Abstract
PSF (gene name SFPQ) is a member of a small family of proteins with dual functions in RNA biogenesis and DNA repair. PSF and PSF-containing complexes stimulate double-strand break repair in cell free systems, most likely via direct interaction with the repair substrate. Prior in vitro studies are, however, insufficient to demonstrate whether PSF contributes to DNA repair in living cells. Here, we investigate the effect of miRNA-mediated PSF knockdown in human (HeLa) cells. We find that PSF is essential for reproductive viability. To circumvent this and investigate the DNA damage sensitivity phenotype, we established a genetic rescue assay based on co-transfection of PSF miRNA and mutant PSF expression constructs. Mutational analysis suggests that sequences required for viability and radioresistance are partially separable, and that the latter requires a unique N-terminal PSF domain. As an independent means to investigate PSF sequences involved in DNA repair, we established an assay based on real-time relocalization of PSF-containing complexes to sites of dense, laser-induced DNA damage in living cells. We show that relocalization is driven by sequences in PSF, rather than its dimerization partner, p54nrb/NONO, and that sequences required for relocalization reside in the same N-terminal domain that contributes to radioresistance. Further evidence for the importance of PSF sequences in mediating relocalization is provided by observations that PSF promotes relocalization of a third protein, PSPC1, under conditions where p54nrb is limiting. Together, these observations support the model derived from prior biochemical studies that PSF influences repair via direct, local, interaction with the DNA substrate.
Keywords: PSF, Radiation sensitivity, Double-strand break repair, Laser-induced DNA damage
1. Introduction
Three human proteins, polypyrimidine tract binding protein-associated splicing factor (PSF/SFPQ), 54 kDa RNA binding protein (p54nrb/NONO), and Paraspeckle Component 1 (PSPC1) make up the Drosophila behavior human splicing (DBHS) family. All three share tandem RNA recognition motifs flanked by an additional region of homology. The additional region of homology, which is predicted to form a coiled-coil domain, promotes formation of heteromeric complexes between each protein and the other two family members [1-3].
All three mammalian DBHS proteins are components of nuclear paraspeckles. Paraspeckles regulate gene expression via retention of adenosine-to-inosine hyperedited mRNAs [4,5], reviewed in [6]). In addition to their role in controlling expression of hyperedited mRNAs, PSF and p54nrb have been reported to participate in a number of other processes relating to mRNA biogenesis, including pre-mRNA 3′ end formation, cyclic AMP signaling [7], and nuclear receptor-dependent transcriptional regulation [8-11]. Interestingly, PSF and p54nrb interact with DNA as well as with RNA. More than 15 years ago, Busch and coworkers purified and characterized a DNA-binding heterodimer of proteins that migrated at 52 kDa and 100 kDa in SDS-PAGE, almost certainly corresponding to the polypeptides later identified as p54nrb and PSF, respectively [12]. Subsequent work has shown that PSF binds directly to DNA, that it accelerates annealing of complementary single-stranded nucleic acids, and that it promotes invasion of supercoiled DNA by complementary oligonucleotides to form D-loops [13,14].
PSF binds directly to the homologous recombination protein, Rad51, and cooperates with it in DNA pairing and strand displacement assays [15]. A different PSF•p54nrb complex promotes nonhomologous end joining in vitro, suggesting its involvement in this other main pathway of DNA double-strand break (DSB) repair in vertebrates [16,17]. A recent report indicates that PSF and p54nrb are transiently recruited to DNA damage sites in human cells, and that release from these sites is regulated by a third protein, matrin 3 [18].
There are many proteins with dual functions in mRNA biogenesis and DNA repair. Examples include the TFIIH complex, which participates both in RNA polymerase II-mediated transcriptional initiation and in nucleotide excision repair (reviewed in [19]), PARP-1, which functions in both promoter/enhancer regulation and DNA single-strand break/base excision repair (reviewed in [20,21]), and Ku protein, which functions in both the control of mRNA expression and in the nonhomologous end joining (NHEJ) pathway of DNA double-strand break (DSB) repair (for examples, [22,23], for reviews [24,25]). It appears that PSF and p54nrb provide yet another example of this phenomenon.
Until recently, experimental support for the role of PSF and p54nrb in DNA repair has derived principally from in vitro studies. We recently reported, however, that attenuation of p54nrb expression in mammalian cells delays DSB repair and sensitizes the cells to ionizing radiation [26], which provides genetic evidence in support of the role of p54nrb in DNA repair in vivo. Here we describe another, complementary set of experiments that implicate the PSF subunit in DNA repair in vivo in human cells. We find that PSF is essential in human cells, which necessitated a different approach toward establishing its function, using a genetic rescue assay. We find that the sequences required to rescue the radiosensitive phenotype correlate with those required for relocalization of PSF-containing protein complexes to sites of laser-induced DNA damage and with a previously defined DNA binding domain [27,28].
2. Materials and Methods
2.1 Cells and Immunostaining
HeLa cells were cultured at 37°C in Dulbecco's Minimal Essential Medium supplemented with 10% FBS, 2 mM glutamine and antibiotics. Plasmid DNAs were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen). Fixation and Immunostaining were performed as described in Supplementary Material.
2.2 Expression clones
Wild type and mutant human PSF, p54nrb, and PSPC1 cDNAs were amplified by PCR and inserted in pDsRed-Monomer-N1 or pAcGFP-N1 (Clontech, Mountain View, CA) as described in Supplementary Material. An alignment highlighting the similarities in the coding sequences of these cDNA is shown in Supplementary Fig. 1. Human Ku80 and XRCC4 were inserted in pENTR/D-TOPO (Invitrogen, Carlsbad, CA). In-frame insertion of mCherry and or EYFP coding sequences was followed by lambda integrase-mediated transfer to pcDNA-DEST40 (Invitrogen).
2.3 miRNA treatment and protein expression detection
The p54nrb miRNA knockdown and control vectors were constructed as previously described [26]. An analogous PSF miRNA knockdown vector was constructed using the oligonucleotides listed in Supplementary Table 1. Plasmids were transfected into HeLa cells and proteins were extracted and analyzed by immunoblotting using anti-p54nrb mouse monoclonal antibody (Clone 3, BD Biosciences Pharmingen), anti-PSF mouse monoclonal antibody (Sigma, clone B92), or anti-actin rabbit antibody (BD Bioscences Pharmingen). Blots were developed using HRP-conjugated anti-mouse IgG antibody.
2.4 Phenotypic rescue of PSF-deficient cells
HeLa cells were co-transfected with PSF miRNA and with miRNA-resistant rescue plasmids expressing wild type or mutant PSF. After 24 h, cells were treated with spectinomycin (to select for miRNA expression) and G418 (to select for PSF rescue construct expression). After a further 24 h, 500 cells were seeded in 6 well plates. Some plates were exposed to 133Cs γ-radiation (4 Gy). Plates were incubated for 10 d to allow colony growth, fixed with 80% ethanol for 15 min, and stained with 0.4% trypan blue. Colonies of >50 cells were counted, and surviving fraction was calculated relative to non-irradiated controls. Data are from two independent experiments.
2.5 Laser irradiation and imaging
Microirradiation was performed with a pulsed Ti:Sapphire laser (800 nm, Mira 900, Coherent Inc.). Laser power was optimized to produce localized DNA damage with minimal background outside the microbeam (Supplementary Fig. 2). Live-cell images were collected using a LSM510 META confocal microscope equipped with 543 nm HeNe and multi-line (458, 488, and 514 nm) argon lasers and a 40X C-Apochromat 1.2 N.A. water immersion lens (Carl Zeiss MicroImaging, Inc.). A heated stage (Carl Zeiss MicroImaging, Inc.) and an objective lens heater (Physitemp) were used to maintain cells at 37 °C. Fixed-cell images were collected using an Olympus BX60 microscope equipped with a 40X UPlanFl 0.75 N.A lens.
For experiments with fixed cells, HeLa cells were grown on glass coverslips. After irradiation, cells were washed, fixed in 4% paraformaldehyde, 0.5% Triton X-100 for 10 min, washed again, and incubated in blocking solution (15% Goat serum, 0.2% Fish skin gelatin (Sigma) and 0.03% NaN3 in PBS) containing 0.5% Triton X-100 for 10 min. Cells were rinsed and incubated with blocking solution without Triton X-100. Immunostaining was performed by incubating with rabbit anti-p54nrb (Abcam, Cambridge MA), mouse monoclonal anti-PSF (Sigma, clone B92), rabbit anti-53BP1 (Novus Biologicals, Littleton Co) or mouse monoclonal anti γ-H2AX (Millipore) in blocking solution for 2 hr at room temperature, washing, and incubating for 1.5 hr with Alexa Fluor 350 or AlexaFluor 488-conjugated species specific secondary antibodies in blocking solution. After washing, a TUNEL reaction was performed using TAMRA-dUTP (Roche) and terminal deoxynucleotidyl transferase according to the manufacturer's protocol.
3. Results
3.1 PSF is required for reproductive viability and contributes to radioresistance in human cells
Prior work has shown that PSF stimulates homologous recombination and nonhomologous end joining reactions in cell-free systems. To determine whether PSF contributes to radioresistance in vivo, we designed a miRNA targeted to the sequences in the 3′ untranslated region of PSF mRNA (Supplementary Table 1). Constructs expressing PSF miRNA or a previously described p54nrb miRNA [26] were transfected into HeLa cells. Transfection efficiency was >80% based on expression of a linked EmGFP transfection marker at 72 h post-transfection (data not shown). The miRNAs reduced PSF expression by about 90% and p54nrb by about 70%, using actin as an internal standard (Fig 1A, 1B). The effect was selective: PSF miRNA had little or no effect on p54nrb expression and vice versa.
Fig. 1.
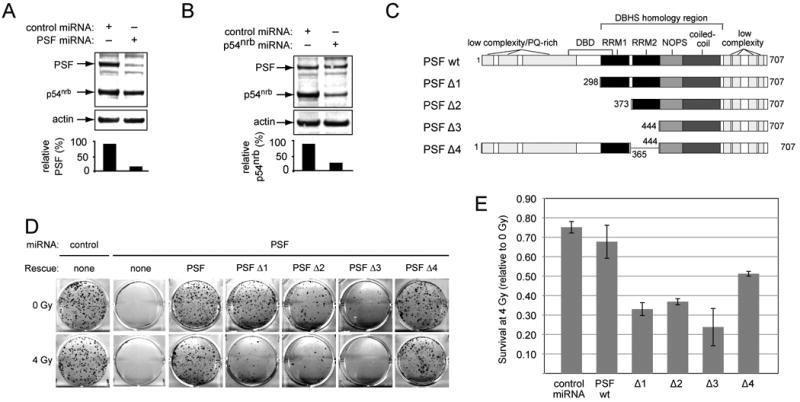
Phenotypic rescue of HeLa cells transfected with PSF miRNA. (A,B) Efficacy of p54nrb and PSF knockdown. HeLa cells were transfected with plasmids expressing hybrid RNAs encoding linked EmGFP and indicated miRNA genes. Extracts were analyzed by immunoblotting for PSF, p54nrb, and β-actin (arrows). Graphs show expression normalized to actin. Bands above and below PSF most likely reflect nonspecific cross-reactivity of antibody with unrelated proteins present in the crude extract. (C) Map of PSF and PSF mutants, showing low complexity regions, two RNA Recognition Motif (RRM) domains, NOPS domain, and coiled-coil domain. Domain boundaries are based on information from the Uniprot, Pfam, Interpro, or SMART databases, except for DNA binding domain (DBD) [27,28] and DHBS homology region (reviewed in [6]. Mutants were created as described in Materials and Methods. (D) Phenotypic rescue. HeLa cells were co-transfected with plasmids expressing indicated miRNAs and expression vectors for wild type or mutant PSF. After 48 h, 500 cells per well from each group were plated in 6 well plates and treated with 0 Gy or 4 Gy of γ-radiation. Plates were incubated for 10 d, fixed, stained, and colonies of >50 cells were counted. (E) Relative survival. Colony numbers from two independent experiments were normalized to survival in the absence of radiation. Error bars indicate range of duplicate observations.
Ideally, radioresistance may be analyzed using stable knockdown cell lines, as we have previously done for p54nrb [26]. In practice, however, we were not able to establish stable human knockdown cell lines using PSF miRNA, consistent with previous suggestions that attenuation of PSF expression may be deleterious in human cells [11]. We therefore established a rescue assay in which PSF miRNA and PSF expression constructs were co-transfected into HeLa cells, which were scored for clonogenic survival, with or without radiation treatment. Rescue constructs lacked the PSF 3′ untranslated region, and so were miRNA resistant. There were no surviving colonies in cells transfected with PSF miRNA alone, whereas co-transfection with a wild-type PSF expression plasmid rescued survival to nearly the same level as with control miRNA (Fig. 1D).
To map the sequences required for rescue of survival, we created a series of deletion mutants lacking progressively increasing amounts of N-terminal sequence (Fig. 1C). PSF has a unique N-terminal domain that is not shared with the other two family members, and that has no evident similarity to other known proteins. In addition, the RRM domains are moderately divergent among family members (66% and 73% identity for RRM1 and RRM2, respectively.) These are therefore candidates to mediate PSF-specific biological activities. We verified the mutant constructs by coupled in vitro transcription-translation, which indicated that all were expressed as polypeptides of expected size (Supplementary Fig. 2A). Moreover, all of the mutants except PSFΔ3 co-immunoprecipitated with p54nrb, albeit at somewhat less than wild-type efficiency (Supplementary Fig. 2B). These results differ somewhat from a prior report that any deletion of PSF sequences ablated the p54nrb interaction, a discrepancy perhaps explained by different methodology (the prior report used GST fusion proteins [1]).
In the absence of radiation treatment, PSFΔ1 and PSFΔ4 restored colony formation to approximately 60 to 80% of values seen with full-length PSF, indicating that the presence of two of the three domains in this region (i.e., two RRMs, or one RRM plus the N-terminal domain) are minimally sufficient to rescue cell viability (Fig. 1D). More severely deleted mutants had much less rescue activity (i.e., PSFΔ2, which retains one of three domains in this region, and PSFΔ3, which retains none).
Because of the putative involvement of PSF in cellular DSB repair, it was of interest to test whether any of the mutants had a radiosensitive phenotype. We compared survival in nonirradiated vs. irradiated (4 Gy) groups. Data were plotted after normalizing to survival in the absence of radiation (Fig. 1E). Cells rescued with PSFΔ1, PSFΔ2, and PSFΔ3 were clearly more radiosensitive than those rescued wild type PSF, with only about half the relative colony formation at 4 Gy. Restoration of the N-terminal and RRM1 sequences in PSFΔ4 restored partial radioresistance. Results suggest that sequences required for radioresistance reside in the N-terminal domain and possibly in RRM1. These overlap with a previously defined DNA binding domain in PSF [27,28].
3.2 Relocalization of PSF and p54nrb to sites of laser-induced DNA damage
As an independent means to investigate PSF sequences involve in DNA repair, we established an assay based on real-time measurement of relocalization to sites of DNA damage. HeLa cells were microirradiated with a pulsed 800 nm Ti:Sapphire laser microbeam [29]. Cells were fixed and stained by TUNEL assay, which detects broken DNA ends, and with antibodies to the DSB markers, γ-H2AX and 53BP1 (Fig. 2A). A distinct stripe of coincident TUNEL, anti-γ-H2AX, and anti-53BP1 staining was seen (Fig. 2A). A similar experiment was then performed using anti-PSF and anti-p54nrb immunostaining. Coincident TUNEL, anti-PSF, and weak anti-p54nrb staining was observed, suggesting that endogenous PSF and p54nrb migrate to damaged regions (Fig. 2B).
Fig. 2.
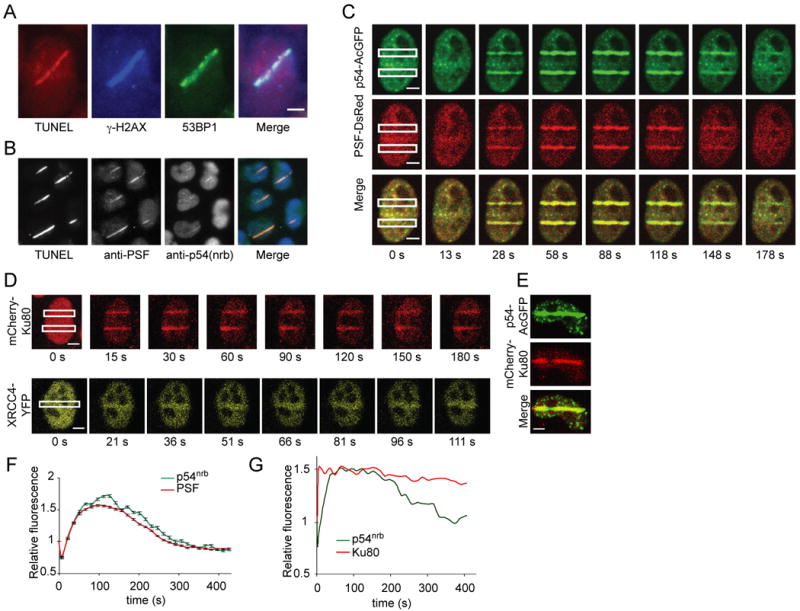
Relocalization of PSF and p54nrb to sites of laser-induced DNA damage. Stripes of DNA damage were induced by laser microirradiation of HeLa cells. Scale bars, 5 μm. (A) DSB formation. Cells were fixed 10 min after irradiation and stained by a TUNEL procedure or with indicated antibodies. (B) Endogenous PSF and p54nrb. Cells were treated and stained as in A except with indicated antibodies. Monochrome images of each channel are shown, together with a merged overlay. (C-E) Live-cell imaging. Cells were transfected with indicated fluorescent protein expression constructs, and after 24 h were subjected to laser microirradiation. Images were collected at indicated intervals. (F) Quantification of PSF and p54nrb. Cells were transfected and treated as in C, and fluorescence intensity in damaged region was measured. Data shown are average of 10 nuclei in the same experiment (G) Quantification of p54nrb and Ku80. Cells were transfected and treated as in (E). Data shown are representative of results obtained in several independent experiments.
To define the time course of PSF and p54nrb accumulation, we performed live cell imaging with HeLa cells expressing fluorescently-tagged proteins. The tagged proteins provide much better signal-to-noise ratio than was seen by immunostaining of endogenous proteins in Fig. 2B. Prior to laser irradiation, fluorescently-tagged PSF and p54nrb were distributed in the nucleoplasm, and to some extent, in discrete foci. The foci were more evident with p54nrb than with PSF, and their number and appearance was consistent with the 13-17 paraspeckles that are expected to be present in HeLa cells [30]. Following irradiation, both proteins rapidly migrated to the damaged region (Fig. 2C). PSF and p54nrb fluorescence were coincident. Two known NHEJ proteins, Ku80 and XRCC4, also migrate to the damaged region (Fig. 2D), and the distributions of Ku80 and p54nrb were spatially coincident (Fig. 2E). AcGFP and DsRed alone showed no relocalization (Supplementary Fig. 3).
The time course of PSF and p54nrb accumulation was identical, but differed from that of Ku (Fig. 2F, 2G). PSF and p54nrb reached a maximum at about 100 s, then slowly declined. Transient association of PSF and p54nrb with DNA damage sites observed here is generally consistent with the time course reported in human U2OS cells using a different laser system [18]. In contrast to the transient association of PSF and p54nrb, Ku reached a maximum within 5 s and remained stable for the ∼7 min duration of the experiment, which is again consistent with previous reports [29,31]. This difference suggests that recruitment of Ku and PSF•p54nrb may be regulated by different mechanisms. that these proteins act at different stages of the DSB repair process, or that they may occupy different locations in the chromatin.
3.3 Sequences required for relocalization of PSF• p54nrb complex
To gain insight into the mechanism of relocalization, we investigated the sequences that were required. We co-transfected HeLa cells with PSF or p54nrb miRNAs and fluorescently-tagged PSF or p54nrb transgenes. The p54nrb miRNA had no effect on relocalization of tagged PSF, whereas the PSF miRNA severely attenuated p54nrb relocalization (Fig. 3A, 3B). These data indicate that sequences in PSF, rather than p54nrb, are responsible for recruitment of the PSF•p54nrb complex to damage sites.
Fig. 3.
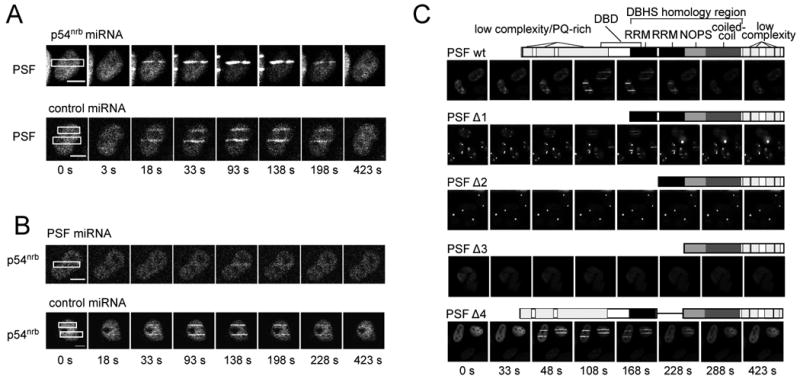
Requirement of PSF N-terminal sequences for relocalization of p54nrb to sites of DNA damage. (A, B) HeLa cells were transfected with miRNA-expressing plasmids and transfected again at 24 h with tagged protein expression vectors as indicated. Laser microirradiation and imaging were performed as in Fig. 2C and 2D. Scale bar is 10 μm. (C) Analysis of PSF mutants. Schematic diagrams are as in Fig. 1. Transfection, laser microirradiation and imaging were performed as in Fig. 2. All constructs are PSF-AcGFP fusions except PSF Δ3, which is PSF-dsRed.
We next sought to identify specific sequences in PSF that mediate relocalization of PSF-containing protein complexes. We expressed each mutant as a fluorescent protein fusion in HeLa cells. The total expression level of each PSF derivative was similar except for PSFΔ3, which appeared to be reduced. Deletion of the N-terminal domain and RRM1 (in PSFΔ1 and PSFΔ2) progressively reduced the ability to relocalize to sites of laser-induced DNA damage (Fig. 3C). Conversely, a mutant (PSFΔ4) that retained the N-terminal domain and RRM1, but was missing RRM2, relocalized to same extent as full-length PSF. Results define a region encompassing residues 1-369 as required for relocalization, coincident with the region required for genetic rescue of the radiosensitive phenotype and overlapping the previously defined DNA binding domain [27,28].
When sequences required for relocalization were partially or fully deleted (in PSFΔ1 and PSFΔ2), but RRM2 was present, the mutant PSF spontaneously accumulated in one or two bright foci. These foci, which are quite different in appearance than paraspeckles, were not seen the mutants lacking RRM2 (PSFΔ3, PSFΔ4). We hypothesize that deletion of an autoinhibitory sequence in the N-terminal region enhances the ability of RRM2 to interact with another partner. In this respect, it is of interest that RRM2 has previously been shown to interact with a transcriptional regulatory protein, FHL2 [32].
3.4 PSF mediates relocalization of PSPC1 when the ratio of PSF to p54nrb expression is altered
To further confirm the role of PSF sequences in relocalization, we investigated the ability of PSF to direct relocalization of the third family member, PSPC1. To confirm that PSF and PSPC1 are capable of interaction in vivo, we performed co-immunoprecipitation experiments using protein extracts of HeLa cells that were metabolically labeled using 35S-methionine. Consistent with our previous results [16], anti-PSF antibody precipitated a polypeptide corresponding to p54nrb and an additional minor band migrating at a position consistent with the predicted 59 kDa size predicted for PSPC1 (Fig. 4A, lane 3). The amount of the 59 kDa species increased in p54nrb miRNA-treated cells (Fig 4A, compare lanes 3 and 2). The identity of this species as PSPC1 was confirmed in a reciprocal co-immunoprecipitation experiment using anti-PSPC1. PSF was present in the resulting immune complexes and the amount was increased in cells that were pre-treated with p54nrb miRNA (Fig. 4B, compare lanes 2 and 3).
Fig. 4.
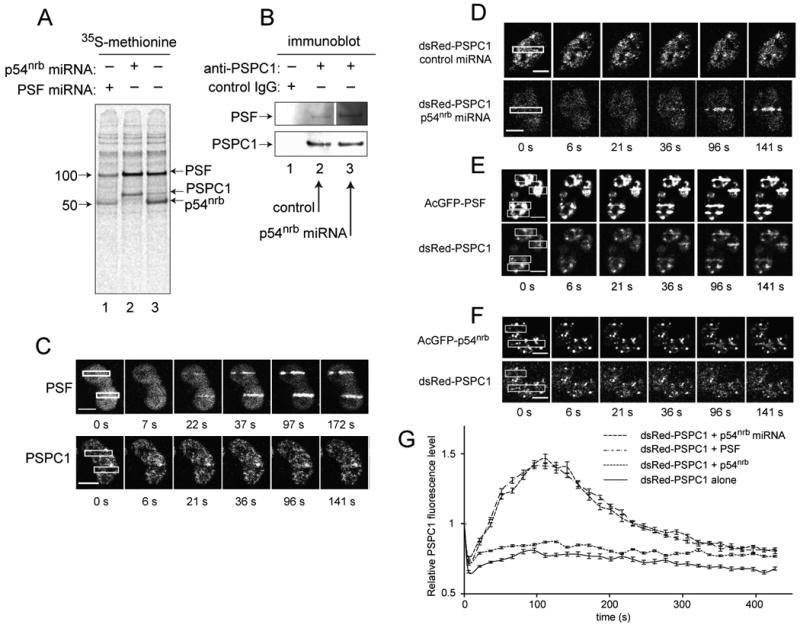
Interaction of PSF with PSPC1 and relocalization of PSF•PSPC1 complex to sites of laser-induced DNA damage. (A) Immunoprecipitation of 35S-methionine-labeled polypeptides with anti-PSF antibody in the presence or absence of p54nrb miRNA. HeLa cells were transfected with indicated miRNA expression vectors, metabolically labeled, and protein extracts immunoprecipitated using anti-PSF antibody. Immune complexes were analyzed by SDS-PAGE and visualized by autoradiography. (B) Immunoprecipitation as in Panel A except that proteins were visualized by immunoblotting. Upper panel, anti-PSF, lower panel, anti-PSPC1. Within each panel, all lanes are from the same exposure of the same gel. (C) Live cell imaging of PSF1 and PSPC1. HeLa cells were transfected with indicated constructs, treated, and analyzed as in Fig 2. Scale bar in all micrographs is 10 μm. (D) Relocalization of PSPC1 in the presence or absence of p54nrb miRNA. (E) Relocalization of PSPC1 in the presence of excess PSF. HeLa cells were co-transfected with plasmids expressing PSF-AcGFP and PSPC1-dsRed, treated, and analyzed as in (C). (F) Relocalization of PSPC1 in the presence of excess p54nrb. (G) Time course of PSPC1 mobilization. Quantification of data in (D), (E), and (F). Average of ten nuclei.
To determine whether PSF was capable of directing the relocalization of PSPC1 to sites of DNA damage, we transfected HeLa cells with fluorescently tagged PSF and PSPC1. In HeLa cells expressing normal levels of p54nrb, PSPC1 showed little or no relocalization (Fig. 4C). By contrast, in HeLa cells transfected with p54nrb miRNA, PSPC1 showed evident relocalization to regions of laser-induced DNA damage (Fig. 4D). To further confirm the role of PSF in directing relocalization of PSPC1, we performed experiments in which PSF or p54nrb was overexpressed by co-transfection with the fluorescently tagged PSPC1. Overexpression of PSF, but not p54nrb, promoted relocalization of PSPC1 to sites of laser-induced DNA damage (Fig. 4E, 4F). We monitored the time course of PSPC1 under each set of conditions (p54nrb depletion or PSF overexpression) and found that it was similar to PSF itself, with peak accumulation at about 120 s and a steep decline by 300 s (Fig. 4G). Together, these data suggest that formation of a PSF•PSPC1 complex in HeLa cells is favored when the PSF:p54nrb ratio is altered (i.e., when p54nrb is attenuated or when PSF is overexpressed). Under these conditions, PSF sequences mediate relocalization of the PSPC1 complex.
4. Discussion
PSF (gene name SFPQ) has long been implicated in DNA repair, based on its interaction with repair proteins and by the ability of the purified protein to stimulate model DNA repair reactions in vitro. The present study presents three novel findings: (i) PSF is essential for reproductive viability of human (HeLa) cells, (ii) genetic rescue of PSF knockdown cells with hypomorphic deletion mutants (i.e., mutants that do not fully rescue clonogenic survival) is associated with a significantly radiosensitive phenotype, and (iii) PSF sequences are responsible for recruitment of PSF-containing complexes to sites of dense DNA damage. Results are consistent with the proposed role of PSF in DNA double-strand break repair in vivo in human cells.
While this paper was in review, another report appeared indicating that attenuation of PSF expression sensitizes mouse cells to DNA damaging agents [33]. The mouse cell results differ in that PSF was not essential for clonogenic viability. This parallels the situation with Ku protein, is also essential for somatic cell viability in human cells but not in rodents [34].
Table I summarizes the relationship between PSF sequences required for dimerization, clonogenic survival, radioresistance, and relocalization. As shown in Supplementary Figure 2, a single RRM (either RRM1 or RRM2) and C-terminal sequences (including the coiled-coil motif) are sufficient for dimerization, consistent with prior reports [8,35]. These sequences alone are not sufficient for the other functions of PSF, which require, in addition, the unique N-terminal domain and possibly RRM1. The finding that PSF (not p54nrb or PSPC1) is responsible for recruitment to regions of damaged DNA is consistent with prior reports that isolated PSF, in the absence of these partners, binds DNA and mediates pairing and Rad51-dependent strand exchange [14,15]. The sequences required for radioresistance and DNA-damage dependent relocalization sequences overlap a previously described DNA binding domain [27,28]. This DNA binding domain has no evident homology to other known proteins, and the details of the PSF-DNA interface have yet to be fully defined.
Table 1. Summary of mutational analysis.
| Constructa | N-terminal | RRM1 | RRM2 | NOPS/coiled-coil | Dimerizationb | Viabilityc | Radioresistanced | Relocalizatione |
|---|---|---|---|---|---|---|---|---|
| PSF wt | • | • | • | • | yes | 100% | yes | yes |
| PSF Δ1 | • | • | • | yes | 58% | no | slight | |
| PSF Δ2 | • | • | yes | 22% | no | no | ||
| PSF Δ3 | • | no | 11% | no | no | |||
| PSF Δ4 | • | • | • | yes | 81% | partial | yes |
Refer to Fig. 1C for schematic
Estimated based on Supplementary Figure 2.
Mean of two independent experiments, Fig. 1 and data not shown
Based on Fig. 1, Panel E
Based on Fig. 3
Our data are consistent with a mechanism of radioresistance that involves physical movement of PSF-containing complexes to DNA damage sites. Prior work indicates that isolated PSF and PSF-containing complexes bind constitutively and nonspecifically to DNA in vitro. The finding that relocalization is DNA damage-dependent in vivo suggests that some additional level of regulation is at work. It could be that DNA damage-induced chromatin decondensation facilitates binding to internal DNA sequences [35]. Alternatively or in addition, interaction with Rad51 or other DNA repair proteins could mediate selective recruitment to DNA damage sites in vivo [15].
Our data do not exclude an alternative possibility that attenuation of PSF expression affects the DNA damage response indirectly, via effects on RNA biogenesis (i.e., by suppressing expression of other DNA damage response genes). A solely indirect mechanism of action seems unlikely, however, for two reasons: (1) it could not account for the in vitro data showing that purified protein enhances end-joining and early steps of homologous recombination in cell-free systems, and (2) a recent report showed that attenuation of PSF expression in mouse cells did not influence levels of the 70 kDa Ku protein subunit or any of a number of other repair proteins that were tested [33].
The apparent dual role of PSF in DNA repair and RNA biogenesis is intriguing. Some evidence indicates that Ku protein itself has dual roles in DNA repair and RNA biogenesis (reviewed in [23]; see [24] for earlier references). PSF undergoes methylation, phosphorylation, and sumoylation within the N-terminal and RRM1 regions [36-39], and it is possible that such modifications might influence the balance between DNA and RNA binding. Additionally, interaction of VL30 retroelement RNA with the PSF•p54nrb complex mediates a switch from a DNA to a RNA binding form [28]. It will be of interest to determine whether there are other endogenous RNAs that mediate this switching behavior, and if so whether therapeutic RNAs might be designed to block DNA interaction and thus sensitize tumor cells to ionizing radiation.
Supplementary Material
Acknowledgments
We thank the Medical College of Georgia Imaging Core Facility and Office of Biostatistics and Bioinformatics for their services. This work was supported by a US Public Health Service grant to WSD (CA98239) and by the NIH Common Fund (EY018244).
Abbreviations
- RRM
RNA recognition motif
- PSF
polypyrimidine tract binding protein-associated splicing factor
- DSB
double-strand break
- DBHS
Drosophila behavior human splicing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peng R, Dye BT, Perez I, Barnard DC, Thompson AB, Patton JG. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA. 2002;8:1334–1347. doi: 10.1017/s1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuwahara S, Ikei A, Taguchi Y, Tabuchi Y, Fujimoto N, Obinata M, Uesugi S, Kurihara Y. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol Reprod. 2006;75:352–359. doi: 10.1095/biolreprod.106.051136. [DOI] [PubMed] [Google Scholar]
- 4.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amelio AL, Miraglia LJ, Conkright JJ, Mercer BA, Batalov S, Cavett V, Orth AP, Busby J, Hogenesch JB, Conkright MD. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci U S A. 2007;104:20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishitani K, Yoshida T, Kitagawa H, Ohta H, Nozawa S, Kato S. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem Biophys Res Commun. 2003;306:660–665. doi: 10.1016/s0006-291x(03)01021-0. [DOI] [PubMed] [Google Scholar]
- 10.Dong X, Sweet J, Challis JR, Brown T, Lye SJ. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol Cell Biol. 2007;27:4863–4875. doi: 10.1128/MCB.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang WW, Zhang LX, Busch RK, Farres J, Busch H. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem J. 1993;290(Pt 1):267–272. doi: 10.1042/bj2900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhmedov AT, Bertrand P, Corteggiani E, Lopez BS. Characterization of two nuclear mammalian homologous DNA-pairing activities that do not require associated exonuclease activity. Proc Natl Acad Sci U S A. 1995;92:1729–1733. doi: 10.1073/pnas.92.5.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhmedov AT, Lopez BS. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000;28:3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morozumi Y, Takizawa Y, Takaku M, Kurumizaka H. Human PSF binds to RAD51 and modulates its homologous-pairing and strand-exchange activities. Nucleic Acids Res. 2009;37:4296–4307. doi: 10.1093/nar/gkp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor•p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 17.Udayakumar D, Bladen CL, Hudson FZ, Dynan WS. Distinct pathways of nonhomologous end joining that are differentially regulated by DNA-dependent protein kinase mediated phosphorylation. J Biol Chem. 2003;278:41631–41635. doi: 10.1074/jbc.M306470200. [DOI] [PubMed] [Google Scholar]
- 18.Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- 19.Beck BD, Hah DS, Lee SH. XPB and XPD between transcription and DNA repair. Adv Exp Med Biol. 2008;637:39–46. doi: 10.1007/978-0-387-09599-8_5. [DOI] [PubMed] [Google Scholar]
- 20.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst) 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Giffin W, Torrance H, Rodda DJ, Prefontaine GG, Pope L, Hache RJ. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 23.Woodard RL, Lee K, Huang J, Dynan WS. Distinct roles for Ku protein in transcriptional reinitiation and DNA repair. J Biol Chem. 2001;276:15423–15433. doi: 10.1074/jbc.M010752200. [DOI] [PubMed] [Google Scholar]
- 24.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Kuhne WW, Kulharya A, Hudson FZ, Ha K, Cao Z, Dynan WS. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009;37:6746–6753. doi: 10.1093/nar/gkp741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urban RJ, Bodenburg YH, Wood TG. NH2 terminus of PTB-associated splicing factor binds to the porcine P450scc IGF-I response element. Am J Physiol Endocrinol Metab. 2002;283:E423–427. doi: 10.1152/ajpendo.00057.2002. [DOI] [PubMed] [Google Scholar]
- 28.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci U S A. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 31.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dye BT, Patton JG. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- 33.Rajesh C, Baker DK, Pierce AJ, Pittman DL. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2010 Sep 2; doi: 10.1093/nar/gkq738. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci U S A. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 37.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 38.Zhong N, Kim CY, Rizzu P, Geula C, Porter DR, Pothos EN, Squitieri F, Heutink P, Xu J. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J Biol Chem. 2006;281:20940–20948. doi: 10.1074/jbc.M601935200. [DOI] [PubMed] [Google Scholar]
- 39.Buxade M, Morrice N, Krebs DL, Proud CG. The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J Biol Chem. 2008;283:57–65. doi: 10.1074/jbc.M705286200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


