Abstract
Transient transfection of isolated Brugia malayi embryos by biolistics has proven to be useful in defining promoter structure and function in this parasite. However, isolated transfected embryos are developmentally incompetent. A method of producing developmentally competent transfected parasites is therefore needed. We report that L3 parasites can be chemically transfected in situ in the peritoneal cavity of a gerbil with a construct consisting of a secreted luciferase reporter gene containing a promoter, the 3' untranslated region and first intron derived from the B. malayi 70kDa heat shock protein gene. The in situ chemically transfected parasites are developmentally competent, producing adult parasites with an efficiency similar to that obtained from implanted untreated L3s. Cultured adult parasites and progeny microfilariae (mf) derived from L3s transfected with this construct secreted luciferase into the culture medium. When the transfected mf were fed to mosquitoes and the resulting L3s collected, the L3s also secreted luciferase into the culture medium. Progeny mf from transgenic adult parasites contained transgenic DNA, and the transgenic mRNA produced in these parasites was found to be correctly cis- and trans-spliced. In situ chemical transformation thus results in developmentally competent transfected B. malayi in which the transgenic sequences remain transcriptionally active in all life cycle stages and are present in the subsequent generation.
Keywords: Filariasis, Transfection, HSP70, Transgenesis, Gaussia, Luciferase
1. Introduction
Nematodes are the most common parasitic organisms infecting humans (Warren and Mahmoud, 1985). Although the intestinal dwelling nematodes are the most prevalent nematode parasites of humans, it is the tissue dwelling filarial worms that produce the most severe pathology. The three most common human filarial parasites are Brugia malayi, Wuchereria bancrofti and Onchocerca volvulus. The first two are parasites of the lymphatic and circulatory system, and result in lymphatic pathologies leading to elephantiasis. Onchocerca volvulus resides in the connective tissue and dermis and results in onchocerciasis or river blindness. It has been estimated that B. malayi and W. bancrofti combined infect approximately 120 million individuals worldwide (http://www.who.int/lymphatic_filariasis/epidemiology/en), while O. volvulus infects approximately 37 million people (African Programme for Onchocerciasis Control, 2005).
The two most common human filariae, O. volvulus and W. bancrofti, are obligate parasites of humans. This has made it extremely difficult to obtain material from these organisms. In contrast, B. malayi can be successfully cultured in a variety of surrogate animal hosts (Denham and Fletcher, 1987; Mak et al., 1990; Nelson et al., 1991; Weil et al., 1992). Accordingly, B. malayi has become the model organism of choice for the study of the filarial parasites. The B. malayi genome was recently sequenced, assembled and annotated (Ghedin et al., 2007). The completion of the B. malayi genome sequence opened up new avenues of research into how these parasites regulate their gene expression. This question is central to understanding how this parasite has adapted to life in two very different host environments (the vertebrate and insect) and how it survives active attack by the host's immune system. With the appropriate tools, this information also has the potential to link gene polymorphisms to particular phenotypes (e.g. drug resistance). Given the difficulty in carrying out classical genetic studies in these organisms (due to the complexity of their life cycles and lack of easily scored phenotypic markers), reverse genetic approaches involving the introduction of exogenous DNA molecules offer the most promising avenue for the study of gene structure and regulation in these parasites.
Isolated B. malayi embryos can be reproducibly transiently transfected using bombardment with DNA-coated gold particles (Shu et al., 2003). This method has been used to map the core promoter sequences of a number of different B. malayi genes (Shu et al., 2003; Higazi et al., 2005; de Oliveira et al., 2008; Liu et al., 2009) and to demonstrate that synthetic genes containing an ecdysone response element recognized by the B. malayi homologue of the ecdysone receptor are responsive to ecdysone in vivo (Tzertzinis et al., 2010). Biolistic transient transfection of isolated embryos has also been used to explore the sequences necessary and sufficient to support trans-splicing of B. malayi premRNAs in vivo (Higazi and Unnasch, 2004; Liu et al., 2007, 2010). However, because isolated embryos are incapable of further development, this method cannot be used to study gene expression and regulation in other life cycle stages or to study phenotypes that are expressed only by intact parasites (e.g. drug resistance). Similarly, while biolistics can be used to transfect intact B. malayi of other life cycle stages (Higazi et al., 2002), this results in damage to the cuticle of the transfected parasites that ultimately proves fatal.
In the experiments described below, we demonstrate that molting L3 B. malayi parasites are amenable to DNA transfection by standard chemical methods in vivo. Such transfected parasites are developmentally competent and a reporter gene encoded by the exogenous DNA used to transfect these parasites is maintained and is transcriptionally active throughout all stages of the parasites' lifecycle. The development of a method of producing developmentally competent transfected parasites is a first step in employing transgenesis to study gene function and regulation in all life cycle stages of B. malayi.
2. Materials and methods
2.1. Preparation of pBmHSP70/GLuc
pBmHSP70/GLuc was constructed using the pGLuc-Basic vector (New England Biolabs, Ipswich, MA, USA) as a backbone. As a first step in this process, the promoter from the B. malayi gene encoding the 70 kDa heat shock protein (HSP) homologue (consisting of the 658 nucleotides located upstream of the start codon of the BmHSP70 opening reading frame (ORF)) was amplified by PCR from B. malayi genomic DNA, as previously described (Shu et al., 2003). The resulting amplicon was cloned into the pPCR2.1 cloning vector (Invitrogen, Carlsbad, CA, USA), and its DNA sequence confirmed. The insert from a clone whose sequence was confirmed was then sub-cloned into the EcoR l site of pGLuc-Basic. The 3' untranslated region (UTR) from the BmHSP70 gene (corresponding to the nucleotides located in positions +1 to +241 relative to the stop codon of the BmHSP70 ORF) was then amplified by PCR with appropriate primers engineered to contain synthetic Xbal sites at their 5' ends. The sequences of the primers used to amplify this fragment were 5' GGGTCTAGAATGACTTTTTGATCGGAAT 3' and 5' GGGTCTAGAACAATATTCACAAGGACCA 3'. The resulting amplicon was cloned into the PCR 2.1 TA cloning vector and the cloned DNA sequence confirmed. The insert was sub-cloned into the Xbal site of the construct containing the BmHSP70 promoter described above.
The sequence corresponding to positions 258–263 in the GLuc ORF of the construct described above were then mutated to form a synthetic Spel restriction site, using the Gene Tailor in vitro mutagenesis system (Invitrogen) as previously described (Liu et al., 2009). The sequence corresponding to the first intron of the BmHSP70 gene was then amplified from B. malayi. The intron sequence was amplified using primers containing synthetic Spel sites at their 5' ends. The sequences of the primers used to amplify the intron sequence were 5' GGACTAGTGTACAATTGTTATTGGTTTATTTT 3' and 5' GGACTAGTTATCCATCTCTTTCTAAGGAAA 3'. The latter primer was designed so that the last six nucleotides of the intron were replaced by the synthetic Spel site. The resulting amplicon was then cloned into the PCR 2.1 TA cloning vector and the DNA sequence of several clones determined. The insert of one of the clones whose sequence was confirmed was excised from the TA vector and cloned into the synthetic Spel site of the construct described above. This resulted in the insertion of the intron sequence (lacking its 3' 6 nucleotides) into the GLuc ORF, flanked by two synthetic Spel sites. The synthetic Spel sites were then mutated back to their corresponding native sequences, using the Gene Tailor in vitro mutagenesis kit, as described above. The sequence of the construct was deposited in the Genbank sequence database with the accession number HQ388295.
2.2. Biolistic transfection of embryos
Biolistic transfection of embryos was carried out essentially as previously described (Shu et al., 2003). In brief, plasmid DNA from each construct was precipitated on to the surface of 0.6 μm gold particles (BioRad Hercules, CA, USA) at a loading ratio of 16 μg of DNA per mg of gold. Embryos were isolated from gravid female parasites (obtained from the Filariasis Research Reagent Resource Center (FR3) at the University of Georgia, USA) by dissection, placed in 30 μL of tissue culture medium and transfected in a PDS 1000/He Biolistic apparatus (BioRad, Hercules, CA, USA). The embryos were maintained under a vacuum of 12 inches of mercury (40.64 kPa) during the bombardment and were transfected with 500 μg of coated gold particles per bombardment at a pressure of 1100 PSI (7,584.5 kPa). Following bombardment, the embryos were maintained in RPMI tissue culture medium containing 25 mM HEPES, 20% FCS, 20 mM Glucose, 24 mM sodium bicarbonate, 2.5 mg/mL amphotericin B, 10 units/mL penicillin, 10 units/mL streptomycin and 40 mg/mL gentamicin, at 37°C and 5% CO2 for 48 h. Following culture, the embryos were harvested by centrifugation at 3000 g for 3 min. The embryos were resuspended in 100 μL passive lysis buffer (Promega, Madison, WI, USA) and homogenized in a glass micro-homogenizer. The homogenates were centrifuged at 3000 g for 3 min and the supernatant collected. A total of 20 μL of the supernatant was mixed with 100 μL of LAR II luciferase assay solution (Promega) in a glass tube. The number of relative light units (RLU) produced was then measured in a Berthold single tube luminometer (Berthold Detection Systems, Pforzheim, Germany). The program utilized for RLU measurement consisted of a delay of 5 s after substrate addition, followed by an 8 s measurement period.
2.3. Transfection of L3s in vitro
A total of 200 infective L3s per transfection (obtained from the FR3) were placed in individual wells of 24 well tissue culture plate in 600 μL of tissue culture medium consisting of Minimal Essential Medium Alpha (Invitrogen) containing 1000 units/mL of penicillin-streptomycin (Invitrogen) 10 μg/mL gentamycin (Sigma Chemical) 2μg/mL ciprofloxin (Bayer) and 1μg/mL ceftazidime (Glaxo Wellcome). A solution consisting of 6 μg of the transfecting plasmid DNA in 240mM CaCl2 in a total volume of 150 μL was then prepared. The solution was added dropwise while mixing to 150 μL of 2 × HBS (50 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4 (pH 7.1)) while bubbling air through the mixture, to prepare a calcium phosphate - DNA co-precipitate. The mixture was then incubated at room temperature for 30 min to complete precipitate formation. The mixture containing the precipitate (total volume 300 μL) was then added dropwise to an individual well containing the L3s while swirling to evenly distribute the precipitate among the larvae. The plate was then incubated at 37°C and 5% CO2 for up to 10 days. On day 5 following the addition of the precipitate suspension, ascorbic acid was added to a final concentration of 15 mg/mL to each well to induce molting (Rajan et al., 2003). Firefly luciferase activity was assayed in homogenates prepared from the transfected L3s as described in Section 2.2.
2.4. Transfection of L3s in vivo, recovery and analysis of recovered parasites for reporter gene activity
A calcium phosphate - DNA co-precipitate was prepared as described above. A total of 300 μL of this suspension was mixed with 600 μL of RPMI 1640 medium (Invitrogen) containing 250 L3s. The mixture was then injected into the peritoneal cavity of a naïve jird. The animals were held undisturbed for a period of 120 days. The animals were then subjected to peritoneal lavage employing 5 mL of PBS to recover microfilariae (mf). The lavage was repeated at 7 day intervals. At day 150 p.i., the animals were euthanized and adult parasites recovered from the peritoneal cavity.
Recovered parasites (adults and mf) were cultured from periods ranging from 2–6 days in RPMI tissue culture medium containing 25 mM HEPES, 20% FCS, 20 mM Glucose, 24 mM sodium bicarbonate, 2.5 mg/mL amphotericin B, 10 units/mL penicillin, 10 units/mL streptomycin and 40 mg/mL gentamicin, at 37°C and 5% CO2. GLuc activity was measured in the culture medium using the renilla assay reagents provided in the Stop and Glo luciferase assay kit (Promega). A total of 20 μL of the culture medium was mixed with 100 μL of LAR II luciferase assay solution (Promega) and 100 μL of Stop and Glo reagent in a glass tube. The number of relative RLUs produced was then measured in a Berthold single tube luminometer. The program utilized for RLU measurement consisted of a delay of 5 s after substrate addition, followed by an 10 s measurement period. Preliminary experiments utilizing purified GLuc (New England Biolabs, Ipswich, MA, USA) demonstrated that this assay was capable of detecting GLuc activity at the low attomolar range, with a linear range extending over at least five orders of magnitude (Supplementary Fig. S1). The specificity of the activities detected in the culture supernatants were confirmed by a 5 min pre-incubation of culture supernatants with an anti-GLuc antibody (New England Biolabs) at a final dilution of 1/5000 prior to substrate addition. Preliminary experiments demonstrated that the antibody, when used under these conditions, was capable of complete inactivation of purified GLuc over at least a four order of magnitude range (Supplementary Fig. S2). In experiments to determine the prevalence of transgenic F1 mf, pools of 50 mf each were assayed. The prevalence of transgenic mf was determined from the number of positive pools using the Poolscreen algorithm. Poolscreen was originally developed to calculate the prevalence of parasite-infected vectors from results obtained from screening pools of vector insects. It utilizes the proportion of negative pools and the pool size to calculate a prevalence of positive organisms together with an associated 95% confidence interval (Katholi et al., 1995).
2.5. Production of L3s from transgenic mf
Microfilariae collected by peritoneal lavage were pelleted by centrifugation at 670 g for 15 min, and resuspended in uninfected canine blood to yield concentrations of 5,000 mf/mL, 7,500 mf/mL and 10,000 mf/mL. Adult mosquitoes (Aedes aegypti, black-eyed Liverpool strain) were allowed to feed on the microfilaremic blood through artificial feeders for 3 h. Blood was collected in heparin tubes and used on the day of collection. Each group of mosquitoes was allowed to feed on a 5 mL quantity of blood. Fourteen days after the blood meal, mosquitoes were collected using standard methods (www.filariasiscenter.org). Briefly, mosquitoes were stunned by placing them at −20°C for 1 min, crushed with a mortar and pestle, and rinsed over mesh screens using Hanks' Balanced Salt Solution (HBSS). Brugia malayi larvae were then allowed to migrate out of mosquitoes while the mesh screen was soaked in warm HBSS (~37°C). Larvae were then transferred to RPMI-1640 containing 100 mg/mL streptomycin, 100 U/mL penicillin and 40 μg/mL gentamicin.
2.6. Freezing and revival of transgenic mf
Microfilariae obtained from peritoneal lavage of infected jirds were counted and divided into aliquots of 1×105/mL in liquid nitrogen freezing vials (Nunc). The mf were pelleted at 2300 g for 15 s and resuspended in 1 mL of cell culture freezing medium (Invitrogen). The vials were placed at −80°C for 24 h and then transferred to liquid nitrogen.
To revive the frozen mf, the vials were removed from the liquid nitrogen and agitated in a 37°C water bath until completely thawed. The mf were allowed to settle to the bottom of the vial and excess freezing medium removed. A total of 1 mL of RPMI 1640 tissue culture medium was added to the vial to resuspend the mf, and the parasites were transferred to a six well tissue culture plate. Parasite viability was assessed by microscopic examination. The mf were then cultured at 37°C and 5% CO2 for 48 h, and the medium assayed for GLuc activity, as described above. Developmental competency of the cryopreserved transgenic mf was confirmed by feeding thawed mf to mosquitoes and assaying for the recovery of larvae, as described in Section 2.5.
2.7. PCR and reverse transcriptase (RT)-PCR assays of transgenic parasites
Nested PCR assays were utilized to detect the presence of the GLuc gene in transfected parasites. DNA was prepared using the Qiacube automated DNA preparation apparatus (Qiagen, Valencia, CA, USA) and the DNAeasy blood and tissue DNA extraction kit (Qiagen). Nested PCRs utilized Taq polymerase from Invitrogen, and employed the reaction components recommended by the manufacturer. First stage PCRs to detect the GLuc gene employed 1 μg of DNA prepared from the transgenic parasites (obtained from 5000 mf or 100 L3s) and two primers derived from the GLuc ORF (GLuc 772c 5' GCCCTGATCTGCATCGCTGTGG3' and GLuc1560nc 5' TTTGAGGCAGCCAGTTGTGCAGTC 3'). Nested PCRs utilized 5 μL of the amplicon from the first reaction as a template, and primers GLuc920c (5' TCAAAGAGATGGAAGCCAATG3') and GLuc1427nc (5' ACCTGTGCGATGAACTGCTC) 3'. Cycling conditions in both reactions consisted of 30 cycles of 30 s at 94°C, 30 s at 52°C and 1 min at 72°C. Products were analyzed by electrophoresis on a 1% agarose gel.
Hemi-nested splice leader (SL) mediated RT-PCRs to detect GLuc containing mRNA were carried out essentially as previously described (Liu et al., 2007). The initial RT-PCR included 1 μg of total RNA from the transgenic parasites, prepared as previously described (Liu et al., 2007) and utilized a 5' primer derived from the SL1 sequence of B. malayi (5' GGTTTAATTACCCAAGTTTGAG 3') and GLuc1560nc. Reaction conditions consisted of 50°C for 30 min and 95°C for 15 min, followed by 30 cycles consisting of 30 s at 94°C, 30 s at 52°C and 1 min at 72°C. Hemi-nested PCRs utilized 5 μL of the product from the first reaction, the SL1 primer and GLuc1427nc. Cycling conditions consisted of 30 s at 94°C, 30 s at 52°C and 1 min at 72°C. Products were analyzed by electrophoresis on a 1% agarose gel.
3. Results
3.1. Development of a method to produce developmentally competent transgenic B. malayi L3s
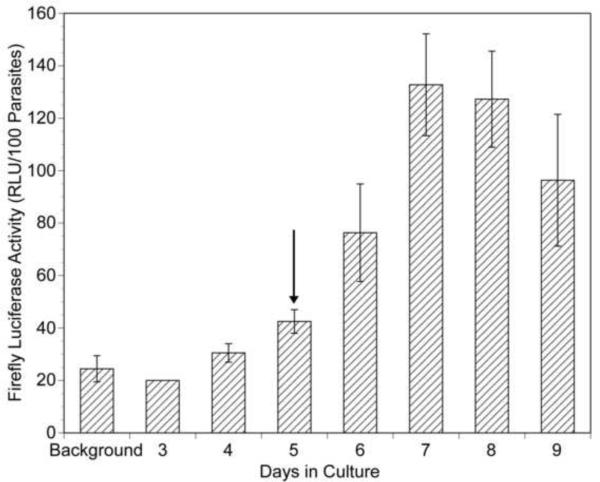
As described Section 1, biolistics has been successfully used to transiently transfect isolated B. malayi embryos. Biolistics and intrauterine microinjection were also successfully used to transiently transfect adult female parasites (Higazi et al., 2002) while biolistic bombardment was successful in transfecting L3s (unpublished data). However, both biolistics and microinjection resulted in damage to the cuticle of the parasites, and implantation of these parasites into naïve jirds resulted in death of the worms (data not shown). Attempts to transfect intact B. malayi of various life cycle stages utilizing a variety of less invasive methods (such as electroporation and chemical transfection methods) proved unsuccessful. Based upon these preliminary studies, we hypothesized that the mature cuticle might represent a barrier to chemical transfection. We further hypothesized that if this was the case, molting parasites might prove more susceptible to such chemical transfection methods. Studies by Rajan and colleagues have demonstrated that molting of L3 to L4 parasites could be induced in culture by the addition of ascorbic acid to the culture medium (Rajan et al., 2003). We therefore hypothesized that B. malayi might be susceptible to chemical transfection methods during the L3 to L4 molt. To test this hypothesis, L3s were maintained in culture in the presence of calcium phosphate - DNA co-precipitates prepared using the construct pBmHSP70 (−659 to −1)/luc (Shu et al., 2003). This construct consists of the BmHSP70 promoter driving the expression of a firefly luc reporter gene. pBmHSP70 (−659 to −1)/luc has previously been found to result in high levels of firefly luc activity when used to biolistically transfect B. malayi embryos (Shu et al., 2003). No firefly luc activity was seen in homogenates prepared from the cultured L3s maintained for 4 days in the presence of the calcium phosphate - BmHSP70 (−659 to −1)/luc precipitates (Fig. 1). However, firefly luc activity began to be detectable in L3 homogenates after 6 days in culture (1 day after ascorbic acid was added to the culture to induce molting) at levels which were approximately three times background. The firefly luc activity increased over the following days, reaching a peak of approximately five times background in homogenates of L3s harvested after 7–8 days in culture, a point when morphological evidence of molting (i.e. cast cuticles) began to appear in the culture medium (data not shown). While the level of firefly luc activity detected was much lower than that seen in isolated embryos biolistically transfected with the same construct (which average approximately 1000× background (Shu et al., 2003)), the data suggested that cultured L3s were capable of being transfected when induced to molt in culture.
Fig. 1.
Firefly luciferase (luc) activity in homogenates of cultured larvae exposed to calcium phosphate – pBmHSP70(−659 to−1)/luc co-precipitates. Larvae were cultured in the presence of calcium phosphate - pBmHSP70(−659 to−1)/luc co-precipitates for varying periods and homogenates assayed for firefly luciferase activity as described in Materials and methods. Each culture contained 200 L3s. “Background” represents the activity of 200 L3s cultured in the absence of the transfecting DNA. RLU, relative light units. The vertical arrow indicates the day upon which ascorbic acid was added to the cultures to induce molting (Rajan et al., 2003). Columns represent the mean and error bars indicate the S.D. from multiple assays (2–6) run at each time point.
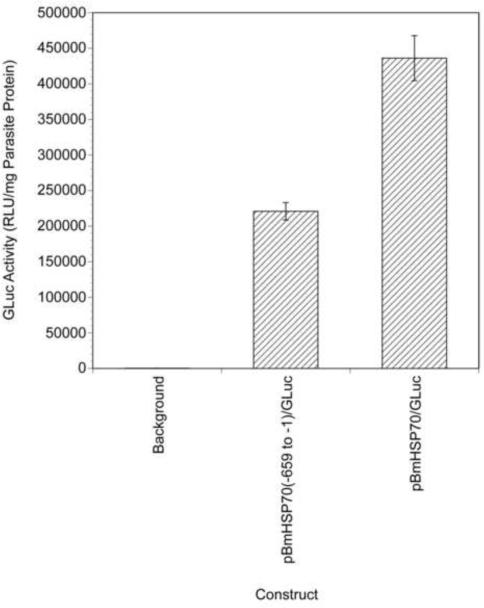
While firefly luc has proven to be a useful reporter gene in B. malayi (Shu et al., 2003), assaying for this reporter is destructive, requiring parasites to be homogenized prior to assaying for activity. This renders firefly luc unsuitable for use as a marker for monitoring for transfection in viable parasites. In contrast, the luc produced by G. princeps (GLuc) represented a potentially useful marker for monitoring transfection in viable parasites. GLuc utilizes coelenterazine as a substrate and exhibits a higher specific activity than firefly luciferase, making it a very sensitive reporter for transfection (Tannous et al., 2005). Furthermore, GLuc is secreted from transfected cells (Wurdinger et al., 2008), meaning that lysis of the transfected organism is not necessary to assay for reporter activity. GLuc has been shown to be active and secreted from many different cell types and organisms, including Schistosoma mansoni (Cheng and Davis, 2007). To determine whether GLuc might be used as a non-destructive reporter for transfected B. malayi, the BmHSP70(−659 to −1) promoter was cloned upstream of the GLuc reporter gene in the vector pGLuc-Basic resulting in the construct pBmHSP70(−659 to −1)/GLuc. This construct was used to biolistically transfect B. malayi embryos, and the presence of GLuc in the tissue culture medium assayed after 48 h in culture. High levels of GLuc activity were detected in the culture media of these transiently transfected embryos (approximately 700 times background levels; Fig. 2) suggesting that GLuc could serve as a reporter for assaying for transfection in viable B. malayi.
Fig. 2.
Gaussia princeps luciferase (GLuc) activity in the culture media of embryos biolistically transfected with pBmHSP70(−659 to −1)/GLuc and pBmHSP70/GLuc. Isolated embryos were biolistically transfected with the two constructs and cultured for 48 h. The culture medium was assayed for GLuc activity as described in Materials and methods. The embryos were harvested and their protein content determined using the Bradford method (Bradford, 1976). Activities are expressed as the number of relative light units (RLU) from the medium normalized to the amount of parasite protein in each well. Columns indicate the mean and error bars the S.D.s of six independent transformations.
Most mRNAs in B. malayi (including BmHSP70) are trans-spliced, and previous studies have shown that trans splicing in this organism is dependent upon a trans splicing motif (TSM), which in the case of the BmHSP70 gene is encoded in the first intron (Higazi and Unnasch, 2004; Liu et al., 2007). Furthermore, studies of transgenic gene expression in the related parasitic nematode parasite Strongyloides stercoralis have demonstrated that proper temporal and spatial expression requires sequences encoded in the 3' UTR (Li et al., 2006). It was thus felt that an optimal construct for expression of transgenic sequences throughout the B. malayi lifecycle would exhibit as many features of a native B. malayi gene as possible. To prepare such a construct, the 3' UTR of the native BmHSP70 gene was cloned downstream of the GLuc ORF in pBmHSP70(−659 to −1)/GLuc. A comparative sequence analysis of 100 intron-exon boundaries present in native B. malayi genes was then conducted to identify the sequences that most often flanked introns. This analysis revealed that the most common dinucleotide flanking the 5' end of an intron was AA, while the most common dinucleotide flanking the 3' end of an intron was GT (Supplementary Fig. S3). Based on this analysis, the sequence of the GLuc ORF was searched for the sequence AAGT. This sequence was found to be present at positions 262–265 of the GLuc ORF. The first intron of the native BmHSP70 gene (which encoded the TSM) was then inserted following position 263 of the GLuc ORF of the construct described above, as described in Materials and methods. This resulted in a construct designated pBmHSP70/GLuc, which contained the native BmHSP70 promoter inserted 5' of the GLuc ORF, the first intron of the native BmHSP70 gene inserted into the GLuc ORF, and the native BmHSP70 3' UTR inserted 3' of the GLuc ORF (Fig. 3). pBmHSP70/GLuc was then tested for its ability to produce GLuc activity when used to transfect B. malayi embryos. pBmHSP70/GLuc transfected embryos were found to secrete approximately twice as much GLuc as embryos transfected with pBmHSP70(−659 to −1)/GLuc, which lacked the first intron and 3' UTR (Fig 2).
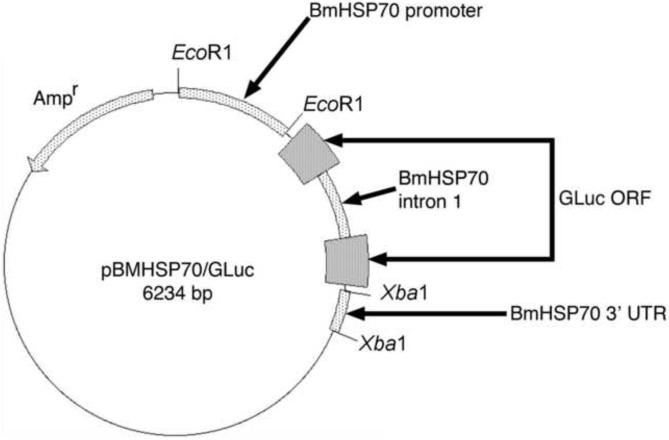
Fig. 3.
Map of the reporter plasmid pBmHSP70/Gluc. ORF, opening reading frame; UTR, untranslated region.
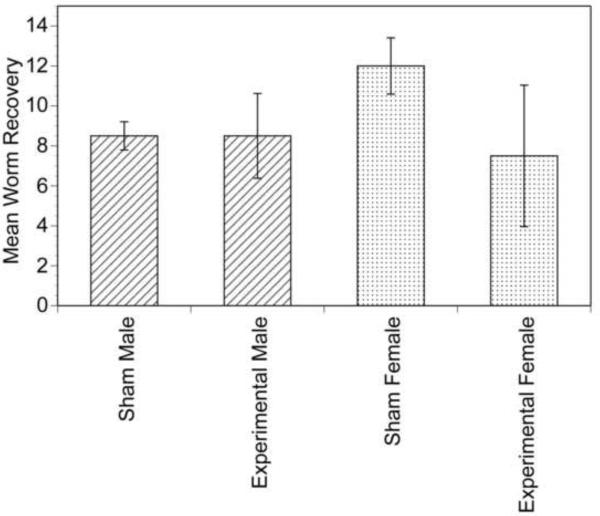
pBmHSP70/GLuc was then used to transfect L3s in culture as described above. After 7 days in culture, the parasites were injected into the peritoneal cavity of naïve gerbils and severe combined immunodeficiency (SCID) mice. Unfortunately, parasites were not recovered from any of these animals when the animals were necropsied 140 days after injection of the larvae (data not shown). Gerbils injected with transfected cultured L4s necropsied at 2 months p.i. revealed the presence of stunted, developmentally impaired larval parasites (data not shown). This suggested that although the cultured L4s appeared morphologically normal and exhibited high motility, they were incapable of further development. Aliquots of 250 uncultured L3s each were then mixed with calcium phosphate-pBmHSP70/GLuc co-precipitates and the mixture injected directly into naïve gerbils, bypassing the culture step. When these animals were necropsied 140 days later, adult parasites were recovered from all animals. The number of adult parasites recovered from animals injected with the L3s together with calcium phosphate-pBmHSP70/GLuc co-precipitates did not differ significantly from that recovered from animals injected with 250 untreated L3s, suggesting that the co-precipitates did not adversely affect the viability or developmental potential of the L3s (Fig. 4). Consistent with this conclusion, large numbers of mf were also recovered from the peritoneal cavity of each animal.
Fig. 4.
Recovery of adult parasites from gerbils co-injected with L3s and calcium phosphate - pBmHSP70/Gluc co-precipitates or L3s alone. Sham = animals injected with L3s alone and experimental = animals co-injected with L3s and calcium phosphate - pBmHSP70/GLuc co-precipitates. Columns represent the mean recovery and error bars the S.D.s of two animals per group.
3.2. Stability and activity of transgenic sequences in subsequent life cycle stages
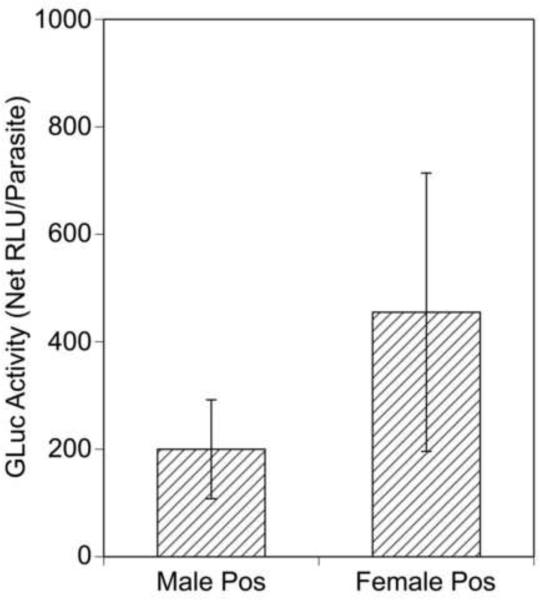
Recovered adult parasites were then cultured individually for 4 days and the culture medium assayed for GLuc activity. In conducting these assays, samples exhibiting GLuc activity levels that were greater than the mean plus 2 S.D.s of control assays of media from untransfected parasites were considered to be positive. Using this criterion, 27/40 (67%) of the individual adult females and 13/22 (60%) of the individual adult males were found to secrete detectable levels of GLuc into their culture medium. In general, the adult female parasites secreted more GLuc into their medium than did the males, although this difference was not statistically significant (Fig. 5).
Fig. 5.
Gaussia princeps luciferase (GLuc) activity secreted by transfected individual adult parasites. Individual adult parasites were cultured for 4 days and the culture medium assayed for GLuc activity as described in Section 2.4. Bars show the means and error bars the S.D.s of the GLuc activity (relative light units (RLU) minus background) in the culture medium of those individual adult parasites deemed positive (pos) for GLuc activity (defined as exhibiting RLU greater than the mean plus 2 S.D. of the RLU measured in media collected from untransfected individual adult parasites cultured in parallel).
Microfilariae represent the first developmental stage following sexual reproduction in B. malayi. It was therefore of interest to determine whether mf produced by these adults contained the transgenic sequences, as this would indicate that the transgenic sequences were capable of being inherited. To investigate this question, aliquots of the mf recovered from the peritoneal cavity of the animals were cultured for varying times, and the tissue culture medium assayed for GLuc activity. The progeny mf were found to secrete low but detectable levels of GLuc into the medium after 2 days in culture. The activity detected increased approximately three-fold after an additional 4 days, indicating continued production of GLuc from the cultured parasites (Fig. 6). Pre-incubation of the tissue culture medium with an anti-GLuc antibody eliminated the GLuc activity from these samples, confirming the specificity of the assay (Fig. 6).
Fig. 6.
Gaussia princeps luciferase (GLuc) activity in progeny microfilariae (mf) recovered from animals co-injected with L3s and calcium phosphate - pBmHSP70/GLuc co-precipitates. Progeny mf were recovered from animals co-injected with L3s and calcium phosphate - pBmHSP70/GLuc co-precipitates by peritoneal lavage. The mf were divided into aliquots of 5000 and cultured for 2–6 days (d). The resulting medium was then assayed for GLuc activity. Sham = media from mf recovered from animals injected with L3s alone, +DNA = media from mf recovered from animals co-injected with L3s and calcium phosphate - pBmHSP70/GLuc co-precipitates, and +DNA & Ab = media from mf recovered from animals co-injected with L3s and calcium phosphate - pBmHSP70/GLuc co-precipitates pre-incubated with anti-GLuc antibody. Columns represent the mean and error bars the S.E.M. calculated from three individual aliquots of mf per group. RLU, relative light units.
To estimate the proportion of transgenic mf in the F1 population, aliquots containing 50 mf each were cultured for 4 days and the culture medium in each well was assayed for GLuc activity. Using a cut-off of the mean plus 2 S.D.s of control assays employing media from wells containing 50 untransfected mf, 20/96 (21%) of the pools were positive, leading to a predicted prevalence of transgenic mf in the F1 population of 0.47% (95% confidence interval 0.28% – 0.68%).
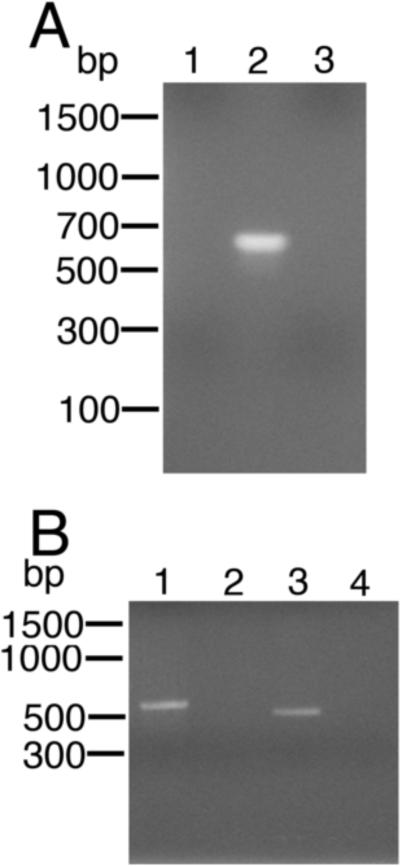
If these progeny mf contained transgenic sequences, both the DNA and mRNA corresponding to the GLuc gene should be detectable in these parasites. To confirm this, a nested PCR was carried out on DNA isolated from the progeny mf to detect the GLuc genomic sequences, while a SL mediated hemi-nested RT-PCR was conducted to detect mRNA sequences derived from the GLuc gene. The nested PCR (employing GLuc primers spanning the insertion site for the intron in BmHSP70/GLuc) produced an amplicon of approximately 600 bp, near the expected size of 624 nucleotides (Fig. 7A). Sequence analysis of this amplicon confirmed that it was derived from BmHSP70/GLuc. Similarly, the RTPCR produced an amplicon of approximately 500 bp (Fig. 7B), consistent with the expected size of the amplicon derived from a correctly cis- and trans-spliced mRNA (471 bp). Sequence analysis of this product confirmed that it was derived from a correctly cis- and trans-spliced mRNA produced from BmHSP70/GLuc (data not shown).
Fig. 7.
PCR and spliced leader (SL) reverse transcriptase (RT)-PCR analysis of progeny microfilariae (mf) recovered from animals co-injected with L3s and calcium phosphate - pBmHSP70/Gluc co-precipitates. A) PCR analysis of DNA extracted from progeny mf utilizing primers derived from the GLuc open reading frame (ORF). PCRs were carried out as described in Materials and methods. Lane 1 = PCR using DNA extracted from mf recovered from an animal injected with L3s alone. Lane 2 = PCR using DNA extracted from mf recovered from an animal co-injected with L3s and calcium phosphate - pBmHSP70/GLuc co-precipitates. Lane 3 = PCR negative control. B) Hemi-nested SL mediated RT-PCR analysis of mRNA extracted from mf utilizing primers derived from the GLuc ORF and the B. malayi SL. Lane 1 = SL mediated RT-PCR positive control (RNA extracted from embryos transfected with pBmHSP70(E1-I1-E2-I2-E3)/luc and amplified using primers derived from the BmHSP70 gene as previously described (Liu et al., 2007)). Lane 2 = RNA extracted from mf recovered from an animal injected with L3s alone. Lane 3 = RNA extracted from mf recovered from an animal co-injected with L3s and calcium phosphate - BmHSP70/GLuc co-precipitates. Lane 4 = RT-PCR negative control.
Given that the transgenic sequences appeared to be inherited by the progeny mf, it was of interest to determine whether these mf could be induced to develop to L3s, thus completing the life cycle. To accomplish this, the mf were mixed with blood and fed to A. aegypti mosquitoes. After 14 days, approximately 200 L3s were obtained from the mouthparts of these mosquitoes by dissection. The L3s were cultured for 48 h and the medium assayed for GLuc activity. The medium from the L3s contained low levels of GLuc activity which was inhibited by the addition of the anti-GLuc antibody, suggesting that the transgene was still present and active in these parasites (Fig. 8).
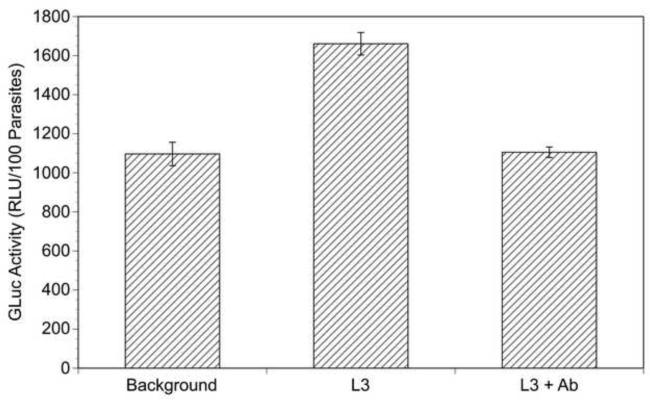
Fig. 8.
Gaussia princeps luciferase (GLuc) activity secreted by L3s derived from transgenic microfilariae (mf). L3s were generated from progeny transgenic mf and assayed for secreted GLuc activity as described in Section 2.4. Each culture contained 100 L3s. Background = activity from media in which untransfected L3s were cultured. L3 = activity from media from L3s produced from transgenic mf. L3 + antibody (Ab) activity from media from L3s produced from transgenic mf pre-incubated with anti-GLuc antibody. Columns represent means and error bars S.D.s of duplicate experiments. RLU, relative light units.
One important outcome of any stable transfection system would be the establishment of transfected parasite lines. Because B. malayi is difficult and expensive to maintain (involving obligate animal hosts and a mosquito vector) it would be useful to develop a method for long-term storage of such parasite lines. As a first step in meeting this goal, transgenic mf isolated from the animals described above were frozen in liquid nitrogen and stored for a period of 2 weeks. The aliquots were then removed from the liquid nitrogen, thawed and the viability of the parasites assessed by microscopic examination. The mf were then cultured for 48 h and the culture medium assayed for GLuc activity. Approximately 95% of the cryopreserved mf were viable upon thawing and the revived mf secreted GLuc activity into their tissue culture medium (Fig. 9). This activity was eliminated by pre-incubation of the culture medium with the anti-GLuc antibody (Fig. 9). Aliquots of transfected and non-transfected mf were also thawed and fed to mosquitoes to determine whether the transfection process had impaired the ability of the cryporeserved mf to develop normally. L3s were recovered from mosquitoes fed upon the non-transfected mf at a rate of 8.5 L3s per 100 surviving mosquitoes, while L3s were recovered from mosquitoes fed upon the transfected mf at a rate of 8.9 L3s per 100 surviving mosquitoes, suggesting that transfected and non-transfected cryopreserved mf were equally developmentally competent.
Fig. 9.
Gaussia princeps luciferase (GLuc) activity secreted by cryopreserved transgenic microfilariae (mf). Cryopreserved Non-Transfected mf = activity from media in which cryopreserved and thawed non-transgenic mf were cultured. Cryopreserved transfected mf = activity from media in which cryopreserved and thawed transgenic mf were cultured. Cryopreserved Transfected mf + antibody (Ab) activity from media in which cryopreserved and thawed transgenic mf were cultured that was pre-incubated with anti-GLuc Ab.
4. Discussion
The data presented above suggest that B. malayi L3s can be induced to take up transgenic sequences, and that these do not affect the ability of the L3s to develop into the adult stage. The transgenic sequences appear to be inherited by progeny mf produced by these adult parasites and are detectable in L3s developing from these mf. However, this process appears to be much less efficient than biolistic introduction of transgenic sequences into isolated embryos. For example, assays of embryos biolistically transfected with pBmHSP70/GLuc secreted levels of GLuc into the culture medium which were approximately 700 times the background level. In contrast, the maximum amount of GLuc secreted by the chemically transfected intact parasites was only five times background. There are several possible explanations for this difference. First, it is possible that the chemical transfection process introduces far fewer copies of the transgenic DNA into each parasite cell than does biolistic transfection. It would be reasonable to expect that the level of reporter gene activity present would be at least in part dependent upon the number of gene copies present in each cell. Second, it is possible that only a small proportion of the cells in the L3s are transformed by chemical methods. Third, it is possible that only a small proportion of the parasites themselves take up or inherit the exogenous DNA. The latter is perhaps less likely than the others, as the data presented above suggest that approximately 60% of the individual adult parasites assayed appeared to secrete GLuc into their tissue culture medium, suggesting that quite a large proportion of the L3s had taken up the exogenous DNA.
It is possible that changes in the co-injection procedure might result in an increase in the efficiency of the in vivo transfection method. For example, alternative chemical or biological methods of transfection may be more efficient than the calcium-phosphate - DNA coprecipitates reported here. For example, lipofection with DNA-containing micelles might prove to be effective in transfecting molting L3s. Similarly, pantropic lentiviral or retroviral systems might also increase the efficiency of the transfection process. Such pantropic viral systems have been successfully used to transfect trematodes (Kines et al., 2006). Finally, increasing the effective concentration of the calcium-phosphate - DNA co-precipitates using microchambers (Abraham et al., 1993) to confine the L3s in the peritoneal cavity of the jird might also increase the efficiency of the transfection process.
The studies presented above demonstrate that GLuc can be used as a sensitive reporter to non-destructively identify transfected B. malayi. However, GLuc cannot be considered an efficient selectable marker for transformants, as detection of GLuc activity requires incubation of the individual parasites in culture for a relatively long period (at least 48 h). Suitable selectable markers would include those that permit positive selection of transfected parasites by survival (e.g. drug resistance) or by direct inspection (either manual or automated) of transfected parasites (e.g. fluorescent proteins (Higazi et al., 2002; Lok and Massey, 2002)).
The data presented above provide no indication concerning the form in which the transgenic sequences are maintained in B. malayi. However, in many other nematodes, including Caenorhabditis elegans, transgenic sequences are often maintained as large extrachromosomal arrays (Mello et al., 1991). These arrays, while heritable, are not inherited in a Mendelian fashion. Transgenic methods that result in stable integration of the transgenic sequences into the nuclear genome are therefore desirable for the production of stable parasite lines. Several techniques have been reported to encourage integration of transgenic sequences, including the use of heterologous transposons as vectors (Morales et al., 2007) pseudotyped retroviruses (Kines et al., 2008) and including homologous genomic DNA as a carrier during transfection (Schlager et al., 2009). Utilizing the in vivo transfection method described here in conjunction with one of these systems to develop an integrative transfection method is a logical next step in the effort to develop stable transgenic lines of B. malayi.
Supplementary Material
Acknowledgements
We gratefully acknowledge the assistance of T.V. Rajan (University of Connecticut, USA), who provided us with invaluable insights into the biology of B. malayi and the molting process. We also thank Dr. Naomi Lang-Unnasch for critically reading the manuscript. Parasite material was provided by the Filariasis Research Reagent Resource Center (FR3) at the University of Georgia, USA, under a contract from the National Institute of Allergy and Infectious Diseases, USA. This work was supported by a grant from the National Institutes of Health, USA to TRU (project #R01AI048562).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: Nucleotide sequence data reported in this paper are available in the GenBank™ database under the accession number HQ388295.
Note: Supplementary data is associated with this article
References
- Abraham D, Lange AM, Yutanawiboonchai W, Trpis M, Dickerson JW, Swenson B, Eberhard ML. Survival and development of larval Onchocerca volvulus in diffusion chambers implanted in primate and rodent hosts. J Parasitol. 1993;79:571–582. [PubMed] [Google Scholar]
- African Programme for Onchocerciasis Control Final communiqué of the 11th session of the Joint Action Forum (JAF) of African Programme for Onchocerciasis Control (APOC); Paris, France. 6–9 December 2005; APOC, Ouagadougou, Burkina Faso; 2005. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cheng G, Davis RE. An improved and secreted luciferase reporter for schistosomes. Mol. Biochem. Parasitol. 2007;155:167–171. doi: 10.1016/j.molbiopara.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira AD, Katholi CR, Unnasch TR. Characterization of the promoter of the Brugia malayi 12kDa small subunit ribosomal protein (RPS12) gene. Int. J. Parasitol. 2008;38:1111–1119. doi: 10.1016/j.ijpara.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham DA, Fletcher C. The cat infected with Brugia pahangi as a model of human filariasis. Ciba Found. Symp. 1987;127:225–235. doi: 10.1002/9780470513446.ch15. [DOI] [PubMed] [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, Angiuoli SV, Creasy T, Amedeo P, Haas B, El-Sayed NM, Wortman JR, Feldblyum T, Tallon L, Schatz M, Shumway M, Koo H, Salzberg SL, Schobel S, Pertea M, Pop M, White O, Barton GJ, Carlow CK, Crawford MJ, Daub J, Dimmic MW, Estes CF, Foster JM, Ganatra M, Gregory WF, Johnson NM, Jin J, Komuniecki R, Korf I, Kumar S, Laney S, Li BW, Li W, Lindblom TH, Lustigman S, Ma D, Maina CV, Martin DM, McCarter JP, McReynolds L, Mitreva M, Nutman TB, Parkinson J, Peregrin-Alvarez JM, Poole C, Ren Q, Saunders L, Sluder AE, Smith K, Stanke M, Unnasch TR, Ware J, Wei AD, Weil G, Williams DJ, Zhang Y, Williams SA, Fraser-Liggett C, Slatko B, Blaxter ML, Scott AL. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higazi TB, Merriweather A, Shu L, Davis R, Unnasch TR. Brugia malayi: Transient transfection by microinjection and particle bombardment. Exp. Parasitol. 2002;100:95–102. doi: 10.1016/S0014-4894(02)00004-8. [DOI] [PubMed] [Google Scholar]
- Higazi TB, Unnasch TR. Intron encoded sequences necessary for trans splicing in transiently transfected Brugia malayi. Mol. Biochem. Parasitol. 2004;137:181–184. doi: 10.1016/j.molbiopara.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Higazi TB, DeOliveira A, Katholi CR, Shu L, Barchue J, Lisanby M, Unnasch TR. Identification of elements essential for transcription in Brugia malayi promoters. J. Mol. Biol. 2005;353:1–13. doi: 10.1016/j.jmb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Katholi CR, Toé L, Merriweather A, Unnasch TR. Determining the prevalence of Onchocerca volvulus infection in vector populations by PCR screening of pools of black flies. J. Infect. Dis. 1995;172:1414–1417. doi: 10.1093/infdis/172.5.1414. [DOI] [PubMed] [Google Scholar]
- Kines KJ, Mann VH, Morales ME, Shelby BD, Kalinna BH, Gobert GN, Chirgwin SR, Brindley PJ. Transduction of Schistosoma mansoni by vesicular stomatitis virus glycoprotein-pseudotyped Moloney murine leukemia retrovirus. Exp. Parasitol. 2006;112:209–220. doi: 10.1016/j.exppara.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Kines KJ, Morales ME, Mann VH, Gobert GN, Brindley PJ. Integration of reporter transgenes into Schistosoma mansoni chromosomes mediated by pseudotyped murine leukemia virus. FASEB J. 2008;22:2936–2948. doi: 10.1096/fj.08-108308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Massey HC, Jr., Nolan TJ, Schad GA, Kraus K, Sundaram M, Lok JB. Successful transgenesis of the parasitic nematode Strongyloides stercoralis requires endogenous non-coding control elements. Int. J. Parasitol. 2006;36:671–679. doi: 10.1016/j.ijpara.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Liu C, de Oliveira A, Higazi TB, Ghedin E, DePasse J, Unnasch TR. Sequences necessary for trans-splicing in transiently transfected Brugia malayi. Mol. Biochem. Parasitol. 2007;156:62–73. doi: 10.1016/j.molbiopara.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chauhan C, Katholi CR, Unnasch TR. The splice leader addition domain represents an essential conserved motif for heterologous gene expression in B. malayi. Mol. Biochem. Parasitol. 2009;166:15–21. doi: 10.1016/j.molbiopara.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chauhan C, Unnasch TR. The role of local secondary structure in the function of the trans splicing motif of Brugia malayi. Mol. Biochem. Parasitol. 2010;169:115–119. doi: 10.1016/j.molbiopara.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok JB, Massey HC., Jr. Transgene expression in Strongyloides stercoralis following gonadal microinjection of DNA constructs. Mol. Biochem. Parasitol. 2002;119:279–284. doi: 10.1016/s0166-6851(01)00414-5. [DOI] [PubMed] [Google Scholar]
- Mak JW, Choong MF, Lam PL, Suresh K. Experimental infection of the leaf-monkeys, Presbytis cristata and Presbytis melalophos with subperiodic Brugia malayi. Acta Trop. 1990;47:223–226. doi: 10.1016/0001-706x(90)90013-p. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales ME, Mann VH, Kines KJ, Gobert GN, Fraser MJ, Jr., Kalinna BH, Correnti JM, Pearce EJ, Brindley PJ. piggyBac transposon mediated transgenesis of the human blood fluke, Schistosoma mansoni. FASEB J. 2007;21:3479–3489. doi: 10.1096/fj.07-8726com. [DOI] [PubMed] [Google Scholar]
- Nelson FK, Greiner DL, Shultz LD, Rajan TV. The immunodeficient SCID mouse as a model for human lymphatic filariasis. J. Exp. Med. 1991;173:659–663. doi: 10.1084/jem.173.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan TV, Paciorkowski N, Kalajzic I, McGuiness C. Ascorbic acid is a requirement for the morphogenesis of the human filarial parasite Brugia malayi. J. Parasitol. 2003;89:868–870. doi: 10.1645/GE-3137RN. [DOI] [PubMed] [Google Scholar]
- Schlager B, Wang X, Braach G, Sommer RJ. Molecular cloning of a dominant roller mutant and establishment of DNA-mediated transformation in the nematode Pristionchus pacificus. Genesis. 2009;47:300–304. doi: 10.1002/dvg.20499. [DOI] [PubMed] [Google Scholar]
- Shu L, Katholi C, Higazi T, Unnasch TR. Analysis of the Brugia malayi HSP70 promoter using a homologous transient transfection system. Mol. Biochem. Parasitol. 2003;128:67–75. doi: 10.1016/s0166-6851(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Tzertzinis G, Egana AL, Palli SR, Robinson-Rechavi M, Gissendanner CR, Liu C, Unnasch TR, Maina CV. Molecular evidence for a functional ecdysone signaling system in Brugia malayi. PLOS NTD. 2010;4:e625. doi: 10.1371/journal.pntd.0000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren KS, Mahmoud AAF. Tropical and Geographical Medicine. Mc Graw-Hill; New York: 1985. [Google Scholar]
- Weil GJ, Li BW, Liftis F, Chandrashekar R. Brugia malayi: antibody responses to larval antigens in infected and immunized jirds. Exp. Parasitol. 1992;74:315–323. doi: 10.1016/0014-4894(92)90155-4. [DOI] [PubMed] [Google Scholar]
- Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO, Tannous BA. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5:171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.