978). The limit of quantitation (LOQ) was approximately 0.8 fmol where the precision and accuracy was 14% and −9.3%, respectively (data not shown). The blank matrix and internal standard samples were found to have no target analytes. Representative extracted ion chromatograms (EIC) for the blank matrix, internal standard, and LOQ are shown in Figure 3 A, B, and C, respectively.
Figure 3.
Extracted ion chromatograms for unlabeled/native CA12-derived peptide m/z 570.3→ 684.8 (top panels) and its corresponding SIL peptide m/z 572.3 → 687.8 (bottom panels). (A) blank matrix (B) SIL internal standard (C) LC-SRM LOQ (D) IP-LC-SRM LOQ (E) PRC3 cell lysate sample detecting endogenous CA12 peptide 1 (F) PRC3 cell lysate sample detecting endogenous CA12 peptide 5 m/z 837.4 → 1019.4 (top panel) and its corresponding SIL peptide m/z 840.4 → 1025.4.
Antigen capture and recovery of CA12-FLAG standard
The effectiveness of IP experiments is an important factor for detection of low abundance proteins. We developed an optimized sample preparation protocol with the objective of maximizing the recovery of the target. To determine the optimal conditions for efficient capture and elution of the target protein we used CA12-FLAG standard as a test antigen and monitored the capture and elution efficiency by LC-SRM. We chose a bead system with high antibody coupling efficiency and low non-specific binding and based on the information available in the literature,31, 32 we selected Protein G magnetic beads for immunoprecipitation of the CA12 antigen prior to analysis by LC-SRM. To test the optimal buffer for elution of CA12-FLAG standard from the antibody-coated magnetic beads, the antigen was eluted from the beads using four different elution buffers (buffer 1: 50 mM hydrochloric acid; buffer 2: 5% acetic acid; buffer 3: 0.2 M sodium carbonate, pH 11; and buffer 4: 4 M urea/50 mM hydrochloric acid). All of the four elution buffers tested yielded recovery of antigen within the first 40 µL of buffer used. The second elution with eluants did not displace any more protein standard. The overall mean recovery across different elution buffers varied from 40 to 82%, Table 2. The results indicated that acidified urea buffer provided the most efficient elution of the antigen with mean percent recovery of 82% while the other commonly used buffers, in IP assays, were not as effective. To determine if the amount of antibody used is adequate for complete capture of endogenous CA12, higher amounts of anti-CA12-polyclonal-antibody-coated Protein G magnetic beads at 100 and 200 µL were incubated with CA12-FLAG standard and 4 M urea/50 mM HCl was used for elution. The amounts of protein recovered using three different levels of antibody were comparable with each other (data not shown), suggesting that even the lowest amount of antibody is adequate for the complete precipitation of endogenous CA12. Therefore, the ratio of 2 µL of antibody per sample with 50 µL of magnetic beads was adopted for all subsequent experiments.
Table 2.
Comparison of the amount of CA12-FLAG recovered after capture on anti-CA12-antibody-coated Protein G magnetic beads and elution under different conditions.
| Elution Condition | IP #1 - CA12-FLAG recovery (%) | IP #2 - CA12-FLAG recovery (%) |
|---|---|---|
| 50 mM HCl | 72 | 66 |
| 5% acetic acid | 56 | 60 |
| 0.2 M sodium carbonate, pH 11.0 | 47 | 33 |
| 4 M urea, 50 mM HCl | 86 | 80 |
Note. Recovery (%) was calculated according to the following formula: (amount measured after IP/amount measured before IP) × 100%.
Accuracy and precision of IP-LC-SRM analysis
Having confirmed the reproducibility of LC-MS measurements, we next evaluated accuracy (recovery) and reproducibility of the antibody capture protocol. Known amounts of CA12-FLAG were spiked into reagent blank and the CA12-FLAG was captured three independent times at four different concentrations. After elution with 4 M urea/50 mM HCl, samples were processed and analyzed by LC-SRM to quantitate recoveries. For intra-batch accuracy and precision, a single capture was performed at four different concentrations of CA12-FLAG ranging between 652 and 82 fmol and triplicate analyses were performed on each of the samples. The inter-batch accuracy and precision of the method was assessed by three independent captures and analyses at all four levels. These results are shown in Table 3. Overall, the intra-batch CVs for the target peptide were within 17% and the mean accuracy of the measurements was within 12% of the expected value. The inter-batch CVs for the three capture experiments ranged from 7.0% to 14%. The biases of the mean for the repeat capture experiments were from 0.1% to 4.9%. In summary, with a maximum of 12% error, the LOQ (Figure 3 D) for the IP-LC-SRM method is about 3.5 fmol of CA12 peptide on-column suggesting that the method is capable of quantitation of as low as 152 fmol of CA12-FLAG in cell lysate. The limit of detection (LOD) was estimated to be in the very low fmol amounts of protein (signal-to-noise ratio of 3 for light peptide). The established method was then applied to analysis of endogenous CA12 expressed in PRC3 cells.
Table 3.
Intra- and inter-assay accuracy and precision for IP-LC-SRM analysis of CA12-FLAG target protein spiked into pRC3 cell lysate blank matrix and captured with anti-CA12-antibody-coated Protein G magnetic beads. The intra- batch mean back-calculated amount, accuracy (percent relative error, %RE), and precision (percent coefficient of variation, %CV) are shown for quantitation of peptide 1 m/z 570.3 → 684.8 performed on three separate days. Each measurement (independent capture) was performed in triplicate (n = 3). The inter-batch mean, accuracy, and precision were calculated using all measurements from the independent captures (n = 9).
| CA12 fmol of peptide on column | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3.8 (163)a | 7.5 (326)a | 15 (652)a | |||||||
| Batch 1 |
Batch 2 |
Batch 3 |
Batch 1 |
Batch 2 |
Batch 3 |
Batch 1 |
Batch 2 |
Batch 3 |
|
| Analysis 1 | 4.1 | 4.0 | 3.9 | 7.8 | 7.7 | 7.1 | 14 | 15 | 15 |
| Analysis 2 | 4.7 | 4.7 | 3.0 | 7.3 | 7.0 | 7.8 | 16 | 16 | 13 |
| Analysis 3 | 3.7 | 3.6 | 3.7 | 9.1 | 7.3 | 9.8 | 15 | 16 | 14 |
| Intra-batch analysis | |||||||||
| Mean concentration | 4.2 | 4.1 | 3.5 | 8.06 | 7.3 | 8.2 | 15 | 16 | 14 |
| Accuracy (%RE) | 12 | 8.6 | −6.0 | 7.5 | −2.4 | 9.6 | 1.0 | 4.6 | −5.5 |
| Precision (%CV) | 12 | 14 | 13 | 12 | 4.8 | 17 | 7.5 | 2.8 | 7.6 |
| Inter-batch analysis | |||||||||
| Mean concentration | 3.9 | 7.9 | 15 | ||||||
| Accuracy (%RE) | 4.8 | 4.9 | 0.1 | ||||||
| Precision (%CV) | 14 | 12 | 7.0 | ||||||
fmol of protein used per spike-and-recovery experiment; the sample with the least amount of CA12-FLAG (82 fmol) gave > 30% difference between the calculated and specified amount, therefore, it was excluded from the analysis.
Measurement of endogenous CA12 levels in RCC cells
A total of three PRC3 cell lysate samples were analyzed for their endogenous CA12 concentrations using this optimized IP-LC-SRM assay. To correct for the recovery yield (~82%, obtained for CA12-FLAG standard), a correction factor of 1.2 was used to multiply the measured values to obtain the calculated amounts present in PRC3 cells. For the three samples, the mean amount of CA12 was found to be at 180 fmol per 8.35 mg of total extracted proteins, and the RSD was 4.6%. Representative LC-SRM traces from endogenous CA12 peptides and SIL standards acquired from one of the cell lysate samples are illustrated in Figure 3 E and F.
Conclusion
The traditional proteomic-based route to protein identification using non-targeted (LC-MS/MS followed by database search) or LC-SRM analysis of a sample produced by in-gel digestion of a band corresponding to CA12 molecular weight failed to detect and identify CA12 protein in RCC PRC3 cells. In order to determine whether it was due to low abundance or non-expression of the protein, we developed a more sensitive and specific proteomic platform consisting of magnetic bead-based antibody enrichment step prior to LC-SRM. To enhance sensitivity of this platform we carefully optimized a method for capture and recovery of the target protein to yield maximum recovery while minimizing nonspecific binding. Using a computational prediction method, we selected surrogate peptide- precursor ions and corresponding fragment ions- transitions for our candidate protein. Synthesis and characterization of the peptide standards allowed us to optimize instrument parameters for the LC-SRM assay.
We demonstrated that this IP-LC-SRM assay is capable of measuring CA12 peptides with high selectivity and specificity; complexity of the samples was greatly reduced after IP resulting in less interference from biological matrix. The analytical performance of the method was validated and the assay was found to be precise and accurate down to 0.8 fmol of CA12 peptide on-column. Although, the analytical LOQ was found to be in the low fmol level, the actual amount of spiked CA12-FLAG that can be measured with this technique was approximately 5-fold higher. This result suggests that the sensitivity of this method critically depends on the specificity of the antibody used; the use of high affinity monoclonal antibodies will likely increase the LOQ levels. Additionally, we showed that the flexibility of this methodology allows for capture from large sample volumes allowing for further enrichment by volume reduction extending analysis to low fmol range sufficient for quantitation of clinically relevant biomarkers. Our future studies will focus on comparative analysis of this potential biomarker in both cancer and non-cancer cell lines as well as clinical samples.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health/National Cancer Institute (NIH/NCI) Grant 1RO1 CA122591 and by Korean Research WCU grant R31-2008-000-10086-0. We also thank Dr. Tomas Rejtar from the Barnett Institute for helpful discussions. Contribution Number 952 from the Barnett Institute.
References
- 1.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Hanash S. Mass Spectrom Rev. 2005;24:413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 3.Wu L, Han DK. Expert Rev Proteomics. 2006;3:611–619. doi: 10.1586/14789450.3.6.611. [DOI] [PubMed] [Google Scholar]
- 4.Arnaud CH. Chemical & Engineering News. 2006;84:17–25. [Google Scholar]
- 5.Blow N. Nature Methods. 2008;5:741–747. doi: 10.1038/nmeth0108-109. [DOI] [PubMed] [Google Scholar]
- 6.Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, Buhlmann P, Bjorck L, Domon B, Aebersold R. Mol Cell Proteomics. 2008;7:1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, Carr SA. Mol Cell Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rifai N, Gillette MA, Carr SA. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 9.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 10.Le Blanc JC, Hager JW, Ilisiu AM, Hunter C, Zhong F, Chu I. Proteomics. 2003;3:859–869. doi: 10.1002/pmic.200300415. [DOI] [PubMed] [Google Scholar]
- 11.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Proc Natl Acad Sci U S A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yocum AK, Chinnaiyan AM. Brief Funct Genomic Proteomic. 2009;8:145–157. doi: 10.1093/bfgp/eln056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 14.Anderson L, Hunter CL. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum D, Colangelo C, Williams K, Gerstein M. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gygi SP, Rochon Y, Franza BR, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rochefort H. Breast Cancer Res Treat. 1990;16:3–13. doi: 10.1007/BF01806570. [DOI] [PubMed] [Google Scholar]
- 18.van Dalen A. Anticancer Res. 1996;16:2345–2349. [PubMed] [Google Scholar]
- 19.Westley B, Rochefort H. Cell. 1980;20:353–362. doi: 10.1016/0092-8674(80)90621-2. [DOI] [PubMed] [Google Scholar]
- 20.Hondermarck H. Mol Cell Proteomics. 2003;2:281–291. doi: 10.1074/mcp.R300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 22.Tureci O, Sahin U, Vollmar E, Siemer S, Gottert E, Seitz G, Parkkila AK, Shah GN, Grubb JH, Pfreundschuh M, Sly WS. Proc Natl Acad Sci U S A. 1998;95:7608–7613. doi: 10.1073/pnas.95.13.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov SV, Kuzmin I, Wei MH, Pack S, Geil L, Johnson BE, Stanbridge EJ, Lerman MI. Proc Natl Acad Sci U S A. 1998;95:12596–12601. doi: 10.1073/pnas.95.21.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao SY, Lerman MI, Stanbridge EJ. BMC Dev Biol. 2009;9:22. doi: 10.1186/1471-213X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hulick PZM, Margulis V, Skates S, Hamel M, Dahl DM, Michaelson DM, Liebermann T, Signoretti S, Carney W, Wood C, Iliopoulos O. Clinical Protoemics. 2009;5:9. [Google Scholar]
- 26.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 27.Harlow ELD. 1988 [Google Scholar]
- 28.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 29.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breci LA, Tabb DL, Yates JR, 3rd, Wysocki VH. Anal Chem. 2003;75:1963–1971. doi: 10.1021/ac026359i. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson GB, Platt FM. Anal Biochem. 1991;199:219–222. doi: 10.1016/0003-2697(91)90093-9. [DOI] [PubMed] [Google Scholar]
- 32.Whiteaker JR, Zhao L, Zhang HY, Feng LC, Piening BD, Anderson L, Paulovich AG. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.