Abstract
Homology-dependent repair of DNA double-strand breaks (DSB) initiates by the 5′-3′ resection of the DNA ends to create single-stranded DNA (ssDNA), the substrate for Rad51/RecA binding. Long tracts of ssDNA are also required for activation of the ATR-mediated checkpoint response. Thus, identifying the proteins required and the underlying mechanism for DNA end resection has been an intense area of investigation. Genetic studies in Saccharomyces cerevisiae show that end resection takes place in two steps. Initially, a short oligonucleotide tract is removed from the 5′ strand to create an early intermediate with a short 3′ overhang. Then in a second step the early intermediate is rapidly processed generating an extensive tract of ssDNA. The first step is dependent on the highly conserved Mre11-Rad50-Xrs2 complex and Sae2, while the second step employs the exonuclease Exo1 and/or the helicase-topoisomerase complex Sgs1-Top3-Rmi1 with the endonuclease Dna2. Here we review recent in vitro and in vivo findings that shed more light into the mechanisms of DSB processing in mitotic and meiotic DSB repair as well as in telomere metabolism.
1. DNA end-resection: Why and when, who and how?
Cells have developed elaborate mechanisms to detect and repair a wide variety of DNA lesions, including DNA double strand breaks (DSBs), one of the most cytotoxic forms of DNA damage. DNA double-strand breaks can arise accidentally during normal cell metabolism, by exposure to DNA damaging agents, or as intermediates in programmed genome rearrangements. The timely detection and accurate repair of DSBs is integral to the maintenance of genome integrity. Highly conserved proteins are recruited to DSBs for checkpoint activation and subsequent repair. Their importance is evident by the inherited defects resulting in sterility, developmental disorders, immune deficiencies and predisposition to cancer caused by deficiencies in these factors [1].
Two mechanistically distinct pathways have evolved to repair mitotic DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ). As their names imply the first relies on the presence of an undamaged homologous duplex to serve as a template for repair of the broken chromosome while the latter involves the religation of the DSB ends with little or no homology. A key feature of HR is the preservation of the genetic material, as the donor sequence is usually the sister chromatid. On the other hand, NHEJ can be accompanied by gain or loss of nucleotides at the junction and is therefore considered mutagenic [2].
The repair of DSBs by HR requires the processing of the ends to yield 3′ single-stranded DNA (ssDNA) tails or overhangs, which are substrates for the Rad51/RecA strand exchange protein. The transition from DSB to ssDNA is also required for activation of the ATR-mediated checkpoint response [3]. Insofar as the resected DNA ends are inhibitory to NHEJ, DNA end-resection is the first step that differentiates HR and NHEJ and therefore constitutes a critical control point in repair pathway choice. Consequently, end resection is cell cycle regulated to ensure that it takes place during the S and G2 phases of the cell cycle when the presence of the sister chromatid provides a template for accurate repair by HR [4–6].
Recent genetic studies by several laboratories have elucidated the molecular details of DSB end resection in Saccharomyces cerevisiae and a two-step mechanism that includes nucleases and helicases has been proposed [7, 8]. Shortly after the DSB is formed, the highly conserved Mre11-Rad50-Xrs2 (MRX) complex is recruited to DNA ends to exert architectural and catalytic functions. The MRX complex provides the Mre11 nuclease which cooperates with Sae2 to catalyze the first step in DSB processing, the removal of a short oligonucleotide from the 5′ end [7–9]. The initial step is essential for meiotic DSB processing, in which DSBs are formed by the topoisomerase Spo11. Upon DNA cleavage, Spo11 remains covalently attached to the 5′ ends of the break presenting a block to resection. Removal of Spo11 from meiotic DSB ends involves a Sae2 and MRX-dependent endonucleolytic step that releases Spo11 bound to a short (10–40 nt) oligonucleotide [10].
Several studies suggest that the action of MRX-Sae2 in the initiation of end resection is the rate-limiting step for DSB processing. Based on quantitative measurement of ssDNA intermediates formed in vivo at a defined DSB, Zierhut et al [11] reported that initiation of resection likely occurs with a stochastic, slower rate compared to the rate measured for processive resection far from the break. In agreement with these findings, it was reported that initiation of 5′ processing is about three orders of magnitude slower than the 5′ processing rate once initiated [12]. In cells lacking MRX the rate of resection 28 kb away from the break is not significantly different to wild-type cells [8]. Similarly, a prominent stabilization of cut fragments produced at an inducible DSB was reported in the mre11Δ mutant, but resection was still detected 7kb away from the DSB [7]. Collectively, these observations are in support of a model in which the MRX-Sae2 cleavage step accelerates the rate of resection initiation.
The short 3′ ssDNA tails formed after MRX-Sae2 cleavage are subject to extensive resection in a second step executed via two parallel pathways. One is dependent on the 5′-3′ exonuclease, Exo1, while the other depends on the concerted action of the Sgs1-Top3-Rmi1 complex (hereafter referred to as STR) with the Dna2 endonuclease [7, 8, 13]. The extensively resected ssDNA tracts formed vary in length from a few hundred nucleotides to tens of kilobases depending on the availability and location of the homologous template and correlate with the kinetics of repair [14]. The formation of long ssDNA tracts might only occur when the preferred template for repair (sister chromatid) is not available, for example, when both sister chromatids are cleaved by HO endonuclease. Long ssDNA tracts are required, however, to activate the G2/M checkpoint and it has been suggested that extensive resection serves to ensure the fidelity of repair [8, 14]. MRX-Sae2-dependent, minimally-processed intermediates accumulate in the absence of both STR and Exo1, but can support mitotic gene conversion, albeit inefficiently [7, 8]. De novo telomere addition, a rare DSB repair event in wild type cells, becomes frequent in the absence of long range resection by Exo1 and STR-Dna2 [14–16].
The helicase-nuclease ensemble for DNA resection seems to be a general theme in DNA-end processing machinery. In Escherichia coli both the dominant and the back-up end-processing activities, the RecBCD complex and RecQ-RecJ, respectively, employ helicases and nucleases [17–19]. DNA end resection in Bacillus subtilis by AddAB bears a close resemblance to that catalyzed by RecBCD [20]. In archaea, studies performed with purified Pyrococcus furiosus proteins, suggest that end resection is executed by the bipolar helicase HerA and the 5′-3′ exonuclease NurA [21]. The Mre11-Rad50 complex stimulates end resection by HerA-NurA, similar to findings with eukaryotic systems (see below). Finally, a more recent study in Mycobacterium smegmatis identified the AdnAB complex as a novel helicase-nuclease end resection complex [22]. A role for a RecQ family helicase in resection appears to be conserved in human cells and in Xenopus extracts. The mammalian Sgs1 homolog, BLM, functions in a parallel pathway with Exo1 to promote DSB resection [13], while the Xenopus WRN RecQ helicase catalyzes unwinding of DNA ends followed by 5′-3′ degradation of the single-strand tails by the Xenopus DNA2 nuclease [23].
2. In vitro reconstitution of end resection
Three groups have recently reconstituted elements of the resection process in vitro with purified S. cerevisiae proteins [24–26]. Two of the studies focused on the Sgs1-Dna2 pathway and reported that purified Sgs1, Dna2 and RPA constitute a minimal set of proteins required for resection of linear duplex DNA in an ATP-dependent manner [24, 26]. Sgs1 and Dna2 interact physically and replacement of Sgs1 with Srs2 or Pif1 helicase could not support degradation of the template DNA, suggesting functional interaction as well. Moreover, resection of the linear substrate was dependent on the helicase activity of Sgs1 and the nuclease activity of Dna2, in agreement with the in vivo results [7, 8, 24, 26]
The directionality of Sgs1-Dna2 promoted end resection is 5′ to 3′, with only limited processing of the 3′ strand in the vicinity of the DNA break. Since Dna2 can degrade 5′ and 3′ strands in vitro, both studies investigated the mechanism that establishes strand bias in end resection. It is interesting that RPA serves this role. Not only was RPA required for the Sgs1-mediated DNA unwinding step, but also enhanced the 5′ to 3′-degradative capacity of Dna2 while repressing 3′ to 5′ degradation, thus enforcing the 5′ strand specificity of the resection machinery [24, 26]. It is possible that Dna2 is responsible for the limited degradation observed for the 3′ strand, which might happen just before RPA can bind to the short 3′ overhang to establish the strand bias.
Addition of Top3-Rmi1 stimulated resection by increasing the affinity of Sgs1 for DNA. This stimulatory role was most evident in suboptimal conditions when the Top3-Rmi1 complex became effectively essential for recruiting Sgs1 to the DNA [24]. Notably, the catalytic activity of Top3, which is indispensable for double Holliday junction dissolution, is not required for the stimulation of DSB resection both in vitro and in vivo [26].
Both of the above studies addressed the role of the MRX complex in the reconstituted end-resection assays. Addition of purified MRX complex stimulated the Sgs1-Dna2-RPA end resection two- to four- fold by promoting Sgs1-mediated DNA unwinding [24, 26]. Moreover, MRX interacts with Sgs1 and Dna2 raising the possibility that MRX potentiates resection by recruiting Sgs1 and Dna2 to the DNA ends [24, 26]. Resection of 3′ ssDNA tailed substrates by Sgs1-Dna2 was independent of MRX, suggesting that in addition to acting as a scaffold, the MRX complex stimulates Sgs1-Dna2 resection by creating DNA substates that efficiently recruit Sgs1-Dna2 [26].
In vitro reconstitution of the Exo1-dependent pathway was the focus of the third study [25]. Using a linear dsDNA substrate, Nicolette et al. [25] reported 5′-3′ degradation catalyzed by Exo1, in agreement with previous findings [27, 28]. Addition of purified MRX-Sae2 resulted in a concentration dependent stimulation of Exo1-mediated degradation; at high concentrations Exo1 exhibited MRX-Sae2 independent activity, but at limiting concentrations MRX-Sae2 strongly stimulated the Exo1 activity (60 to 300-fold). MRX or Sae2 alone could stimulate Exo1 activity, but exhibited synergistic stimulation when added simultaneously [25]. In contrast, addition of Sae2 did not alter the efficiency of Dna2-mediated 5′ strand processing in the presence of MRX [26]. Unlike Sgs1-Dna2 mediated resection, RPA was not only dispensable but inhibitory to Exo1 activity in vitro, and MRX-Sae2 could partially overcome the RPA inhibition [25].
The MRX-Sae2 stimulation was largely dependent on the nuclease activity of Exo1, but only marginally dependent on the nuclease activity of Mre11, suggesting that Exo1 is responsible for most of the resection observed in these reactions. Exo1 seems to support 5′ degradation both as an exonuclease and a 5′ flap endonuclease and MRX-Sae2 stimulated both activities [25]. Collectively, these results suggest that MRX-Sae2 affect the recruitment of Exo1 to the DNA ends, similar to the MRX-mediated Sgs1 recruitment as proposed by the other two studies [24, 26]. Unlike Sgs1-Dna2, no stable protein-protein interactions could be detected between MRX or Sae2 and Exo1. Nevertheless, MRX, Sae2 and Exo1 formed complexes in the context of DNA, as evidenced by the cooperative binding to oligonucleotide substrates when all proteins were present. Strand-specific UV cross-linking verified the 5′ strand association of Exo1, which was strongly enhanced by the presence of MRX-Sae2 [25]. It therefore seems that MRX-Sae2 enables Exo1 recruitment to the DNA end by creating a specific DNA structure that allows higher affinity binding of Exo1. Notably, Sae2 was found specifically UV cross-linked to the 3′ strand, independently of MRX and Exo1 [25]. This observation raises the possibility that like the fission yeast Ctp1, which inhibits removal of 3′-linked Top1, Sae2 protects the 3′ strand from degradation [29].
To test whether limited 5′ strand degradation was responsible for the Exo1 stimulation by MRX-Sae2 the Paull group reconstituted end resection in two steps. First, the DNA substrate was pretreated with MRX-Sae2 followed by removal of protein. The processed substrate was then incubated with Exo1 alone. Indeed, Exo1-mediated resection was more efficient when the substrate was pre-resected by MRX-Sae2, suggesting that the limited 5′ strand processing can promote further processing by Exo1. Nevertheless, addition of MRX-Sae2 in the second reaction further stimulated Exo1-mediated resection, suggesting that both the structural and catalytic activities of MRX-Sae2 contribute to enhance Exo1-mediated resection [25].
Along the same lines, Eid et al. [30] recently reported a physical and functional interaction between the mammalian homologs of Sae2 and Exo1, CtIP and EXO1, respectively. They reported that CtIP binds directly to EXO1 in the absence or presence of DNA damage. Unlike their budding yeast counterparts, purified CtIP inhibited exonucleolytic processing by EXO1 on both nicked and linearized plasmids with 3′ overhangs, suggesting that CtIP restrains long range resection by Exo1 [30].
3. Putting the in vitro and in vivo observations together
The in vitro studies described above suggest a stimulatory role of the MRX complex in promoting DSB processing through the Sgs1-Dna2 and Exo1 pathways. Under certain circumstances, however, the MRX complex is essential for DSB resection in vivo. There are several possible explanations for the more important role for MRX demonstrated by genetic assays. First, the clipping of ends by MRX-Sae2 is essential for removal of Spo11 and possibly other modifications to the DNA ends; only substrates with ‘clean’ ends were used for the in vitro assays. Second, the requirement for MRX recruitment of STR-Dna2 and/or Exo1 is likely to be more stringent in vivo than in the reconstitued systems. Finally, there are other proteins present in cells that can bind to DSBs, blocking access to the resection machinery and thus increasing the requirement for the MRX-Sae2 activities. Such a factor is the Ku heterodimer, which forms a ring that binds DNA by sliding onto DSB end through its opening. Once bound, Ku protects the ends from degradation and mediates recruitment of downstream NHEJ factors [2]. Two recent in vivo studies investigated the interplay between Ku and the resection machinery in budding yeast [9, 31].
Shim et al used chromatin immunoprecipitation assays to investigate recruitment of the resection machinery at an inducible, unrepairable DSB [9]. Similar to the in vitro results, they reported that MRX facilitates association of Exo1 and Dna2 to DSBs and the nuclease activities of Mre11 and Sae2 are dispensable for the recruitment. Unlike Sae2, CtIP was required for the efficient recruitment of EXO1 at laser induced-DNA damage; depletion of either MRE11 or CtIP equally impaired EXO1 localization to DSB sites [30]. Whether this reflects differences in the sensitivity of the applied techniques or a particular requirement for MRN/CtIP trimming of ends for EXO1 recruitment at break sites with more complex damage than at endonuclease-induced DSB remains unanswered. In the absence of the MRX complex the Ku heterodimer accumulates in excess at DSB ends [9, 32]. Following loss of Ku in mre11Δ mutants, recruitment of Dna2 to the DSB was modestly increased, but recruitment of Exo1 was almost fully restored and correlated with increased resection. The in vivo findings were further supported by the in vitro finding that the presence of MRX neutralized the inhibitory effect of Ku on Exo1-mediated 5′ degradation of duplex DNA ends [9].
In an independent study we investigated the importance of the MRX-Sae2 cleavage in preventing the inhibitory action of Ku, as monitored by survival in response to ionizing radiation (IR) [31]. In mre11 nuclease defective (mre11-nd) and sae2Δ mutants, the MRX complex can form, allowing recruitment of Sgs1-Dna2 and/or Exo1, but the first step in resection is compromised resulting in radiation sensitivity. Loss of Ku suppressed the IR sensitivity in a manner dependent on both Exo1 and Sgs1 [31]. In other words, the requirement for the first step in resection can be bypassed when Ku is absent because access of Sgs1 and Exo1 is allowed to restore resection. Under these conditions a cooperation between the first and second step of resection can be established, which resembles the behavior observed in vitro. Collectively, these results suggest that in addition to acting as a scaffold, the MRX-Sae2 complex serves to create a processed intermediate that is stimulatory to the Exo1- and Sgs1-Dna2-mediated resection as evidenced by the in vitro studies, and furthermore can no longer be bound by Ku, thus negating its inhibitory role.
The more severe phenotype of mre11-nd mutants in fission yeast and mice suggests that the first step in resection is more limiting in these organisms [33, 34]. Considering that Ku exacerbates the resection defect of the mre11-nd mutant in budding yeast, an attractive hypothesis is that the presence of Ku is more dominating in these other organisms, raising the barrier to initiate resection. Alternatively, it is possible that the STR-Dna2 resection pathway, which seems to contribute more to the IR resistance of the mre11-nd mutant than Exo1, has more limited functions in end-resection in fission yeast and mice (much like in budding yeast meiosis) [9, 31, 35].
4. How redundant are the Exo1- and Sgs1-mediated pathways?
Most studies to date that address the mechanism of mitotic DSB resection report redundancy between the Exo1 and Sgs1-Dna2 pathways, but recent work on resection of meiotic DSBs and telomeres suggests otherwise.
In Saccharomyces cerevisiae meiotic DSB formation is tighly coupled with resection and several lines of evidence suggested a role for Exo1 in the ‘hyperresection’ of meiotic breaks observed in dmc1Δ mutants [36, 37]. Moreover, Manfrini et al. [36] showed that Sgs1-Dna2 contribute to the dmc1Δ hyperresection when Exo1 is absent. Two recent studies addressed the role of Exo1 and/or Sgs1 in physiological meiotic DSB resection. Using different assays to measure 5′ strand resection of DSB ends formed at a meiotic hotspot, both studies reported a significant role for Exo1 [38, 39]. The mean length of ssDNA formed at meiotic DSBs is 800 nucleotides (nt), but deletion of EXO1 or expression of a nuclease-dead Exo1 isoform reduced it to 270nt [39]. Thus, Exo1 is required for meiotic DSB processing and, as in mitosis, it is dispensable for the initial resection, which is presumably dependent on MRX-Sae2 [38, 39]. Consistent with previous studies, Zakharyevich et al. showed that the extent of resection did not change over time, suggesting the initial break formation and removal of Spo11 are tightly coupled to Exo1-dependent resection of the 5′ ends [39]. Although the nuclease activity of Exo1 is essential for end resection, it is not required for the Exo1 function in promoting meiotic crossovers [38, 39]. Thus, even the short overhangs produced by MRX-Sae2 can be used effectively for meiotic joint molecule formation and subsequent resolution to form crossovers.
The use of a meiotic null sgs1 allele showed that Sgs1 does not contribute significantly to DSB resection during meiosis [39]. From a teleological standpoint, resection during meiosis when ~150–200 DSBs are formed should be tightly regulated, so it would appear more advantageous for the cell to employ as few pathways as possible to resect meiotic DSBs. Finally, as discussed above, the absence of the STR-Dna2 pathway during meiosis could explain why mre11-nd mutants, which rely on the STR-Dna2 pathway for survival in response to IR and camptothecin, have such a strong meiotic resection defect [31, 35].
Telomeres are also resected in a 5′-3′ fashion, giving rise to the single stranded G-tails, central intermediates in modulating telomere homeostasis since they serve as substrates for extension by telomerase [40]. It was recently shown that similar machineries resect DSBs and telomeres. More specifically, MRX and Sae2 were reported to act in the same pathway, whereas Sgs1 functions in conjuction with Dna2 [41]. The mre11Δ mutant, but not mre11-nd, exhibits short telomeres and mrx null mutants are defective in 5′ C-strand degradation, suggesting that, as with DSBs, the MRX complex provides the scaffold to support telomeric C-strand degradation [41–43]. A sae2Δ sgs1Δ double mutant was found to recapitulate the mre11Δ behavior with short, unresected telomeres [41]. These observations suggest that Sae2 and Sgs1 constitute two distinct but partially complementary pathways for telomere processing controlled by the MRX complex. The activity of Exo1 at telomeres is blocked by Ku and this might explain the more severe telomere end processing defect observed for the sae2Δ sgs1Δ double mutant compared with an endonuclease-induced DSB [7, 44].
The contribution of the Exo1-mediated pathway is more evident in the resection of ‘uncapped’ telomeres. The telomeric G-tails are bound by Cdc13, an essential ssDNA binding protein necessary for telomerase recruitment and end protection [45]. In cdc13 mutants the telomeres become ‘uncapped’ and the C-strand is extensively resected in a manner partially dependent on Exo1 [46]. Recent studies designed to identify the additional activities resecting uncapped telomeres, showed that Sgs1 contributes to resection. Elimination of both Sgs1 and Exo1 prevented resection at loci further than 5 kb from the telomere, and this was most evident in cells lacking the checkpoint protein Rad9. However, resection close to the telomere was still detected in exo1Δ sgs1Δ and exo1Δ sgs1Δ rad9Δ mutants, presumably due to MRX-Sae2 activity. Remarkably, the exo1Δ sgs1Δ rad9Δ triple mutant is able to grow in the complete absence of Cdc13 [47].
5. Conclusions
During the last few years remarkable progress has been made in our understanding of the molecular mechanism and control of DNA end processing in mitotic and meiotic DSB repair as well as in telomere metabolism. However, many questions remain. Are these mechanisms conserved in higher eukaryotes? What are the targets of the resection machinery that are cell-cycle regulated? How redundant are the pathways for long-range resection in mitotic DSB repair? How much resection is required during sister chromatid recombination? Does the length of resection affect mitotic crossover levels? Due to the critical role of DSB resection in determining repair pathway choice and DNA damage signaling one cannot overstate the importance of answering these questions in order to understand the mechanisms that maintain genomic integrity.
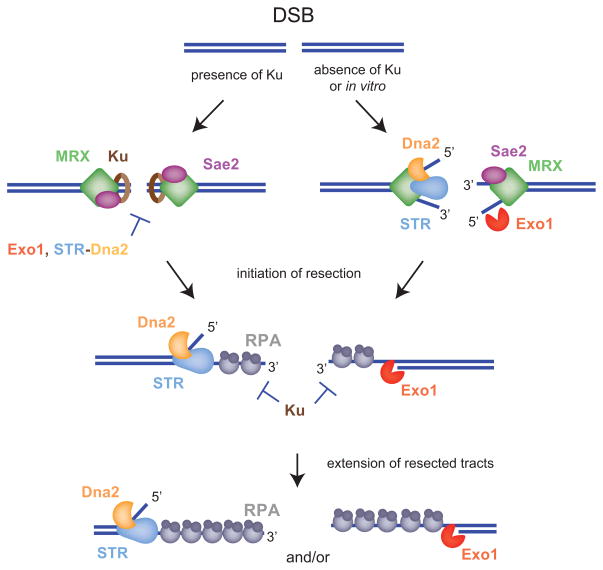
Figure 1. Models for the initiation and extension of DSB-end resection.
The Mre11-Rad50-Xrs2 (MRX) complex rapidly binds to the DSB to perform a variety of functions including DSB sensing by the checkpoint machinery, tethering of the DSB ends and end-processing in preparation for HR. Genetic studies in Saccharomyces cerevisiae suggest that resection initiation requires MRX-Sae2 catalyzed removal of short oligonucleotides from the 5′ ends. The intermediate formed is then extensively processed by two parallel pathways dependent on either Sgs1-Top3-Rmi1 (STR) with Dna2 or Exo1 (left-hand side model). The recent in vitro studies confirm the genetic observations but also propose that MRX acts as a scaffold to efficiently recruit the extensive resection machinery (right-hand side model). Collaboration between the two steps in vivo can be unmasked by loss of the Yku70-Yku80 (Ku) heterodimer, which can also bind DSB ends and prevents the efficient recruitment of Exo1 and Sgs1 by MRX-Sae2.
Acknowledgments
We thank W. K. Holloman for discussions and critical reading of the manuscript. Research performed in my laboratory that is cited in this review was supported by a grant from the National Institutes for Health (GM041784).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 2.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 3.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 4.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 13.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 16.Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet. 2010;6:e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amundsen SK, Smith GR. Interchangeable parts of the Escherichia coli recombination machinery. Cell. 2003;112:741–744. doi: 10.1016/s0092-8674(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 18.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handa N, Ichige A, Kobayashi I. Contribution of RecFOR machinery of homologous recombination to cell survival after loss of a restriction-modification gene complex. Microbiology. 2009;155:2320–2332. doi: 10.1099/mic.0.026401-0. [DOI] [PubMed] [Google Scholar]
- 20.Yeeles JT, Dillingham MS. The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair (Amst) 2010;9:276–285. doi: 10.1016/j.dnarep.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinha KM, Unciuleac MC, Glickman MS, Shuman S. AdnAB: a new DSB-resecting motor-nuclease from mycobacteria. Genes Dev. 2009;23:1423–1437. doi: 10.1101/gad.1805709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao S, Toczylowski T, Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008;36:6091–6100. doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 29.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010;11:962–968. doi: 10.1038/embor.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D, Topper LM, Wilson TE. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics. 2008;178:1237–1249. doi: 10.1534/genetics.107.083535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manfrini N, Guerini I, Citterio A, Lucchini G, Longhese MP. Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. J Biol Chem. 2010;285:11628–11637. doi: 10.1074/jbc.M110.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keelagher RE, Cotton VE, Goldman AS, Borts RH. DNA Repair (Amst) 2010. Separable roles for Exonuclease I in meiotic DNA double-strand break repair. [DOI] [PubMed] [Google Scholar]
- 39.Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis promote double-strand-break resection and resolution of double-Holliday Junctions into crossovers. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.11.032. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 41.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- 43.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 2010;6:e1000966. doi: 10.1371/journal.pgen.1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shore D, Bianchi A. Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 2009;28:2309–2322. doi: 10.1038/emboj.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zubko MK, Guillard S, Lydall D. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics. 2004;168:103–115. doi: 10.1534/genetics.104.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ngo HP, Lydall D. Survival and growth of yeast without telomere capping by Cdc13 in the absence of Sgs1, Exo1, and Rad9. PLoS Genet. 2010;6:e1001072. doi: 10.1371/journal.pgen.1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]