Abstract
HLA-DR0401, 0403 and 0405 are associated with variable T1D susceptibilities when linked with a common HLA-DQ8 (DQA1*0301/DQB1*0302). It is unknown how the modest differences within the peptide binding regions of DR4 subtypes lead to distinct autoimmune risks. Since all Class II HLA molecules share the same intracellular compartments during biosynthesis, it is possible that DQ and DR compete with one another to bind and present antigenic peptides. As such, it is reasonable to hypothesize that a strong DR4 self-peptide binder down-modulates DQ8 epitope presentation more than a weak one. In this study, we first examined the binding of the peptides derived from two putative beta-cell autoantigens – GAD65 and insulin. Protective DR0403 bound similar number of self-peptides as susceptible DR0401 while highly susceptible DR0405 bound substantially less self-peptides than rest two molecules. Kinetic assays were used to further compare the stability of peptide:DR complexes formed between DR0401, 0403 and selected GAD65 peptides, which also bound DQ8. Two peptides with naturally processed DQ8 epitopes bound protective DR0403 with longer half-life and lower dissociation rate than susceptible DR0401, confirming DR0403 as a better peptide competitor than DR0401. The distinguishing peptide binding features of DR0401, DR0403, and DR0405 highlighted in this study help to explain the hierarchy of genetic associations between T1D and these DR4 subtypes. The enhanced peptide competition of DR0403 leads to a down-modulation of DQ8 epitope presentation, as compared to weak competitors such as DR0401 and DR0405, and therefore contributes to disease protection.
Keywords: Type 1 Diabetes, Antigen presentation, HLA-DR4, HLA-DQ8
1. Introduction
Type 1 Diabetes (T1D) is a metabolic disorder caused by autoimmunity that destroys insulin producing pancreatic beta-cells [1–3]. Although environmental mediators are involved in diabetogenesis, the family clustering of disease onset indicates that strong inherited factors participate in diabetes etiology [4, 5]. Approximately 40% of the familial aggregation of T1D is attributed to allelic variation of Class II HLA loci on chromosome 6p21 [6, 7]. A single amino acid difference at position 57 of DQ β-chain (non-Asp β57 for susceptibility vs. Asp β57 for protection) remarkably changes disease susceptibility [8, 9]. In Caucasians, DQB1*0302 is the most prevalent non-Asp β57 susceptible allele in T1D patients. This allele encodes a β-chain for HLA-DQ8, which is responsible for the generation and activation of CD4+ T cells that recognize beta-cell derived autoantigens.
However, the disease predisposition conferred by DQB1*0302 is likely modulated by its closely linked DR loci [10–12]. A previous study reported that DQ8/DR0401 transgenic mice had 3-fold lower incidence (23%) of spontaneous diabetes than DQ8 single transgenic setting (75%) [12]. In humans, when DQ8 is linked with one of the DR4 subtypes (DRA1*0101/DRB1*0401, 0402, 0403, 0404 and 0405), there is a hierarchy of association with the onset of disease [13, 14]. One recent study from the T1D Genetic Consortium indicated that DQB1*0302-DRB1*0405 (OR = 11.37, P = 4 × 10−5) and DQB1*0302-DRB1*0401 (OR = 8.39, P = 6 × 10−36) were associated to T1D with the highest risks, while DQB1*0302-DRB1*0403 was protective (OR= 0.27, P = 0.017) [15]. Since all three haplotypes share a common DQB1*0302, this variable association suggests that certain underlying features allow them to “modify” the susceptibility of DQB1*0302 with variable degrees. However, the mechanisms of this differential modification have not ever been investigated.
Among many yet confirmed interpretations, a peptide competition model proposed that protective Class II HLA alleles compete for diabetogenic peptides with susceptible ones so that the autoreactive T cell responses are diminished or arrested due to insufficient self-antigen presentation [16]. Using Ag-specific T hybridomas from immunized HLA transgenic mice as readouts to evaluate Ag presentation [17], it was demonstrated that the co-expression of DR0401 reduced the epitope presentation by DQ8 [18], suggesting peptide competition (or epitope stealing) between DR0401 and DQ8. However, it is not known if other DR4 subtypes would compete with DQ8 for peptides differently from DR0401. This present study aimed to investigate the interaction between self-peptides and those DR4 subtypes. We used both indirect (equilibrium) and direct (kinetic) peptide binding assays to reveal how the polymorphisms within the peptide binding regions of HLA-DR0401, 0403 and 0405 lead to different capacities for binding self-peptides. These observations suggested a mechanism through which DQ8 epitope presentation is modulated by various DR4 subtypes, leading to different degrees of T1D predisposition.
2. Materials and methods
2.1. Peptides and Recombinant HLA-DR proteins
Partially (12 amino acid residues) overlapped peptide 20-mer panels covering human Glutamic Acid Decarboxylase 65 (GAD65), preproinsulin (PPI) and influenza A/Puerto Rico/8/34 (H1N1) M1 matrix protein (M1MP) were purchased from Mimotopes (Clayton Victoria, Australia). Biotinylated peptides were purchased from Genescript (Piscataway, NJ). Recombinant cDNAs for HLA-DRA1*0101, DRB1*0401, DRB1*0403 and DRB1*0405 fused with leucine zipper sequences were constructed as previously described [19]. The chimeric cDNAs were cloned into the Schneider expression vectors pRmHa-3 and co-transfected with selection plasmid pUChsneo into Schneider cells S-2 by calcium phosphate. High expression cell clones were selected with 2 mg/mL Geneticin (Invitrogen, Carlsbad, CA) supplemented selection medium. Cells were expanded and grown to a density of 6x106 cells/mL. CuSO4 was added at a concentration of 1 mM to induce the production of soluble class II molecules. The DR0401 molecules were purified by L243 affinity chromatography. Eluted HLA-DR recombinant proteins were dialyzed into 200 mM pH6.0 sodium phosphate buffer. For this study, all purified recombinant proteins were not biotinylated.
2.2. Indirect binding Assay
Biotinylated influenza hemagglutinin derived peptide HA306 (PKYVKQNTLKLAT) was used as a reference peptide for DR0401 (ED50 for HA306 to bind 0.05 µM DR0401=0.035 µM from a 16-hour direct binding assay) and DR0405 (ED50 for HA306 to bind 0.05 µM DR0405=0.271 µM) binding assays [20]. Since HA306 was unable to bind DR0403 (ED50 for HA306 to bind 0.05 µM DR0403≫15 µM), as an alternative, biotinylated GAD65-557I (NFIRMVISNPAAT) was used as a reference peptide for DR0403 binding assays (ED50 for 557I to bind 0.05 µM DR0403=0.01 µM). Non-biotinylated HLA-DR proteins were diluted into 150 mM pH5.4 citrate-phosphate buffer containing 7.5 mg/ml of n-Octyl-β-D-Glucopyranoside and 1 mM Pefabloc. The final concentration of DR protein was 0.05 µM. Serially diluted non-biotinylated target peptides (ranged from 0, 0.01, 0.05, 0.1, 0.5, 1, 5 and 10 µM) were incubated with DR protein respectively for 1 hour at 37 °C before adding 0.01 µM of reference peptide. The incubation was stopped after 16 hours by adding equal volume 50 mM pH8.0 Tris-Cl buffer and transferred to ELISA plate coated with anti-HLA-DR monoclonal antibody L243 (10 µg/ml). The ELISA was performed in triplicate using Europium-Streptavidin as detection (Perkin Elmer, Waltham, MA) to determine the binding of reference peptides. The concentrations of target peptides required to inhibit 50% maximal reference peptide binding (IC50) were calculated from regression curves fitted by sigmoidal dose-response equation provided by Prism software (GraphPad, San Diego, CA). Coefficient of variation for europium readings from each peptide cncentration triplicate group was less than 10 % prior to curve generation and IC50 calculation. At least 6 data points were used for curve fitting.
2.3. Kinetic Analysis (direct peptide binding)
Peptide:MHC binding stability was measured as the rate of peptide association and dissociation from target Class II HLA molecules and half-life of peptide:MHC complex as described previously [21, 22]. For dissociation biotinylated target peptide (0.1 µM) mixed with equal amounts of purified DR protein were incubated at 37 °C for 72 hours in 150 mM pH5.4 citrate-phosphate buffer containing 2.5 mg/ml of n-Octyl-B-D-Glucopyranoside and 1 mM Pefabloc. After removal unbound biotinylated peptide by dialysis, the protein concentration was adjusted to 0.05 µM and incubated an additional 72 hours at 37 °C in the presence of 100 µM non-biotinylated cognate peptide. At different time points, a small fraction of sample was removed and stored in −20 °C with the addition of an equal volume of 50 mM pH8.0 Tris-Cl buffer. For association, biotinylated target peptide was mixed with 0.05 µM purified DR protein and incubated at 37 °C for 72 hours, with small fractions of the sample collected and frozen at different time points. The biotinylated peptide that remained bound to the target DR protein in each sample was determined by ELISA. Based on the binding results from different time points, the dissociation curve was simulated by one-phase exponential decay equation provided by Prism software. The dissociation rate constant (Kd), binding half-life (t1/2) and corresponding 95% confidence intervals were also determined from the simulated curve.
2.4. Statistics
To calculate p-values for kinetic results, an F-test was used to evaluate the difference between dissociation rate constant (Kd) of peptide:DR4 complexes.
3. Results
3.1. Binding between candidate peptides and recombinant HLA-DR4 proteins
A panel of GAD65 derived peptides was tested for binding to purified HLA-DR proteins through indirect binding assay. The observed IC50 < 10 uM (a concentration equivalent to 1000-fold labeled reference peptide input, which was 0.01 uM) was assigned as the criterion to determine whether a candidate peptide could successfully bind target DR protein. Out of 72 GAD65-derived peptides, 21 peptides were able to bind DR0401. The best fitted IC50 values of these 21 binding reactions and relevant GAD65 peptide sequences are detailed in Table 1. DR0403 bound 23 GAD65 peptides. Only 8 peptides were able to bind DR0405. The peptide binding assay results from preproinsulin showed that 2 peptides bound DR0401 and DR0403 (Table 2). No insulin peptide was able to bind DR0405. To assess whether the weak binding nature of DR0405 was restricted to GAD65 and insulin (auto-antigens), we also tested the binding of peptides derived from Influenza A M1 Matrix Protein – a foreign antigen unrelated to T1D. Out of 30 peptides covering the entire sequence of MP1 (252 residues), 12 peptides bound DR0401 and 18 peptides bound DR0403, while only 3 peptides were able to bind DR0405 (Table 3). Together, the binding results from two T1D-related self-antigens and one foreign antigen suggested that DR0405 was intrinsically less capable of binding peptides than DR0401 and DR0403. On the other hand, both DR0401 and DR0403 bound a broad range of peptides. Since different reference peptides were used to calculate IC50 values for these two DR4 subtypes, a direct comparison of binding affinity between the peptides and DR0401 or DR0403 was not possible.
Table 1.
HLA-DR4 binding results of 72 GAD65-derived 20-mer peptides.
| I.D |
Sequence |
IC50 (uM) |
||
|---|---|---|---|---|
| 0401† |
0403§ |
0405† |
||
| p1 | 1MASPGSGFWSFGSEDGSGDS20 | - | - | - |
| p2 | 9WSFGSEDGSGDSENPGTARA28 | - | - | - |
| p3 | 17SGDSENPGTARAWCQVAQKF36 | - | - | - |
| p4 | 25TARAWCQVAQKFTGGIGNKL44 | - | - | - |
| p5 | 33AQKFTGGIGNKLCALLYGDA52 | - | - | - |
| p6 | 41GNKLCALLYGDAEKPAESGG60 | - | - | - |
| p7 | 49YGDAEKPAESGGSQPPRAAA68 | - | - | - |
| p8 | 57ESGGSQPPRAAARKAACACD76 | - | - | - |
| p9 | 65RAAARKAACACDQKPCSCSK84 | - | - | - |
| p10 | 73CACDQKPCSCSKVDVNYAFL92 | 0.35 | 3.55 | - |
| p11 | 81SCSKVDVNYAFLHATDLLPA100 | 0.67 | - | - |
| p12 | 89YAFLHATDLLPACDGERPTL108 | 0.54 | - | - |
| p13 | 97LLPACDGERPTLAFLQDVMN116 | - | - | - |
| p14 | 105RPTLAFLQDVMNILLQYVVK124 | - | - | - |
| p15 | 113DVMNILLQYVVKSFDRSTKV132 | 5.08 | 0.30 | - |
| p16 | 121YVVKSFDRSTKVIDFHYPNE140 | - | - | - |
| p17 | 129STKVIDFHYPNELLQEYNWE148 | - | - | - |
| p18 | 137YPNELLQEYNWELADQPQNL156 | - | - | - |
| p19 | 145YNWELADQPQNLEEILMHCQ164 | - | - | - |
| p20 | 153PQNLEEILMHCQTTLKYAIK172 | - | 4.81 | - |
| p21 | 161MHCQTTLKYAIKTGHPRYFN180 | - | - | - |
| p22 | 169YAIKTGHPRYFNQLSTGLDM188 | 1.04 | 10.00 | 0.03 |
| p23 | 177RYFNQLSTGLDMVGLAADWL196 | 0.34 | 1.34 | 0.13 |
| p24 | 185GLDMVGLAADWLTSTANTNM204 | 9.68 | 2.67 | - |
| p25 | 193ADWLTSTANTNMFTYEIAPV212 | 0.69 | 0.55 | - |
| p26 | 201NTNMFTYEIAPVFVLLEYVT220 | 0.82 | 0.90 | 0.15 |
| p27 | 209IAPVFVLLEYVTLKKMREII228 | - | 4.49 | - |
| p28 | 217EYVTLKKMREIIGWPGGSGD236 | - | - | - |
| p29 | 225REIIGWPGGSGDGIFSPGGA244 | - | - | - |
| p30 | 233GSGDGIFSPGGAISNMYAMM252 | 0.35 | - | - |
| p31 | 241PGGAISNMYAMMIARFKMFP260 | - | - | - |
| p32 | 249YAMMIARFKMFPEVKEKGMA268 | - | 10.00 | 0.39 |
| p33 | 257KMFPEVKEKGMAALPRLIAF276 | - | - | - |
| p34 | 265KGMAALPRLIAFTSEHSHFS284 | 0.31 | 0.44 | - |
| p35 | 273LIAFTSEHSHFSLKKGAAAL292 | 0.96 | - | - |
| p36 | 281SHFSLKKGAAALGIGTDSVI300 | - | - | - |
| p37 | 289AAALGIGTDSVILIKCDERG308 | - | - | - |
| p38 | 297DSVILIKCDERGKMIPSDLE316 | - | - | - |
| p39 | 305DERGKMIPSDLERRILEAKQ324 | 0.51 | - | - |
| p40 | 313SDLERRILEAKQKGFVPFLV332 | - | - | - |
| p41 | 321EAKQKGFVPFLVSATAGTTV340 | 0.05 | 1.87 | - |
| p42 | 329PFLVSATAGTTVYGAFDPLL348 | 2.07 | 1.27 | - |
| p43 | 337GTTVYGAFDPLLAVADICKK356 | - | 1.28 | 5.82 |
| p44 | 345DPLLAVADICKKYKIWMHVD364 | - | - | - |
| p45 | 353ICKKYKIWMHVDAAWGGGLL372 | 2.85 | - | - |
| p46 | 361MHVDAAWGGGLLMSRKHKWK380 | - | - | - |
| p47 | 369GGLLMSRKHKWKLSGVERAN388 | 1.09 | 0.42 | - |
| p48 | 377HKWKLSGVERANSVTWNPHK396 | - | 0.19 | - |
| p49 | 385ERANSVTWNPHKMMGVPLQC404 | - | - | - |
| p50 | 393NPHKMMGVPLQCSALLVREE412 | - | - | - |
| p51 | 401PLQCSALLVREEGLMQNCNQ420 | - | - | - |
| p52 | 409VREEGLMQNCNQMHASYLFQ428 | - | - | - |
| p53 | 417NCNQMHASYLFQQDKHYDLS436 | - | - | - |
| p54 | 425YLFQQDKHYDLSYDTGDKAL444 | - | - | - |
| p55 | 433YDLSYDTGDKALQCGRHVDV452 | 10.00 | - | - |
| p56 | 441DKALQCGRHVDVFKLWLMWR460 | - | - | - |
| p57 | 449HVDVFKLWLMWRAKGTTGFE468 | - | 8.20 | - |
| p58 | 457LMWRAKGTTGFEAHVDKCLE476 | - | - | - |
| p59 | 465TGFEAHVDKCLELAEYLYNI484 | - | - | - |
| p60 | 473KCLELAEYLYNIIKNREGYE492 | - | - | - |
| p61 | 481LYNIIKNREGYEMVFDGKPQ500 | - | - | - |
| p62 | 489EGYEMVFDGKPQHTNVCFWY508 | - | - | - |
| p63 | 497GKPQHTNVCFWYIPPSLRTL516 | - | - | - |
| p64 | 505CFWYIPPSLRTLEDNEERMS524 | - | 1.00 | 0.57 |
| p65 | 513LRTLEDNEERMSRLSKVAPV532 | - | 3.24 | - |
| p66 | 521ERMSRLSKVAPVIKARMMEY540 | - | 4.17 | - |
| p67 | 529VAPVIKARMMEYGTTMVSYQ548 | 0.64 | 1.61 | - |
| p68 | 537MMEYGTTMVSYQPLGDKVNF556 | - | - | - |
| p69 | 545VSYQPLGDKVNFFRMVISNP564 | 0.46 | 0.86 | 0.61 |
| p70 | 553KVNFFRMVISNPAATHQDID572 | 0.03 | 0.03 | 0.07 |
| p71 | 561ISNPAATHQDIDFLIEEIER580 | - | - | - |
| p72 | 569QDIDFLIEEIERLGQDL588 | - | - | - |
| Number of peptides with detectable binding reaction | 21 | 23 | 8 | |
Using HA307-319 (PKYVKQNTLKLAT) as reference peptide.
Using 557I (NFIRMVISNPAAT) as reference peptide.
Table 2.
HLA-DR4 binding results of 13 preproinsulin-derived 20-mer peptides.
| I.D |
Sequence |
IC50 (uM) |
||
|---|---|---|---|---|
| 0401† |
0403§ |
0405† |
||
| p1 | 1MALWMRLLPLLALLALWGPD20 | - | - | - |
| p2 | 9PLLALLALWGPDPAAAFVNQ28 | - | - | - |
| p3 | 17WGPDPAAAFVNQHLCGSHLV36 | - | - | - |
| p4 | 25FVNQHLCGSHLVEALYLVCG44 | - | - | - |
| p5 | 33SHLVEALYLVCGERGFFYTP52 | - | - | - |
| p6 | 41LVCGERGFFYTPKTRREAED60 | - | - | - |
| p7 | 49FYTPKTRREAEDLQVGQVEL68 | - | - | - |
| p8 | 57EAEDLQVGQVELGGGPGAGS76 | - | - | - |
| p9 | 65QVELGGGPGAGSLQPLALEG84 | - | - | - |
| p10 | 73GAGSLQPLALEGSLQKRGIV92 | 8.75 | - | - |
| p11 | 81ALEGSLQKRGIVEQCCTSIC100 | - | - | - |
| p12 | 89RGIVEQCCTSICSLYQLENY108 | 0.22 | 1.89 | - |
| p13 | 97TSICSLYQLENYCN110 | - | 0.47 | - |
| Number of peptides with detectable binding reaction | 2 | 2 | 0 | |
Using HA307-319 (PKYVKQNTLKLAT) as reference peptide.
Using 557I (NFIRMVISNPAAT) as reference peptide.
Table 3.
HLA-DR4 binding results of 30 H1N1 Matrix Protein-derived 20-mer peptides.
| I.D |
Sequence |
IC50 (uM) |
||
|---|---|---|---|---|
| 0401† |
0403§ |
0405† |
||
| p1 | 1MSLLTEVETYVLSIVPSGPL20 | 0.05 | 1.49 | - |
| p2 | 9TYVLSIVPSGPLKAEIAQRL28 | 1.36 | 1.16 | - |
| p3 | 17SGPLKAEIAQRLENVFAGKN36 | - | 2.63 | - |
| p4 | 25AQRLENVFAGKNTDLEALME44 | - | - | - |
| p5 | 33AGKNTDLEALMEWLKTRPIL52 | - | - | - |
| p6 | 41ALMEWLKTRPILSPLTKGIL60 | - | - | - |
| p7 | 49RPILSPLTKGILGFVFTLTV68 | - | - | - |
| p8 | 57KGILGFVFTLTVPSERGLQR76 | 0.17 | 0.18 | 1.41 |
| p9 | 65TLTVPSERGLQRRRFVQNAL84 | - | - | - |
| p10 | 73GLQRRRFVQNALNGNGDPNN92 | 2.10 | 0.12 | - |
| p11 | 81QNALNGNGDPNNMDRAVKLY100 | - | - | - |
| p12 | 89DPNNMDRAVKLYRKLKREIT108 | - | - | - |
| p13 | 97VKLYRKLKREITFHGAKEIA116 | 0.26 | 0.49 | - |
| p14 | 105REITFHGAKEIALSYSAGAL124 | 3.01 | 0.12 | - |
| p15 | 113KEIALSYSAGALASCMGLIY132 | - | 0.55 | - |
| p16 | 121AGALASCMGLIYNRMGAVTT140 | - | 0.26 | - |
| p17 | 129GLIYNRMGAVTTESAFGLIC148 | 0.76 | 0.73 | 10.00 |
| p18 | 137AVTTESAFGLICATCEQIAD156 | - | - | - |
| p19 | 145GLICATCEQIADSQHKSHRQ164 | - | - | - |
| p20 | 153QIADSQHKSHRQMVTTTNPL172 | - | 9.45 | - |
| p21 | 161SHRQMVTTTNPLIRHENRMV180 | 2.97 | 1.79 | - |
| p22 | 169TNPLIRHENRMVLASTTAKA188 | 0.46 | 0.04 | - |
| p23 | 177NRMVLASTTAKAMEQMAGSS196 | 0.16 | 0.04 | - |
| p24 | 185TAKAMEQMAGSSEQAAEAME204 | - | 0.81 | - |
| p25 | 193AGSSEQAAEAMEVASQARQM212 | - | - | - |
| p26 | 201EAMEVASQARQMVQAMRAIG220 | - | 1.69 | - |
| p27 | 209ARQMVQAMRAIGTHPSSSTG228 | 2.92 | 0.40 | - |
| p28 | 217RAIGTHPSSSTGLKNDLLEN236 | - | - | - |
| p29 | 225SSTGLKNDLLENLQAYQKRM244 | - | 0.80 | - |
| p30 | 233LLENLQAYQKRMGVQMQRFK252 | 0.82 | - | 4.47 |
| Number of peptides with detectable binding reaction | 12 | 18 | 3 | |
Using HA307-319 (PKYVKQNTLKLAT) as reference peptide.
Using 557I (NFIRMVISNPAAT) as reference peptide.
3.2. Differential peptide competition: DRB1*0401 and DRB1*0403 bound GAD65 peptides with variable affinities and competed for peptides with DQ8 differently
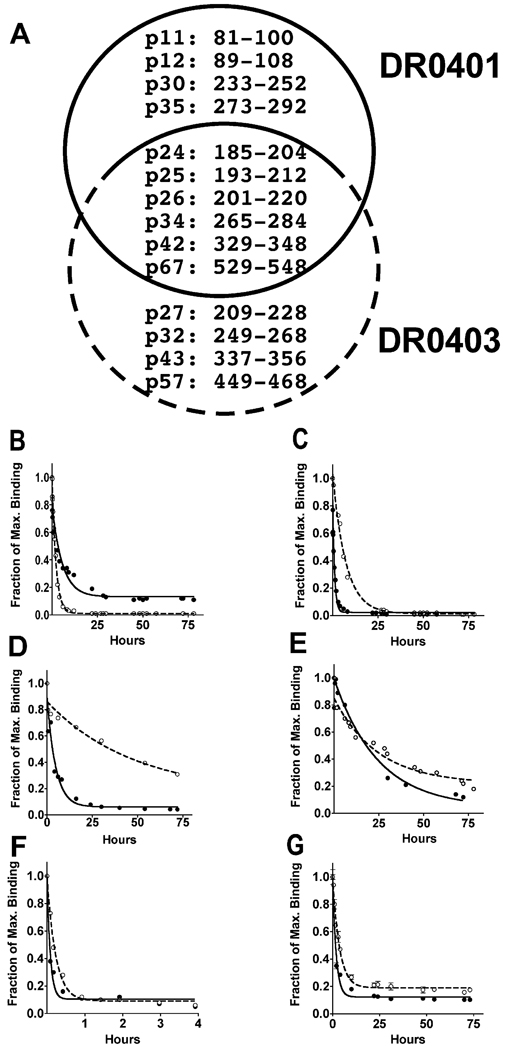
We next chose kinetic assays to examine binding stability between selected GAD65 derived peptides and DR0401 or DR0403. Unlike the competition assay, kinetic assays did not rely on a reference peptide to calculate IC50 values as indicators of relative binding affinity. Instead it measured association rate (Kd, in hour−1), dissociation rate (Kd, in hour−1) and half-life (t1/2, in hour) of a peptide:MHC complex. The difference in binding affinity was determined by a single parameter – time. As shown in supplementary figure 1, we found that Kd was the most meaningful parameter for comparing the relative stability of peptide binding. Based on these results, we decided to focus on dissociation kinetics in our study. We were particularly interested in those peptides also bound DQ8 [23]. A previous study suggested that the presentation of DQ8 epitopes was differentially down-modulated by the co-expression of different DR4 subtypes [18]. Therefore, interrogating peptide binding stability would provide biochemical evidence to explain those results. Combining our DR0401 and DR0403 peptide binding results in Table 1 and a previous DQ8 peptide binding study [23], we found 14 GAD65 peptides that bound DQ8 could also bind DR0401 and/or DR0403 (Fig. 1A). Six peptides bound DQ8, DR0401 and DR0403. Two peptides, GAD65p26 and GAD65p67 actually contained naturally processed DQ8 T epitopes [24]. The dissociation curves showed that GAD65p25, GAD65p26, GAD65p34, GAD65p42 and GAD65p67 dissociated from DR0403 more slowly than from DR0401 (Fig. 1C–G and Table 4). This indicated that these peptides formed more stable complexes with DR0403 than with DR0401. On the other hand, GAD65p24 bound DR0401 with higher stability than DR0403 (Fig. 1B), indicated by a lower dissociation rate and higher half-life. Taken together, kinetic measurements revealed relatively higher binding affinity between DR0403 and most selected GAD65 derived peptides than DR0401 – differences that could not be observed using the indirect binding assay. No kinetic assays were performed for the insulin derived peptides since none of the insulin peptides that bound DR0401 and/or DR0403 were known to bind to bind to DQ8. For example, 9–23BIns a well studied DQ8-restrcited insulin epitope was not bound by DR0401 or DR0403 [25, 26].
Fig. 1.
Kinetic stability of peptide:MHC complex composed by DR0401 or DR0403 with selected GAD65 derived peptides. (A) DQ8 bound GAD65 peptides also bind DR0401 (within black line circle) and/or DR0403 (within blue line circle). The dissociation of GAD65p24 (B), p25 (C), p26 (D), p34 (E), p42 (F) and p67 (G) from HLA-DR0401 (black circles, solid lines) and HLA-DR0403 (open circles, dotted lines) was measured as the remaining amount of biotinylated target peptides against experimental time. The dissociation curves were generated with a single-exponential decay fit [Equation: Y= Span*exp(−Kd*X)+Plateau].
Table 4.
Binding kinetics of HLA-DR0401 and DR0403.
| Peptide |
at1/2 (hour) |
bKd |
|||
|---|---|---|---|---|---|
| 0401 |
0403 |
0401 |
0403 |
p (F-test) |
|
| GAD65p24 | 3.92 (2.87, 6.16) | 1.51 (1.40, 1.65) | 0.0030 (0.0019, 0.0040) | 0.0076 (0.0070, 0.0083) | < 0.0001 |
| GAD65p25 | 0.70 (0.57, 0.91) | 5.07 (4.48, 5.83) | 0.0165 (0.0127, 0.0203) | 0.0023 (0.0020, 0.0026) | < 0.0001 |
| GAD65p26 | 3.30 (2.31, 5.80) | ≫ c33.40 | 0.2098 (0.1195, 0.3001) | 0.0208 (0.0001, 0.0640) | 0.0047 |
| GAD65p34 | 13.13 (10.30, 18.11) | 22.72 (14.86, 48.22) | 0.0528 (0.0382, 0.0672) | 0.0305 (0.0144, 0.0466) | 0.0728 |
| GAD65p42 | 0.06 (0.04, 0.09) | 0.16 (0.13, 0.20) | 0.1952 (0.1267, 0.2636) | 0.0727 (0.0574, 0.0879) | 0.0001 |
| GAD65p67 | 1.29 (1.13, 1.50) | 2.51 (2.25, 2.83) | 0.5377 (0.4621, 0.6133) | 0.2762 (0.2446, 0.3079) | < 0.0001 |
Half life of peptide:MHC complex, in hour
Dissociation constant, the best fit value 95 % CI
Unable to estimate the half life from the dissociation curve since the dissociation is not completed by 72 hours when the assay was stopped.
4. Discussion
It has long been established that a diabetes susceptible HLA haplotype is best defined by a combination of HLA-DQ and -DR alleles [10, 11]. The effort to dissect the roles of individual DQ and DR allele to T1D development is extremely difficult. However, a hierarchy of risks associated with a panel of DQ8-DR4 haplotypes provides opportunities to investigate the differential contribution of these individual DR4 subtypes such as DR0401, 0403 and 0405. Since these haplotypes share a common DQ8, the variable genetic risks associated with these haplotypes are unambiguously due to DRB1*04 alleles themselves. They could either direct distinct autoimmune T cell responses independent to DQ8 or affect DQ8-restricted autoreactive T cell responses through down-modulating DQ8 epitope presentation in variable degrees. An essential task to address these questions is to dissect peptide binding properties – a study that had not been systemically conducted in the context of variable risks for T1D development.
Naturally, these three DR4 subtypes differ from each other by amino acid residues at position 57, 71, 74 and 86 of their beta-chains (Fig. 2). These polymorphisms are critical to shape the DR peptide binding pockets (P1-β86; P4-β71; P9-β57) and they are intrinsic factors leading to differential peptide binding activities, especially the residues at β86 and β57 -- two important anchoring positions [27]. Most Class II HLA molecules encode an Aspartic Acid at β57 , which interacts with an Arginine at α76 (for HLA-DR) or α79 (for HLA-DQ) through an intramolecular salt-bridge [25] -- a feature that has been considered to stabilize peptide binding. Class II MHC heterodimers without this feature have been characterized as poor peptide binders such as T1D susceptible HLA-DQ2 and DQ8 in humans as well as H2-Ag7 in NOD mice [21, 28]. Our results indicated that DR0405, with a Serine instead of an Aspartic Acid at β57, bound a very limited spectrum of peptides. The observation significantly contrasted to the results of DR0401 and DR0403 – both bearing an Asp at β57. Since peptides from more than one protein were studied, the weak binding nature of DR0405 was not an atypical phenomenon. Our estimation is that this distinction will also apply for other antigens in general. The limited spectrum of self-peptide binding for DR0405 reiterates that the ability of the MHC molecule to bind a large set of self-peptides is not a prerequisite for being a diabetes susceptible allele. It also suggests that DR0405 may be less capable of competing for autoantigens with DQ8 than DR0403 and DR0401 simply because it doesn’t have many candidate peptides to compete for. In addition, previous studies have shown that DRB1*0405 is a prevalent allele in Japanese T1D populations [29–31], in which DRB1*0405 is in closely linkage disequilibrium with DQB1*0401 – one of the β57 Asp alleles that are generally regarded as diabetes resistant DQ alleles. This implies that without the predisposition of a non-Asp β57 DQ allele, DRB1*0405 may also be predisposing and directly contribute to the progression of diabetes like other non-Asp β57 alleles such as DQ2, DQ8 and H2-Ag7.
Fig. 2.
The peptide binding groove sequence alignment of DR4 β-chains. The amino acid sequences of DRB1*0403 and DRB1*0405 peptide binding region were aligned to DRB1*0401. The dashes represent amino acid residues identical to DRB1*0401 reference.
The distinction between DR0401 and DR0403 was subtle in indirect binding assay results. Despite that fact that DR0403 differs from DR0401 with three residues along the peptide binding groove, including a Valine (instead of a Glycine) at the position of β86 – a feature that doesn’t allow DR0403 to accommodate big side-chain residues at P1, the peptide binding capability of DR0403 is not inferior to DR0401 at all. In fact, the number of peptides bound by DR0403 is slightly higher than DR0401 for all three antigens studied (Table 1, 2 and 3). Several epitopes (more than 50%) are overlapped for the two alleles. The major finding was the differential binding stability of DR0401 vs. DR0403 when they formed complexes with those peptides which also bound to DQ8. The majority of them (5 out of 6) bound DR0403 with enhanced stability. Two of these peptides encompassed naturally processed DQ8 epitopes -- GAD65208–217 and GAD65539–548 [24]. As demonstrated previously, the presentation of these two epitopes by DQ8 was diminished by the co-expression of DR0401 and the down-modulation was enhanced if antigen presenting cells expressed DR0403 and/or DR0406 instead of DR0401 (It should be noted that the peptide binding ability of DR0403 and DR0406 is likely to be the same because the peptide binding region sequences of these two alleles are identical) [18]. However, that study lacked any direct result showing DR0403 could compete for peptide with DQ8 better than DR0401. In this study, we provide evidence that DQ8 and multiple DR4 subtypes can bind to a shared set of epitopes and that among these, the protective DR0403 subtype binds better to most of these epitopes than DR0401. The enhanced binding of DR0403 provides additional support for the peptide competition hypothesis [16], which emphasizes the relative potentials of DR4 molecules to out-compete DQ8 for epitopes binding and diminish DQ8-restricted autoreactive CD4+ T cell responses. Due to the limited knowledge of DQ8 epitopes, especially the T cell response results, our investigation only focused on GAD65 derived peptides. However, the concept of enhanced peptide competition of DR0403 was validated. Given the fact that DR0403 can bind a broad range of peptides, it is unlikely that DQ8-DR4 peptide competition is limited only to GAD65 derived peptides as demonstrated here. It appears reasonable to conclude that the differential T1D predisposition associated with DQ8-DR0403 and DQ8-DR0401 arises (at least in part) because DR0403 is superior to DR0401 in its overall peptide competition potential, leading to down-modulation of DQ8-restricted CD4+ T cell responses. We also attempted to examine whether protective DR0403 was more able to direct negative selection than susceptible DR0401 using peptide loaded tetramer to detect in vitro primed CD4 T cells from DR0401 and DR0403 subjects. A small number of samples were examined and GAD65-specific T cells were repeatedly detected in both DR0403 and DR0401 individuals (data not shown). A negative selection model was not likely capable of explaining why DR0403 was so protective to T1D while DR0401 was susceptible. However, we were not able to rule out the possibility that the DR0403-restricted T cells detected in DR0403 donors were less pathogenic than DR0401-restricted T cells found in DR0401 donors. This leaves some open questions for future study using more DR0403+ subjects and incorporating a better understanding of DR0403-restricted epitopes within other islet auto-antigens once that information becomes available.
Both binding assays we used in this study were restricted to the cell-free circumstance and therefore lacked the effects of HLA-DM and other chaperones. For technical reasons, our indirect binding assay utilized a high affinity reference peptide. Using this reference peptide probably influenced our ability to distinguish peptides with low-affinity binding, which have been shown to be important for insulin in particular [32]. In spite of these limitations our findings provide valuable insights about the behavior of peptides with strong to moderately weak binding.
In summary, this study used two types of assays to differentiate peptide binding features of highly homologous HLA-DR0401, DR0403 and DR0405 in aiming to elucidate their distinguishable association to autoimmune type 1 Diabetes. The weak peptide binding feature of susceptible DR0405 remarkably contrasted to the protective DR0403 and another disease susceptible subtype DR0401. The susceptible DR0401 and protective DR0403 do not differ from each other significantly in number of peptide binding. However, these two DR4 subtypes bind DQ8 epitope with different stability, which can translate into their differential effects in out-competing DQ8 to bind self-antigen derived peptides. Our findings provide a mechanism explaining why the risk associated with the T1D-susceptible non-Asp57b DQ8 allele is modified by different DR4 subtypes, as reported by previous genetic studies. These results suggest that DR0403 (and perhaps other strong Class II peptide binders) modify the risk of autoimmune diabetes development based on the capacity to bind and retain antigenic self-peptides. As a corollary of these findings, DR4 restricted cells are likely to be less pathogenic than DQ8 restricted cells of the same specificity. In this case, therapeutic strategies aimed at T cell deletion should be designed to focus on DQ8 restricted cells.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Gerald Nepom and Dr. Jane Buckner for their advice and critical review of this manuscript. We would also like to thank Ms. Diana Sorus for assisting submission preparation. This work was supported in part by grants to W.K from American Diabetes Association (ADA 7-06-RA-75) and National Institutes of Health (R21 DK077525-02). X.G was supported by Juvenile Diabetes Research Foundation International Postdoctoral Fellowship Award (3-2008-548)
Abbreviation
- T1D
Type 1 Diabetes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest disclosure
The authors declare no competing financial interests.
References
- 1.Castano L, Eisenbarth GS. Type-I diabetes: a chronic autoimmune disease of human, mouse, and rat. Annu Rev Immunol. 1990;8:647–679. doi: 10.1146/annurev.iy.08.040190.003243. [DOI] [PubMed] [Google Scholar]
- 2.Bach JF. Insulin-dependent diabetes mellitus as an autoimmune disease. Endocr Rev. 1994;15(4):516–542. doi: 10.1210/edrv-15-4-516. [DOI] [PubMed] [Google Scholar]
- 3.Rood PP, Bottino R, Balamurugan AN, Fan Y, Cooper DK, Trucco M. Facilitating physiologic self-regeneration: a step beyond islet cell replacement. Pharm Res. 2006;23(2):227–242. doi: 10.1007/s11095-005-9095-6. [DOI] [PubMed] [Google Scholar]
- 4.Wagener DK, Sacks JM, LaPorte RE, Macgregor JM. The Pittsburgh study of insulin-dependent diabetes mellitus. Risk for diabetes among relatives of IDDM. Diabetes. 1982;31(2):136–144. doi: 10.2337/diab.31.2.136. [DOI] [PubMed] [Google Scholar]
- 5.Warram JH, Krolewski AS, Gottlieb MS, Kahn CR. Differences in risk of insulin-dependent diabetes in offspring of diabetic mothers and diabetic fathers. N Engl J Med. 1984;311(3):149–152. doi: 10.1056/NEJM198407193110304. [DOI] [PubMed] [Google Scholar]
- 6.Rich SS. Mapping genes in diabetes. Genetic epidemiological perspective. Diabetes. 1990;39(11):1315–1319. doi: 10.2337/diab.39.11.1315. [DOI] [PubMed] [Google Scholar]
- 7.Concannon P, Erlich HA, Julier C, Morahan G, Nerup J, Pociot F, et al. Type 1 diabetes: evidence for susceptibility loci from four genome-wide linkage scans in 1,435 multiplex families. Diabetes. 2005;54(10):2995–3001. doi: 10.2337/diabetes.54.10.2995. [DOI] [PubMed] [Google Scholar]
- 8.Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329(6140):599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 9.Morel PA, Dorman JS, Todd JA, McDevitt HO, Trucco M. Aspartic acid at position 57 of the HLA-DQ beta chain protects against type I diabetes: a family study. Proc Natl Acad Sci U S A. 1988;85(21):8111–8115. doi: 10.1073/pnas.85.21.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monos DS, Mickelson E, Hansen JA, Baker L, Zmijewski CM, Kamoun M. Analysis of DR and DQ gene products of the DR4 haplotype in patients with IDDM: possible involvement of more than one locus. Hum Immunol. 1988;23(4):289–299. doi: 10.1016/0198-8859(88)90064-x. [DOI] [PubMed] [Google Scholar]
- 11.Sheehy MJ, Scharf SJ, Rowe JR, Neme de Gimenez MH, Meske LM, Erlich HA, et al. A diabetes-susceptible HLA haplotype is best defined by a combination of HLA-DR and -DQ alleles. J Clin Invest. 1989;83(3):830–835. doi: 10.1172/JCI113965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen L, Chen NY, Tang J, Sherwin R, Wong FS. The regulatory role of DR4 in a spontaneous diabetes DQ8 transgenic model. J Clin Invest. 2001;107(7):871–880. doi: 10.1172/JCI11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Undlien DE, Friede T, Rammensee HG, Joner G, Dahl-Jorgensen K, Sovik O, et al. HLA-encoded genetic predisposition in IDDM: DR4 subtypes may be associated with different degrees of protection. Diabetes. 1997;46(1):143–149. doi: 10.2337/diab.46.1.143. [DOI] [PubMed] [Google Scholar]
- 14.Cucca F, Lampis R, Frau F, Macis D, Angius E, Masile P, et al. The distribution of DR4 haplotypes in Sardinia suggests a primary association of type I diabetes with DRB1 and DQB1 loci. Hum Immunol. 1995;43(4):301–308. doi: 10.1016/0198-8859(95)00042-3. [DOI] [PubMed] [Google Scholar]
- 15.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57(4):1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nepom GT. A unified hypothesis for the complex genetics of HLA associations with IDDM. Diabetes. 1990;39(10):1153–1157. doi: 10.2337/diab.39.10.1153. [DOI] [PubMed] [Google Scholar]
- 17.Canaday DH, Gehring A, Leonard EG, Eilertson B, Schreiber JR, Harding CV, et al. T-cell hybridomas from HLA-transgenic mice as tools for analysis of human antigen processing. J Immunol Methods. 2003;281(1–2):129–142. doi: 10.1016/j.jim.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Ge X, Piganelli JD, Tse HM, Bertera S, Mathews CE, Trucco M, et al. Modulatory role of DR4- to DQ8-restricted CD4 T-cell responses and type 1 diabetes susceptibility. Diabetes. 2006;55(12):3455–3462. doi: 10.2337/db06-0680. [DOI] [PubMed] [Google Scholar]
- 19.Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104(12):R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudebout P, Zeliszewski D, Golvano JJ, Pignal C, Le Gac S, Borras-Cuesta F, et al. Binding analysis of 95 HIV gp120 peptides to HLA-DR1101 and -DR0401 evidenced many HLA-class II binding regions on gp120 and suggested several promiscuous regions. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14(2):91–101. doi: 10.1097/00042560-199702010-00001. [DOI] [PubMed] [Google Scholar]
- 21.Hausmann DH, Yu B, Hausmann S, Wucherpfennig KW. pH-dependent peptide binding properties of the type I diabetes-associated I-Ag7 molecule: rapid release of CLIP at an endosomal pH. J Exp Med. 1999;189(11):1723–1734. doi: 10.1084/jem.189.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi RV, Zarutskie JA, Stern LJ. A three-step kinetic mechanism for peptide binding to MHC class II proteins. Biochemistry. 2000;39(13):3751–3762. doi: 10.1021/bi9923656. [DOI] [PubMed] [Google Scholar]
- 23.Wicker LS, Chen SL, Nepom GT, Elliott JF, Freed DC, Bansal A, et al. Naturally processed T cell epitopes from human glutamic acid decarboxylase identified using mice transgenic for the type 1 diabetes-associated human MHC class II allele, DRB1*0401. J Clin Invest. 1996;98(11):2597–2603. doi: 10.1172/JCI119079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman AE, Tisch RM, Patel SD, Parry SL, Olson J, Noble JA, et al. Determination of glutamic acid decarboxylase 65 peptides presented by the type I diabetes-associated HLA-DQ8 class II molecule identifies an immunogenic peptide motif. J Immunol. 1999;163(11):6275–6282. [PubMed] [Google Scholar]
- 25.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol. 2001;2(6):501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 26.Lieberman SM, DiLorenzo TP. A comprehensive guide to antibody and T-cell responses in type 1 diabetes. Tissue Antigens. 2003;62(5):359–377. doi: 10.1034/j.1399-0039.2003.00152.x. [DOI] [PubMed] [Google Scholar]
- 27.Marshall KW, Liu AF, Canales J, Perahia B, Jorgensen B, Gantzos RD, et al. Role of the polymorphic residues in HLA-DR molecules in allele-specific binding of peptide ligands. J Immunol. 1994;152(10):4946–4957. [PubMed] [Google Scholar]
- 28.Ettinger RA, Liu AW, Nepom GT, Kwok WW. Exceptional stability of the HLA-DQA1*0102/DQB1*0602 alpha beta protein dimer, the class II MHC molecule associated with protection from insulin-dependent diabetes mellitus. J Immunol. 1998;161(11):6439–6445. [PubMed] [Google Scholar]
- 29.Kawasaki E, Eguchi K. Is Type 1 diabetes in the Japanese population the same as among Caucasians? Ann N Y Acad Sci. 2004;1037:96–103. doi: 10.1196/annals.1337.014. [DOI] [PubMed] [Google Scholar]
- 30.Murao S, Makino H, Kaino Y, Konoue E, Ohashi J, Kida K, et al. Differences in the contribution of HLA-DR and -DQ haplotypes to susceptibility to adult- and childhood-onset type 1 diabetes in Japanese patients. Diabetes. 2004;53(10):2684–2690. doi: 10.2337/diabetes.53.10.2684. [DOI] [PubMed] [Google Scholar]
- 31.Kawabata Y, Ikegami H, Kawaguchi Y, Fujisawa T, Shintani M, Ono M, et al. Asian-specific HLA haplotypes reveal heterogeneity of the contribution of HLA-DR and -DQ haplotypes to susceptibility to type 1 diabetes. Diabetes. 2002;51(2):545–551. doi: 10.2337/diabetes.51.2.545. [DOI] [PubMed] [Google Scholar]
- 32.Levisetti MG, Lewis DM, Suri A, Unanue ER. Weak proinsulin peptide-major histocompatibility complexes are targeted in autoimmune diabetes in mice. Diabetes. 2008;57:1852. doi: 10.2337/db08-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.