Abstract
According to recent taxonomic reclassification, the primate family Hylobatidae contains four genera (Hoolock, Nomascus, Symphalangus, and Hylobates) and between 14-18 species, making it by far the most species-rich group of extant hominoids. Known as the “small apes”, these small arboreal primates are distributed throughout Southeast, South and East Asia. Considerable uncertainty surrounds the phylogeny of extant hylobatids, particularly the relationships among the genera and the species within the Hylobates genus. In this paper we use parsimony, likelihood, and Bayesian methods to analyze a dataset containing nearly 14 kilobase pairs, which includes newly collected sequences from X-linked, Y-linked, and mitochondrial loci together with data from previous mitochondrial studies. Parsimony, likelihood, and Bayesian analyses largely failed to find a significant difference among phylogenies with any of the four genera as the most basal taxon. All analyses, however, support a tree with Hylobates and Symphalangus as most closely related genera. One strongly supported phylogenetic result within the Hylobates genus is that H. pileatus is the most basal taxon. Multiple analyses failed to find significant support for any singlular genus-level phylogeny. While it is natural to suspect that there might not be sufficient data for phylogenetic resolution (whenever that situation occurs), an alternative hypothesis relating to the nature of gibbon speciation exists. This lack of resolution may be the result of a rapid radiation or a sudden vicariance event of the hylobatid genera, and it is likely that a similarly rapid radiation occurred within the Hylobates genus. Additional molecular and paleontological evidence are necessary to better test among these, and other, hypotheses of hylobatid evolution.
Keywords: gibbons, siamangs, Hylobatidae, evolution, phylogenetics, radiation
1. Introduction
Gibbons and siamangs are hominoid primates of the family Hylobatidae, which is the sister group to Hominidae, the group that includes humans. According to recent taxonomic reclassification, there are between 14-18 species of extant hylobatids (IUCN, 2010; Mootnick, 2006; Thinh et al., 2010), making this the most species-rich group of extant apes. Inhabitants of the rainforests of Southeast, South and East Asia, all hylobatid species are listed on the IUCN Red List of Threatened Species, with most of them endangered (IUCN, 2010). An accurate phylogeny of these apes would enhance our understanding of Asian biogeography, morphological evolution, hominoid paleontology, and contribute to conservation efforts.
Multiple different data types have been studied to address hylobatid phylogenetics, including morphological, vocal, karyotypic, and molecular genetic data (Chatterjee, 2006; Groves, 1972; Haimoff et al., 1982; Hayashi et al., 1995; Matsudaira and Ishida, 2010; Muller et al., 2003; Roos and Geissmann, 2001; Takacs et al., 2005; Thinh et al., 2010; Whittaker et al., 2007). One point of agreement among molecular studies is that hylobatids can be represented by four monophyletic genera: Hylobates, Symphalangus, Nomascus, and Hoolock (Brandon-Jones et al., 2004). However, genetic studies conflict on two key aspects of hylobatid phylogeny. First, there is disagreement on the relationships among the four genera. Several genetic studies forward Nomascus as the most basal taxon (Chatterjee, 2006; Roos and Geissmann, 2001; Thinh et al., 2010), while others, including those based on chromosomal data, give support for basal positioning of Hoolock (Muller et al., 2003; Takacs et al., 2005). Morphological evidence has supported a basal position for Symphalangus (Groves, 1972). Secondly, within Hylobates there is considerable uncertainty regarding the relationships among the different species. One important question relates to the position of the most basal member of Hylobates. H. pileatus has been suggested to be the most basal member in studies based on molecular and morphological data (Geissmann, 2002; Takacs et al., 2005; Whittaker et al., 2007), while other analyses support a H. klossii as the basal member of Hylobates (e.g. Purvis, 1995).
Using molecular phylogenies, there have been different interpretations of the hylobatid evolutionary history. Chatterjee (2006) forwarded a scenario where hylobatids began their radiation in Eastern Indochina at about 10.5 Ma and subsequent waves of radiation occurred predominantly southwardly, with a westward radiation of Hoolock (Chatterjee, 2006). Whittaker et al. (2007) examined Hylobates and suggested a general north to south biogeographic expansion. Thinh et al. (2010) suggested that the intergeneric hylobatid radiation occurred on the Southeast Asian mainland in the Miocene, specifically in the area of the Hengduan mountains. Subsequently, the genera migrated into their present positions on the mainland. This was followed by a southward expansion by Hylobates onto the Sunda shelf, where vicariant speciation mainly occurred.
In the present study, we collected nuclear DNA sequences from the X-linked G6PD locus and the Y-linked ZFY locus, and the mitochondrial loci COII and ND4/5 from a number of hylobatids. These sequences were added to a comprehensive dataset of mitochondrial DNA sequences, including 5 complete mtDNA genomes (Matsudaira and Ishida, 2010). This alignment was analyzed using parsimony, likelihood, and Bayesian phylogenetic methods, and multiple hypotheses of hylobatid phylogeny were tested. A molecular clock analysis was used to estimate the date of speciation events in the evolutionary history of hylobatids. The results are discussed within the context of recently proposed hypotheses of hylobatid evolutionary history.
2. Materials and Methods
2.1. Collection of DNA Sequences
Gibbon and outgroup samples were provided by several individuals and institutions. A list of samples, including identification number, institution, and genes for which sequences are included in this study is shown in Table 1. Many different sample types were used for this study: tissue, whole blood, serum, hair, urine, and tissue culture cell suspensions. Extraction of all samples was performed using the IsoQuick DNA Extraction Kit (ORCA Research; Bothell, WA), a guanidine thiocyanate-based method, according to the manufacturer’s protocols.
TABLE 1.
| Species | ID | Institution | COII | ND4/5 | G6PD | ZFY |
|---|---|---|---|---|---|---|
| Gorilla gorilla | YN90-47, “Rok” | Yerkes R. P. C. | X | |||
| Hoolock leuconedys | HHL303 | GCC | X | X | X | |
| Hoolock leuconedys | 94098 | GCC | X | |||
| Hylobates agilis | HAA402 | GCC | X | X | ||
| Hylobates klossii | Onnie | Taman Safari | X | X | X | |
| Hylobates lar | 851004001 | GCC | X | |||
| Hylobates moloch | HMO806 | GCC | X | X | X | |
| Hylobates muelleri | 8840822 | GCC | X | X | ||
| Hylobates muelleri | 4611 | Gladys Porter Zoo |
X | |||
| Hylobates pileatus | HP116 | GCC | X | X | ||
| Macaca mulatta | NP94 | Yerkes R. P. C. | X | |||
| Macaca mulatta | 637T | Yerkes R. P. C. | X | |||
| Nomascus gabriellae | 851008 A1 | GCC | X | |||
| Nomascus gabriellae | NG602 | GCC | X | X | ||
| Nomascus leucogenys | NL601 | GCC | X | X | X | |
| Pan troglodytes | YB79-99, “Zeb” | Yerkes R. P. C. | X | |||
| Papio papio | 95-25043 | Brookfield Zoo | X | |||
| Pongo abelii | YN86-27, “Dyak” |
Yerkes R. P. C. | X | |||
|
Symphalangus
syndactylus |
SS906 | GCC | X | X |
DNA sequence data were collected from two mitochondrial regions, one X-linked locus, and one Y-linked locus. The first is the 684 bp (base pair) cytochrome oxidase subunit II locus (COII). The second is an 898 bp fragment consisting of sequences from two NADH dehydrogenase complex genes (ND4 and ND5). The third data set is from 4 introns of the X-linked Glucose-6-phosphate dehydrogenase gene (introns 4, 5, 7, & 8, also called D, E, G, & H). The final data set is from the last intron of the Y-linked zinc finger gene (ZFY). Polymerase chain reaction (PCR) was used to amplify target DNA for both regions in preparation for sequencing. For COII, the entire coding sequence was amplified with primers listed in the Supplementary Materials. For the ND4/5 region, primers A through F, described by Hayashi et al. (1995), were used. Because of the possibility of introgression of mitochondrial sequences into the nucleus, multiple independent PCRs with different primer combinations were performed for all samples. Sequences from all amplifications matched exactly, indicating that each of them came from a single population of target DNA. None of the consensus sequences contained stop codons or other anomalous codons, and each was therefore considered to represent the true mitochondrial sequence. For G6PD, amplification was done using primers located within the flanking exons. In some occasions, nested PCR was used. For ZFY, amplification was done using primers located within the flanking exons using nested PCR.
PCR conditions were as follows: up to 10 microliters of DNA (concentration unquantified, from 50 microliters of IsoQuick extracted DNA) was used as a template in a 50 microliter reaction containing 10 mM Tris-HCl pH 8.3, 50 mM KCl, 20-25 pM of each primer, 1.25-2.5 units of polymerase (AmpliTaq, Perkin-Elmer/ABI, Foster City, CA, or Qiagen Taq, Valencia, CA), 1.25-1.5 mM MgCl2, and 0.15-0.2 mM of each dNTP. Reactions were run in either a MJ Research Minicycler or a Perkin-Elmer 9600 thermocycler. Cycling parameters for hylobatids were as follows:
For COII a 3 min. 94°C denaturation was followed by 30 repetitions of a 60 s. 94°C, 60 s. 48°C 60 s. 72°C cycle. For ND4/5 a 3 min. 94°C denaturation was followed by 30 repetitions of a 45 s. 94°C, 60 s. 48°C 60 s. 72°C cycle. For G6PD gibbon samples, a 3 min. 94°C denaturation was followed by 40 repetitions of a 30 s. 94°C, 45 s. 65°C 60 s. 72°C cycle. For the non-gibbon hominoids, the cycle consisted of 30-35 repetitions of 60 s. at 94°C, 80 s. at 65°C, and 130 s. at 72°C. For introns D and E, 4 s. were added to the extension with each cycle. For Papio and Macaca, the cycle consisted of 30-35 repetitions of 30 s. at 93°C, 30 s. at 60-64°C, and 45 s. at 72°C. For ZFY a 3 min. 94°C denaturation was followed by 40 repetitions of a 30 s. 94°C, 45 s. 50°C, 45 s. 72°C cycle. A final cycle of 5-10 m. at 72°C cycle followed.
Fragments were gel-purified and used directly in sequencing reactions. DNA sequencing of double-stranded PCR product was performed using the ABI Prism Dye-Deoxy reaction kit (Applied Biosystems Inc., Foster City CA) according to the manufacturer’s protocol. Unincorporated primers and nucleotides were removed using Centriflex bead matrix columns (Edge BioSystems, Gaithersburg, MD) according to the manufacturer’s protocols. Column eluant was dried down in a Speed-Vac with no heat, resuspended in 3.5 microliters of loading buffer (Formamide, 50 mM EDTA, 30 mg/ml dextran blue) and run on a 4.75 Sequagel (National Diagnostics) or a 6% Long Ranger acrylamide gel (FMC, Rockland, ME) for 12 hours on an ABI 373 automated sequencer.
2.2. Alignment of DNA Sequences
The DNA sequences collected from gibbons were added to the available DNA sequences of hylobatids, Homo sapiens, Pan troglodytes, P. paniscus, Gorilla gorilla, Pongo abelii, P. pygmaues, Macaca mulatta, and Papio from Genbank and the literature (Table 2). This included 5 complete hylobatid mitochondrial genomes (H. agilis, H. pileatus, S. syndactylus, N. siki, and H. lar) and additional species for the mtDNA loci cytochrome b (CytB), cytochrome oxidase II (COII), and the NADH dehydrogenases (ND3, ND4, ND4L, and ND5) (Table 2). Only the protein coding regions of the mtDNA genomes were used because the non-protein coding regions were not straightforward to align. The protein coding genes were aligned using ClustalW (Thompson et al., 1994), separated by gaps in order to preserve the reading frame.
Table 2.
2.3. Phylogenetic Analyses
In the phylogenetic analyses each gene was analyzed separately as well as in a single combined multi-gene alignment of all data. For maximum likelihood and Bayesian analyses, this alignment was analyzed as 5 independent data partitions (mtDNA first, second, and third codon positions, G6PD, and ZFY). This method was chosen based on the results of a recent study (Ren et al., 2009) which showed that likelihood performed better than i) a ‘supermatrix’ method that concatenated partitions into a single alignment analyzed using one set of parameters, and ii) a ‘supertree’ method that separated partitions, estimated a phylogenetic tree for each partition, and used supertree methods to combine independent trees (Ren et al., 2009). In each analysis, Papio and/or Macaca were used as the outgroup taxa to the ingroup, which included both hylobatids and the other hominoids, Homo, Pan, Pongo, and Gorilla.
A parsimony-based phylogenetic analysis was conducted in PAUP* v. 4.0b10 (Swofford, 2001). Again, the three separate loci were analyzed both independently and together. In all cases, two separate character-weighting matrices were used. For mtDNA, a 10:1 weighting scheme was used, with transversions being weighed more heavily than transitions. For G6PD and ZFY, a 2:1 weighting scheme was used, with transversions being weighed more heavily than transitions. These weightings reflect the greater frequency of transitions than transversions in molecular data, with an increased bias in mitochondrial DNA. Papio and Macaca were used as outgroups, and a branch- and-bound tree search was performed to find the most parsimonious tree(s). A parsimony based bootstrap analysis was conducted using 1000 search replicates, using a heuristic search at each replicate of 10 random taxon additions. For the combined alignment, phylogenetic hypothesis testing was done using constraint trees with each genus as basal and subsequently conducting a heuristic search. These constrained trees were compared to the most parsimonious trees using the Templeton (1983) and winning sites tests (Prager and Wilson, 1988) using PAUP*.
A maximum likelihood-based phylogenetic analysis was conducted in RAxML v. 7.0.4 (Stamatakis, 2006; Stamatakis et al., 2008). G6PD, ZFY, and mitochondrial loci were analyzed separately, with the mtDNA partition having three different partitions corresponding to each codon position. A combined alignment of all data using five separate partitions was also analyzed. For all partitions a general time reversible model of evolution with a gamma model for among site rate variation was used (GTRGAMMA), and Papio and Macaca were used as outgroups in all analyses. A maximum likelihood bootstrap analysis with 1000 replicates was also conducted on the combined alignment. Phylogenetic hypothesis testing was done on the combined alignment using constraint trees with each genus as basal and subsequently conducting a maximum likelihood search. These constrained trees were compared to the most likely tree using the SH test (Shimodaira and Hasegawa, 1999) in RAxML.
A Bayesian-based phylogenetic analysis was conducted with an MPI enabled MrBayes v. 3.1.2 (Altekar et al., 2004; Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). Three separate partitions were used for the mtDNA sequence data, each with a general time reversible model of evolution with 6 substitution classes (Nst=6) and a Γ and invariant (Invgamma) model for among site rate variation. For the G6PD data, a model with 2 substitution classes (Nst=2) and a gamma model for among site rate variation was used. For the ZFY data, a model with 6 substitution classes and a gamma model for among site rate variation was used. A Dirichlet prior of 1 was used for the each of the base state frequencies in each of the four partitions. These models were chosen using MrModelTest v. 2 (Nylander, 2004). The site-specific rate model was set to variable, allowing the rate to vary across partitions. All of these parameters were unlinked across the partitions. Macaca mulatta was used as the outgroup (only one outgroup taxon is allowed by MrBayes). Three independent runs were initiated each using a random starting tree; each run consisted of 5 million steps, with sampling every 100 steps, and used 4 chains and a heating parameter of 0.085, allowing for good swapping between chains. The first 20% of each run was discarded as burnin. Convergence was assessed among chains by examining the Potential Scale Reduction Factors (PSRF), effective sample sizes (ESS) (estimated by Tracer v. 1.4.1 (Rambaut and Drummond, 2007)), and the similarity of parameter estimates among runs. A similar analysis was conducted for each independent locus as well. Phylogenetic hypothesis testing was done on the combined alignment using constraint trees with each genus as basal and subsequently conducting a Bayesian analysis. The harmonic mean likelihoods of the constrained trees were compared to the unconstrained trees in a manner similar to Nylander et al. (2004).
2.4. Molecular Clock Analysis
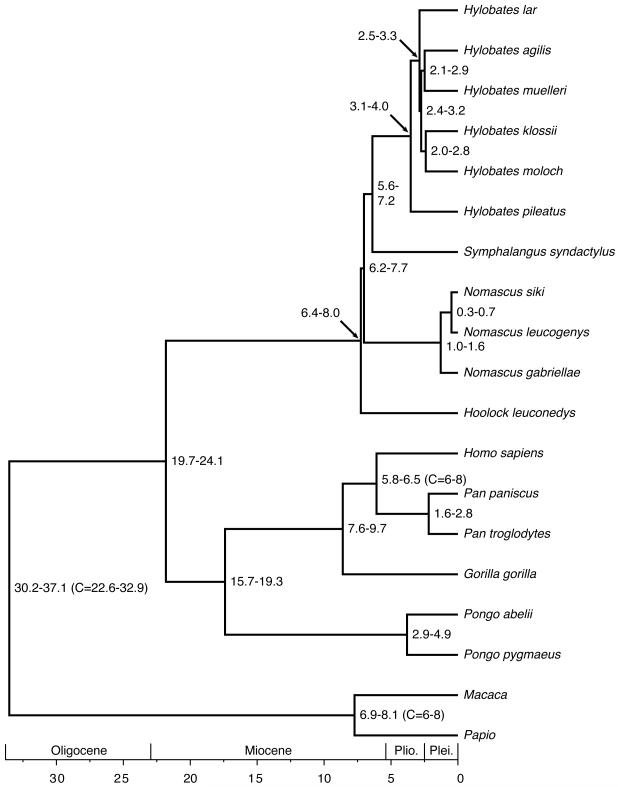
A molecular clock analysis was conducted using the Bayesian MCMC program mcmctree of PAML v. 4.3 (Yang, 2007) using a user-input tree matching the Bayesian consensus tree (Figure 3). This tree was chosen because a completely bifurcating tree is required. The alignment was broken into four separate partitions and analyzed together using the HKY+Γ5 model (Hasegawa et al., 1985; Yang, 1994), a model with different transition/transversion rate ratios (κ), different base frequencies and different gamma shape parameter α for each of the four partitions. Gamma priors were assigned on parameters κ < G(6,2), with mean 3, and α < G(1, 1). Substitution rates are assumed to drift over time independently among the partitions. A 100 Myr time unit was used. The rate at the root is the gamma prior μ < G(1, 0.4). A geometric Brownian motion model was used to accommodate substitution rate variation across lineages (Rannala and Yang, 2007). The rate-drift parameter was assigned the gamma prior σ2 < G(1, 10). The prior of times was generated from the birth-death process with sampling, with parameters λ = μ = 1 and ρ = 0, so that the kernel is uniform (Yang and Rannala, 2006). Two independent runs were conducted, each with a 10,000 step burnin followed by 100,000 steps, with sampling every 5 steps. Convergence was assessed by agreement between two independent runs. The tree was calibrated by assigning divergence times to two independent nodes. A divergence time of 6 – 8 Ma for the Homo/Pan split was based on the presence of the candidate hominins Orrorin tugenensis (~5.8 Ma ago) (Senut et al., 2001), Ardipithecus kadabba (5.2–5.8 Ma ago) (Haile-Selassie et al., 2004), and Sahelanthropus tchadensis (6.8-7.2 Ma ago) (Lebatard et al., 2008). A divergence time of 6 – 8 Ma ago was also assumed for the Macaca/Papio split based on the age of the earliest members of Macaca (5.5 Ma) and paleontological estimates for the divergence of these taxa between 7 – 8 Ma ago (Delson, 1992; Delson, 2000). The root node of the tree (crown catarrhine) was calibrated using a gamma distribution with a shape parameter of 110 and scale parameter of 1/400. This corresponds to a distribution with the 2.5% date at 22.6 Ma, 97.5% date at 32.9 Ma, and mean date of 27.4 Ma. This is based on the date of the earliest crown catarrhine, the hominoid Morotopithecus, from 20.6 Ma ago (Gebo et al., 1997). This distribution generates an upper bound, bracketing a conservatively large estimate for the origin of crown catarrhines. Furthermore, these calibrations are ‘soft’ bounds (Yang and Rannala, 2006), i.e. the date estimates for these nodes have a limited prior probability (2.5%) to be older or younger than the calibrations.
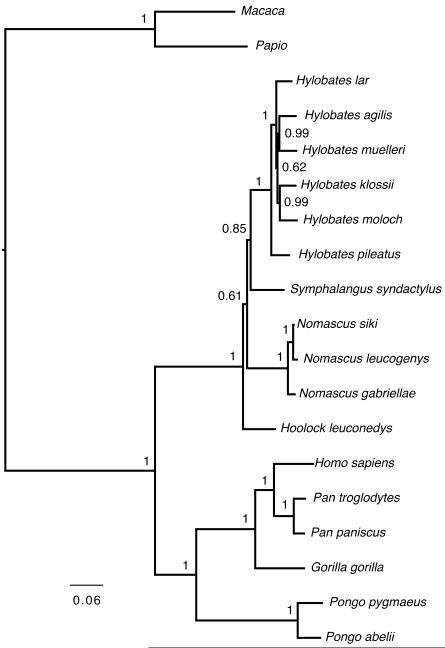
FIGURE 3.
Phylogram estimated using Bayesian phylogenetics. Posterior probability values appear above branches leading to nodes except within most Hylobates nodes, where they appear to the right of the node.
3. Results
The final alignment of mitochondrial coding sequences totaled 10,854 base pairs, the final G6PD alignment totaled 2,191 base pairs, and the ZFY alignment totaled 825 base pairs. Using all analytical methods, the single locus analyses offered little phylogenetic resolution (see Supplementary Materials). For example, in analyses of mtDNA sequences, the position of Hoolock differed among the different optimality criteria; in analyses of the G6PD locus the positions of the genera relative to one another was different among optimality criteria; and in ZFY the limited differences among the species caused little resolution among hylobatids and long branch attraction between the hylobatid and Pongo lineages. Because of the lack of resolution of the single gene analyses, the results from the combined analysis are presented in greater depth.
3.1. Parsimony Results
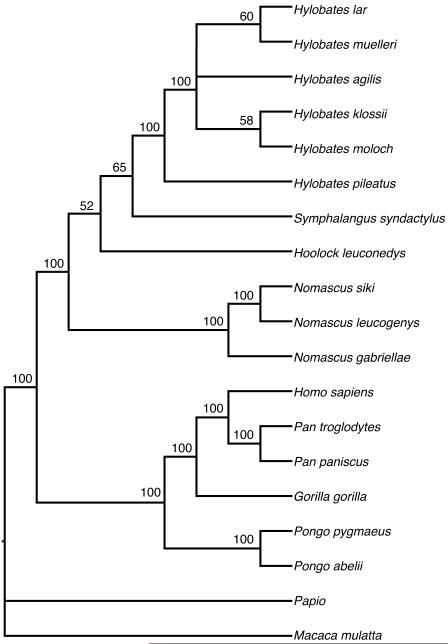
The maximum parsimony analysis resulted in two most parsimonious trees. Both trees agreed in their placement of Nomascus as the most basal taxon, with Hoolock as next most basal, then a Symphalangus + Hylobates clade. Within the Hylobates genus, the two most parsimonious trees differed in their placement of H. agilis. The bootstrap value for the Hoolock node was 52% and for the Symphalangus+Hylobates node was 65%. A bootstrap consensus tree appears in Figure 1. There was strong support for Hylobates as a monophyletic group (100%) and for the placement of H. pileatus as the most basal member of this group. Within the remaining Hylobates species, however, the bootstrap values on all the branches were low (27% – 60%).
FIGURE 1.
Bootstrap consensus cladogram using maximum parsimony.
3.2. Likelihood Results
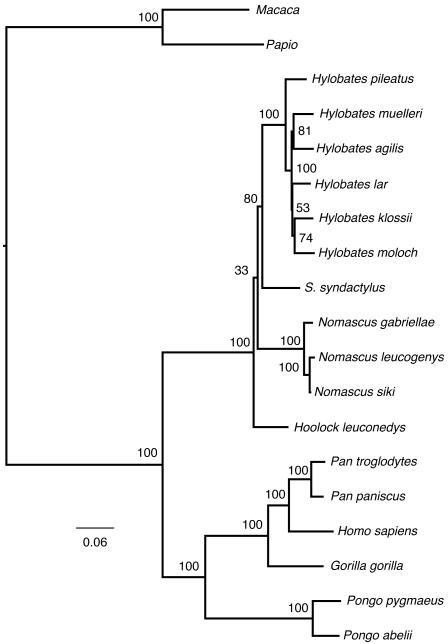
The maximum likelihood tree differed from the most parsimonious trees in the placement of the most basal taxon (Figure 2). In the maximum likelihood tree, Hoolock is the most basal genus, Nomascus is the next most basal, followed by a Symphalangus + Hylobates clade. Within the Hylobates genus, H. pileatus is most basal, but the placement of the remaining species differs from both most parsimonious trees. Bootstrap support for the placement of Hoolock was very low (33%). The placement of Symphalangus was moderately supported (80%). Within Hylobates, the bootstrap values varied.
FIGURE 2.
Bootstrap consensus phylogram using maximum likelihood. Bootstrap values appear above branches leading to nodes except within most Hylobates nodes, where they appear to the right of the node. A cladogram that has collapsed the branches of <50% bootsrap support appears in Supplementary Materials Figure 4.
3.3. Bayesian Results
The Bayesian analysis resulted in a tree similar to the maximum likelihood tree at the genus level (Figure 3). Hoolock was placed as the most basal genus. Nomascus was the next most basal, with a clade credibility value of 61. Symphalangus was sister taxon to Hylobates with a clade credibility value of 85. Within Hylobates, H. pileatus was the most basal species. The placement of the remaining species in the Hylobates genus differed from both the maximum parsimony trees and the most likely tree.
3.4. Hypothesis Testing
Because one of the most critical phylogenetic questions assessed in this study is the placement of the most basal taxon, we conducted further testing among hypotheses placing each of the four genera as the most basal hylobatid. A comparison of the two maximum parsimony trees (placing Nomascus basal) against a tree with Hoolock as most basal showed no statistically significant difference between them (Table 3). Trees placing Symphalangus or Hylobates as most basal were significantly worse than the most parsimonious trees with Nomascus as basal. The likelihood-based SH test found that testing the ML tree against trees with the other genera as basal did not reveal statistically significantly differences among the trees. Similarly, a Bayes factor analysis found no significant differences between trees with Hoolock, Nomascus, Symphalangus or Hylobates as the most basal taxon. These tests show that any resolution among these genera is not statistically robust.
TABLE 3.
| Hoolock basal | Nomascus basal | Symphalangus basal | Hylobates basal | |
|---|---|---|---|---|
| Maximum Parsimony1 |
27,255 (Not Sig. Worse) |
27,251 (BEST) | 27,314 (Sig. Worse) | 27,308 (Sig. Worse) |
| Maximum Likelihood2 |
−62350.566 (BEST) | −62352.505 (Not Sig. Worse) |
−62352.759 (Not Sig. Worse) |
−62351.876325 (Not Sig. Worse) |
| Bayesian3 | −62292.05 (BEST) | −62290.43 (Not Sig. Worse) |
−62291.19 (Not Sig. Worse) |
−62288.21 (Not Sig. Worse) |
Number of tree steps (Templeton Test; Winning Sites Test).
Likelihood values (SH Test).
Harmonic mean likelihood (Bayes factor comparison).
3.5. Molecular Clock Results
A molecular clock was used to estimate divergence dates based using the Bayesian tree. This tree was chosen because a single completely bifurcating tree is required by the methodology and the Bayesian method yielded relatively high posterior probabilities for the placement of the different genera. The molecular clock analyses yielded a date for the origin of Hylobatidae at 21.8 Ma (19.7-24.1 Ma) and for the radiation of the family at 7.3 Ma (6.4-8.0 Ma) (Figure 4). The subsequent radiation of Nomascus and Symphalangus + Hylobates followed shortly thereafter (7.0 Ma and 6.4 Ma). The radiation of Hylobates occurred at 3.5 Ma (3.1-4.0 Ma) (additional dates and parameters are given in the Supplementary Materials). Dates estimated using trees with the other genera as basal gave qualitatively similar results.
FIGURE 4.
Tree scaled to the molecular clock dates of each node. Numbers at nodes indicate the 95% Bayesian credibility interval from the posterior distribution. Three calibrations nodes are indicated with a ‘C=’ within brackets. The dates following the ‘C=’ correspond to the lower (2.5%) and upper (97.5%) bounds of the calibration prior distribution. Other parameter estimates given in Supplementary Materials. All dates are in Ma.
4. Discussion
Combined analyses of the loci offered better phylogenetic resolution among the hylobatids than individual analyses of each gene. While combined analyses of mitochondrial, X-linked, and Y-linked DNA sequence data confirm the existence of the four separate genera of hylobatids, the analyses conflict regarding the relationships among these genera. Parsimony analysis supports Nomascus as the most basal taxon, and some molecular studies have supported this arrangement (Chatterjee, 2006; Roos and Geissmann, 2001; Thinh et al., 2010). Maximum likelihood and Bayesian analyses support Hoolock as the most basal, and both molecular and karyological studies have supported this alternative (Muller et al., 2003; Takacs et al., 2005). Bootstrap values showed moderate support for the particular topology among the hylobatid genera produced by each method. However, hypothesis testing among basal positions for the different genera found few significant differences among these hypotheses. Only a basal position for Symphalangus and Hylobates was rejected using parsimony-based tests; none of the alternatives were rejected using Bayesian- and likelihood-based tests. Within Hylobates, there was some consensus, in that H. pileatus represented the most basal taxon and that H. klossii and H. moloch are sister taxa. The placement of these Hylobates species agrees with other studies (Geissmann, 2002; Takacs et al., 2005; Whittaker et al., 2007). For the other remaining species within Hylobates, there is no consensus. For example, the placement of H. lar differed in each phylogenetic reconstruction.
Although the present study suggests more phylogenetic uncertainty than suggested by other recent analyses (e.g. Matsudaira and Ishida, 2010), it represents a significant advance for three reasons. First, the present dataset is based on a large sample of hylobatids, including members of all 4 genera. Second, the present dataset is the only study that examined regions of three independent genomic compartments. Others have been solely based on mitochondrial sequences. Third, in addition to the phylogenetic analyses, this study included explicit phylogenetic hypothesis testing using parsimony, likelihood, and Bayesian methods.
The persistent phylogenetic uncertainty among hylobatid genera and within Hylobates has a number of potential causes. First, the molecular clock results indicate that the radiation of the genera happened over an interval of less than 1 Ma, and this rapidity of the hylobatid radiation would lead to very short internal branches at the base of the hylobatid phylogeny. Members of the Hylobates genus also experienced a rapid radiation. These radiations have been suggested by others (Muller et al., 2003; Takacs et al., 2005; Thinh et al., 2010; Whittaker et al., 2007). Ancestral polymorphism followed by incomplete lineage sorting tends to create mismatches between gene trees and the species tree to a greater degree when divergence times are shorter and when ancestral population sizes are larger. Rapid radiations may have been coupled with other biogeographic factors including repeated dispersals, repeated forest expansions and contractions, and vicariant speciation, and all of these factors could lead to a complicated and difficult to resolve species phylogeny. Although an accurate phylogeny would assist in the reconstruction of hylobatid biogeographic history and indeed, some have attempted to use gibbon molecular phylogenies in this manner to posit specific dispersals and vicariant speciation events (e.g. Chatterjee, 2006, 2009; Thinh et al., 2010), these methods are completely dependent on having an accurate, well-resolved species tree. Hypothesis tests in the present study show that such a resolved phylogeny is not yet at hand for hylobatids. Therefore, such biogeographic scenarios are premature.
A second scenario that would result in the observed phylogenetic uncertainty is a sudden vicariance model. In this model, the phylogenetic uncertainty among genera of hylobatids is not interpreted as an artifact of ancestral polymorphisms or a complicated dispersal model, but rather is a real signal in its own right, i.e. a hard polytomy. A similar model was proposed for the biogeographic history of Hylobates species (Whittaker et al., 2007) and the model may also apply to the intergeneric radiation of hylobatids. There is paleoclimatological and paleoecological evidence to support the vicariance model. Here it is assumed that the ancestor of hylobatids was widely distributed throughout the rainforests of Southeast, South, and East Asia during the relatively warm and moist Miocene. From the middle Miocene until about 6 Ma, global climate cooled (Zachos et al., 2001). There is evidence for a habitat shift at this time from a nearby site in the Surai Khola area (13 – 1 Ma) of the Nepalese Siwaliks (Corvinus and Rimal, 2001). From about 13 – 9.5 Ma the landscape was dominated by a tropical evergreen forest type that this similar to today’s Southeast Asian rainforests (Corvinus and Rimal, 2001). Subsequently, from about 9.5 – 7.5 Ma there is evidence for moist deciduous forests at the site. Around the Miocene-Pliocene boundary, the habitat was drier and the subtropical environment was varied, with only about 25% evergreen forests (Corvinus and Rimal, 2001). During this cooling and habitat shift, the rainforest habitat of hylobatids might have fragmented or shrunk relatively simultaneously into multiple discontiguous distributions throughout Southeast, South, and East Asia. This model proposes that a large contiguous hylobatid distribution fragmented simultaneously and vicariantly into four descendant populations that later evolved into the different hylobatid genera. Such a simultaneous vicariant speciation event would likely lead to phylogenetic uncertainty among the hylobatid genera. The estimated timing of generic radiation produced by this study supports this model. The biogeography of other Southeast Asian mammals has similarly been interpreted as the result of vicariant events in the Late Miocene and Pliocene (e.g. Gorog et al., 2004; Steiper, 2006).
Within Hylobates, a similar model seems apparent. Within this genus, there was agreement among the analyses at two nodes, H. pileatus as the most basal species and for a sister taxon relationship between H. klossii and H. moloch. There was no agreement for the placement of the other 3 species or the other nodes. The current distribution of H. pileatus suggests that Hylobates may have originated on the mainland while derived members of the genus dispersed west and southward. Once distributed throughout Southeast Asia, cooling may have caused a second round of in situ vicariant speciation. The timing of these nodes roughly coincides with the onset of the Northern Hemisphere Glaciation at 3.2 Ma (Zachos et al., 2001), potentially linking these phenomena. It is noteworthy that there is some agreement that H. klossii and H. moloch are sister taxa, though with varying degrees of support. H. klossii occurs on the Mentawai islands, west of Sumatra, while H. moloch occurs on Java, implying a dispersal event between these locations. Overall, this interpretation supports Whittaker et al. (2007) and suggests that cooling events may have also driven vicariant speciation within Hylobates.
The two biogeographic models make different predictions for future studies of gibbon molecular phylogenetics. A rapid divergence, multiple dispersal, and vicariance model could have potentially led to ancestral polymorphisms that would cause multiple incongruences between gene trees and species trees. However, this model would predict that a robust phylogeny for hylobatids would emerge with additional study. This situation may parallel the situation with New World primates and African hominoids, taxa that required a considerable amount of data from a number of independent loci to resolve a robust phylogeny (Ray et al., 2005; Ruvolo, 1997; Wildman et al., 2009). In situ vicariance events, on the other hand, makes the prediction that additional analyses of hylobatids will not uncover a robust tree. Instead, this model predicts the intergeneric phylogeny of gibbons and many of the intrageneric nodes within Hylobates will remain resistant to the recovery of an accurate and well-supported bifurcating tree. In either case, examining a large number multiple independent loci will be required to assess hylobatid phylogenetics with confidence. Paleontological evidence is also of importance in testing among these hypotheses. Unfortunately, there is a relative lack of hylobatid fossils from critical time periods and localities. Additional paleontological research throughout Southeast, South, and East Asia at Miocene sites that are inferred to be rainforest would greatly improve our understanding of hylobatid evolutionary history.
Supplementary Material
SUPPLEMENTARY MATERIALS FIGURE 1
Maximum parsimony bootstrap tree, maximum likelihood bootstrap tree, and Bayesian tree for the mtDNA dataset. Numbers at the parsimony and likelihood tree nodes are bootstrap values. Numbers at the nodes of the Bayesian tree are credibility values.
SUPPLEMENTARY MATERIALS FIGURE 2
Maximum parsimony bootstrap tree, maximum likelihood bootstrap tree, and Bayesian tree for the G6PD dataset. Numbers at the parsimony and likelihood tree nodes are bootstrap values. Numbers at the nodes of the Bayesian tree are credibility values.
SUPPLEMENTARY MATERIALS FIGURE 3
Maximum parsimony bootstrap tree, maximum likelihood bootstrap tree, and Bayesian tree for the ZFY dataset. Numbers at the parsimony and likelihood tree nodes are bootstrap values. Numbers at the nodes of the Bayesian tree are credibility values.
SUPPLEMENTARY MATERIALS FIGURE 4
Maximum likelihood bootstrap phylogeny where branches with less than <50% bootstrap support are collapsed.
SUPPLEMENTARY MATERIALS TABLE 1
Primers used in this study.
SUPPLEMENTARY MATERIALS TABLE 2
Parameter estimates from the Bayesian molecular clock analysis.
Acknowledgements
We thank Weigang Qiu, Sherry Nelson, Erik Seiffert, and Anne Yoder and for comments and discussion. We are also grateful to George Amato, Adrienne Zihlman, Brookfield Zoo, Gladys Porter Zoo, and Yerkes Primate Center for providing samples and Miranda von Dornum, Julie Heider, and Glenn Maston for research assistance. Jason Hodgson kindly provided a perl script that facilitated data mining. This research was supported by funds from the National Science Foundation (grant SBR-9319021to MR; grant 9731407 to MR and SZ), the L. S. B. Leakey Foundation (grant to SZ) and the Mellon Foundation (funds to SZ). The infrastructure of the Anthropological Genetics Lab at Hunter College was supported by Grant Number RR03037 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Brandon-Jones D, Eudey AA, Geissmann T, Groves CP, Melnick DJ, Morales JC, Shekelle M, Stewart C-B. Asian Primate Classification. Intl J Primatol. 2004;25:97–164. [Google Scholar]
- Chatterjee HJ. Phylogeny and Biogeography of Gibbons: A Dispersal-Vicariance Analysis. Intl J Primatol. 2006;27:699–712. [Google Scholar]
- Chatterjee HJ. Evolutionary Relationships Among the Gibbons: A Biogeographic Perspective. In: Lappan S, Whittaker DJ, editors. The Gibbons. New Perspectives on Small Ape Socioecology and Population Biology. Springer; New York: 2009. pp. 13–36. [Google Scholar]
- Corvinus G, Rimal LN. Biostratigraphy and geology of the Neogene Siwalik Group of the Surai Khola and Rato Khola areas in Nepal. Palaeogeography, Palaeoclimatology, Palaeoecology. 2001;165:251–279. [Google Scholar]
- Delson E. Evolution of Old World Monkeys. In: Jones S, Martin RD, Pilbeam DR, editors. Cambridge Encyclopedia of Human Evolution. Press Syndicate of the University of Cambridge; Cambridge: 1992. pp. 217–222. [Google Scholar]
- Delson E. Cercopithecidae. In: Delson E, Tattersall I, Van Couvering JA, Brooks AS, editors. Encyclopedia of Human Evolution and Prehistory. 2nd Ed. Garland; New York: 2000. pp. 166–171. [Google Scholar]
- Gebo DL, MacLatchy L, Kityo R, Deino A, Kingston J, Pilbeam D. A hominoid genus from the early Miocene of Uganda. Science. 1997;276:401–404. doi: 10.1126/science.276.5311.401. [DOI] [PubMed] [Google Scholar]
- Geissmann T. Taxonomy and Evolution of Gibbons. Evol. Anthro. Suppl. 2002;1:28–31. [Google Scholar]
- Gorog AJ, Sinaga MH, Engstrom MD. Vicariance or dispersal? Historical biogeography of three Sunda shelf murine rodents (Maxomys surifer, Leopoldamys sabanus and Maxomys whiteheadi) Biological Journal of the Linnean Society. 2004;81:91–109. [Google Scholar]
- Groves CP. Systematics and phylogeny of gibbons. In: Rumbaugh DM, editor. Gibbon and Siamang. Karger; Basel: 1972. pp. 1–89. [Google Scholar]
- Haile-Selassie Y, Suwa G, White TD. Late Miocene teeth from Middle Awash, Ethiopia, and early hominid dental evolution. Science. 2004;303:1503–1505. doi: 10.1126/science.1092978. [DOI] [PubMed] [Google Scholar]
- Haimoff EH, Chivers DJ, Gittins SP, Whitten T. A phylogeny of gibbons (Hylobates spp.) based on morphological and behavioural characters. Folia Primatol (Basel) 1982;39:213–237. doi: 10.1159/000156079. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Hayasaka K, Takenaka O, Horai S. Molecular phylogeny of gibbons inferred from mitochondrial DNA sequences: preliminary report. J Mol Evol. 1995;41:359–365. doi: 10.1007/BF00186548. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- IUCN IUCN Red List of Threatened Species. Version 2010.1. 2010 www.iucnredlist.org.
- Lebatard AE, Bourles DL, Duringer P, Jolivet M, Braucher R, Carcaillet J, Schuster M, Arnaud N, Monie P, Lihoreau F, Likius A, Mackaye HT, Vignaud P, Brunet M. Cosmogenic nuclide dating of Sahelanthropus tchadensis and Australopithecus bahrelghazali: Mio-Pliocene hominids from Chad. Proc Natl Acad Sci U S A. 2008;105:3226–3231. doi: 10.1073/pnas.0708015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira K, Ishida T. Phylogenetic relationships and divergence dates of the whole mitochondrial genome sequences among three gibbon genera. Mol Phylogenet Evol. 2010;55:454–459. doi: 10.1016/j.ympev.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Mootnick AR. Gibbon (Hylobatidae) species identification recommended for rescue or breeding centers. Primate Conserv. 2006;21:103–138. [Google Scholar]
- Muller S, Hollatz M, Wienberg J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae) Hum Genet. 2003;113:493–501. doi: 10.1007/s00439-003-0997-2. [DOI] [PubMed] [Google Scholar]
- Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Prager EM, Wilson AC. Ancient origin of lactalbumin from lysozyme: analysis of DNA and amino acid sequences. J Mol Evol. 1988;27:326–335. doi: 10.1007/BF02101195. [DOI] [PubMed] [Google Scholar]
- Purvis A. A composite estimate of primate phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4. 2007 Available from http://beast.bio.ed.ac.uk/Tracer.
- Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst Biol. 2007;56:453–466. doi: 10.1080/10635150701420643. [DOI] [PubMed] [Google Scholar]
- Ray DA, Xing J, Hedges DJ, Hall MA, Laborde ME, Anders BA, White BR, Stoilova N, Fowlkes JD, Landry KE, Chemnick LG, Ryder OA, Batzer MA. Alu insertion loci and platyrrhine primate phylogeny. Mol. Phylogenet. Evol. 2005;35:117–126. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Ren F, Tanaka H, Yang Z. A likelihood look at the supermatrix-supertree controversy. Gene. 2009;441:119–125. doi: 10.1016/j.gene.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roos C, Geissmann T. Molecular phylogeny of the major hylobatid divisions. Mol Phylogenet Evol. 2001;19:486–494. doi: 10.1006/mpev.2001.0939. [DOI] [PubMed] [Google Scholar]
- Ruvolo M. Molecular phylogeny of the hominoids: inferences from multiple independent DNA sequence data sets. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- Senut B, Pickford M, Gommery D, Mein P, Chebol K, Coppens Y. First hominid from the Miocene (Lukeino Formation, Kenya) C. R. Acad. Sci. Ser. II A. 2001;332:137–144. [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Steiper ME. Population history, biogeography, and taxonomy of orangutans (Genus: Pongo) based on a population genetic meta-analysis of multiple loci. J Hum Evol. 2006;50:509–522. doi: 10.1016/j.jhevol.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Mehtods). Version 4. Sinauer Associates; Sunderland, Massachusetts: 2001. [Google Scholar]
- Takacs Z, Morales JC, Geissmann T, Melnick DJ. A complete species-level phylogeny of the Hylobatidae based on mitochondrial ND3-ND4 gene sequences. Mol Phylogenet Evol. 2005;36:456–467. doi: 10.1016/j.ympev.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution. 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- Thinh VN, Mootnick AR, Geissmann T, Li M, Ziegler T, Agil M, Moisson P, Nadler T, Walter L, Roos C. Mitochondrial evidence for multiple radiations in the evolutionary history of small apes. BMC Evol Biol. 2010;10:74. doi: 10.1186/1471-2148-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker DJ, Morales JC, Melnick DJ. Resolution of the Hylobates phylogeny: congruence of mitochondrial D-loop sequences with molecular, behavioral, and morphological data sets. Mol Phylogenet Evol. 2007;45:620–628. doi: 10.1016/j.ympev.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Wildman DE, Jameson NM, Opazo JC, Yi SV. A fully resolved genus level phylogeny of neotropical primates (Platyrrhini) Mol Phylogenet Evol. 2009;53:694–702. doi: 10.1016/j.ympev.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIALS FIGURE 1
Maximum parsimony bootstrap tree, maximum likelihood bootstrap tree, and Bayesian tree for the mtDNA dataset. Numbers at the parsimony and likelihood tree nodes are bootstrap values. Numbers at the nodes of the Bayesian tree are credibility values.
SUPPLEMENTARY MATERIALS FIGURE 2
Maximum parsimony bootstrap tree, maximum likelihood bootstrap tree, and Bayesian tree for the G6PD dataset. Numbers at the parsimony and likelihood tree nodes are bootstrap values. Numbers at the nodes of the Bayesian tree are credibility values.
SUPPLEMENTARY MATERIALS FIGURE 3
Maximum parsimony bootstrap tree, maximum likelihood bootstrap tree, and Bayesian tree for the ZFY dataset. Numbers at the parsimony and likelihood tree nodes are bootstrap values. Numbers at the nodes of the Bayesian tree are credibility values.
SUPPLEMENTARY MATERIALS FIGURE 4
Maximum likelihood bootstrap phylogeny where branches with less than <50% bootstrap support are collapsed.
SUPPLEMENTARY MATERIALS TABLE 1
Primers used in this study.
SUPPLEMENTARY MATERIALS TABLE 2
Parameter estimates from the Bayesian molecular clock analysis.