Abstract
Cell adhesions to the extracellular matrix (ECM) are necessary for morphogenesis, immunity and wound healing1,2. Focal adhesions are multifunctional organelles that mediate cell–ECM adhesion, force transmission, cytoskeletal regulation and signalling1–3. Focal adhesions consist of a complex network4 of trans-plasma-membrane integrins and cytoplasmic proteins that form a <200-nm plaque5,6 linking the ECM to the actin cytoskeleton. The complexity of focal adhesion composition and dynamics implicate an intricate molecular machine7,8. However, focal adhesion molecular architecture remains unknown. Here we used three-dimensional super-resolution fluorescence microscopy (interferometric photoactivated localization microscopy)9 to map nanoscale protein organization in focal adhesions. Our results reveal that integrins and actin are vertically separated by a ~40-nm focal adhesion core region consisting of multiple protein-specific strata: a membrane-apposed integrin signalling layer containing integrin cytoplasmic tails, focal adhesion kinase and paxillin; an intermediate force-transduction layer containing talin and vinculin; and an uppermost actin-regulatory layer containing zyxin, vasodilator-stimulated phosphoprotein and α-actinin. By localizing amino- and carboxy-terminally tagged talins, we reveal talin’s polarized orientation, indicative of a role in organizing the focal adhesion strata. The composite multilaminar protein architecture provides a molecular blueprint for understanding focal adhesion functions.
Modern understanding of cellular function is founded on the revolution in the 1950s to 1970s in visualizing cellular ultrastructure by electron microscopy10,11. Together with the identification of molecular components and their interactions, this has allowed biophysical mechanistic models for organelles such as the actin and microtubule cytoskeletons or the endomembrane transport machinery12–15. In contrast, although there is a wealth of knowledge on the composition, interactions and dynamics of integrin-based focal adhesions, their ultrastructure remains poorly defined. No discernible protein organization pattern has been observed experimentally, either by immunoelectron microscopy6 or by two-dimensional super-resolution light microscopy16. Thus, it is unclear whether focal adhesions are structurally unorganized, or if the relevant structural organization is in the third dimension. Although many cartoon models of focal adhesion protein organization have been proposed based on in vitro protein–protein interaction data1,2, true spatial architecture at the ultrastructural level has been impossible to infer. Thus, a mechanistic understanding of focal adhesion function has remained elusive.
To define focal adhesion molecular architecture, we sought to map the nanoscale organization of focal adhesion proteins. This capability has recently been enabled by advances in super-resolution light microscopy (reviewed in ref. 17). We used iPALM9, which combines photoactivated localization microscopy18 with simultaneous multi-phase interferometry of photons from each fluorescent molecule, to image a high density of specific fluorescence-tagged molecules with three-dimensional nanoscale resolution (Supplementary Fig. 1). We constructed imaging probes with photoactivatable fluorescent proteins (PA-FP, tandemdimer Eos19 or monomeric Eos220) fused to focal adhesion proteins, and expressed them in human osteosarcoma (U2OS; Figs 1–3 and Supplementary Figs 1–8 and 11–19) or mouse embryonic fibroblast (MEF; Supplementary Fig. 10) cells plated on fibronectin-coated coverglasses. With iPALM, PA-FP brightness allows localization accuracy of typically 20 nm (full-width at half-maximum) or better in lateral (xy) dimensions18, and 10–15 nm in the vertical (z) axis9.
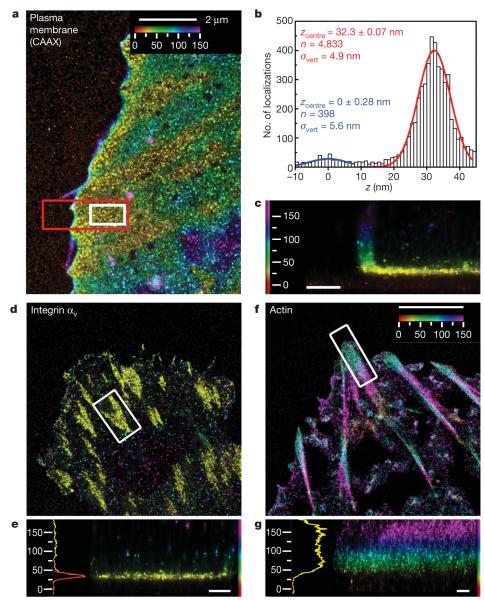
Figure 1. iPALM imaging of a plasma membrane marker, integrin αv and actin.
a–c, Plasma membrane marker CAAX–tdEos. a, Top view; b, histogram and Gaussian fits for the z positions (white box in a) of PA-FP molecules (red) and nonspecific fluorescence adsorbed to substrate (blue); c, side view (red box in a). d, e, Integrin αv–tdEos. d, Top view; e, side view (right), histogram and fits (left). f, g, Actin–mEos2. f, Top view; g, side view (right), histograms and fits (left). The vertical distribution of actin is non-Gaussian, so the focal adhesion peak fit is not shown. Colours in a, c–g indicate the vertical (z) coordinate relative to the substrate (z=0nm, red). Scale bars: 500 nm (c, e, g).
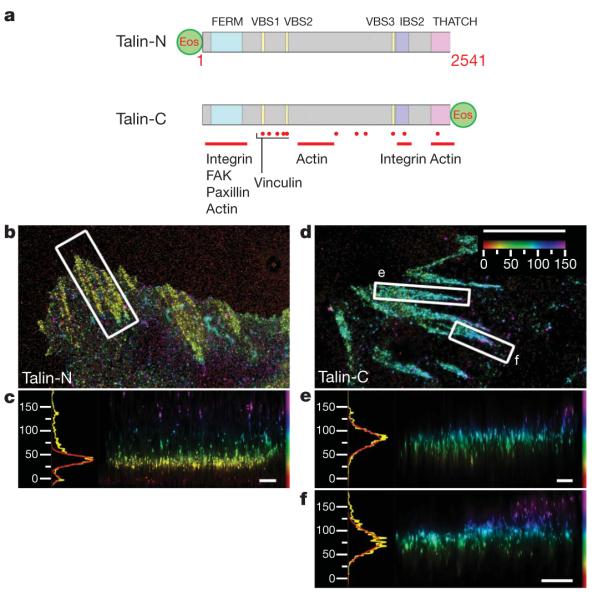
Figure 3. Talin orientation in focal adhesions.
a, Schematic diagram, with important domains and binding sites indicated for Talin PA-FP fusions (FERM, protein 4.1, ezrin, radixin, moesin domain; VBS, vinculin binding sequence; IBS, integrin binding site). Talin-N, N-terminal fusion; Talin-C, C-terminal fusion (Supplementary Table 3). b–f, Top view and side view iPALM images of focal adhesions (white boxes, top-view panels) and corresponding z histograms and fits for talin-N–tdEos (b, c) and talin-C–tdEos (d–f). Colours: vertical (z) coordinate relative to the substrate (z=0nm, red). Scale bars: 5 μm (b, d) and 500 nm (c, e, f).
We first determined the vertical position of the ventral plasma membrane as a reference point for comparative localization with focal adhesion proteins, using PA-FP targeted to the cytoplasmic face of the plasma membrane via fusion with CAAX sequence. The localizations are represented by iPALM rendering (Fig. 1a) with colours indicating the vertical (z) coordinate relative to the coverglass surface (z = 0 nm). Focal adhesions near the cell edge appear as yellow regions where the membrane most closely approaches the substrate, with the ventral plasma membrane contour reflected by the colour gradient. Figure 1c shows the side-view (xz) projection of a focal adhesion area (red box, Fig. 1a), with the leading-edge plasma membrane also apparent. To quantify the localizations in the focal adhesion area (Fig. 1a, white box), vertical coordinate histograms (Fig. 1b) were fitted by a Gaussian with the centre (zcentre) and the width (σvert, the standard deviation of the distribution) shown. The width parameter σvert of ~5 nm demonstrates the spatial resolution, with the positional uncertainty contributed by both the PA-FP brightness limitations18 and the probe size (~4–5 nm for the PA-FP plus a 25-amino-acid linker). The inner plasma membrane zcentre of ~32 nm from the coverglass surface is in good agreement with previous measurements by electron and interference reflection microscopy6.
We next used iPALM to determine the three-dimensional localization of integrin cytoplasmic tails which serve as recruiting sites for focal adhesion proteins. We co-expressed integrin αvPA-FP fusion with untagged integrin β3 to form integrin αvβ3, a fibronectin receptor (Fig. 1d). The vertical position histogram and side-view projection are shown in Fig. 1e. The zcentre and σvert for focal adhesion regions from several cells were quantified, yielding average zcentre=36.8±4.5 nm and average σvert=7.2±1.8 nm (Fig. 4a, b and Supplementary Table 1), indicating a tightly confined integrin αv C-terminal position (Supplementary Fig. 6) close to the inner plasma membrane as expected1,2.
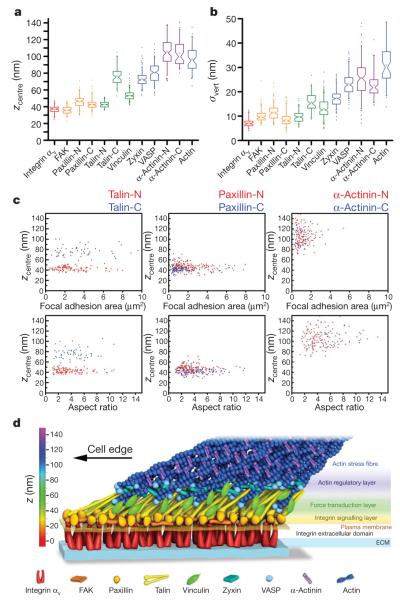
Figure 4. Nanoscale architecture of focal adhesions.
a, b, Peak position (zcentre)(a) and width parameter (σvert)(b) of PA-FP fusions in focal adhesions. Notched boxes, 1st and 3rd quartiles, median and confidence interval; whiskers, 5th and 95th percentiles; +, means, outliers also shown. (See also Supplementary Table 1.) c, zcentre protein positions (nm) versus focal adhesion area (μm2) or aspect ratio for both N- (red) or C- (blue) terminal fusions of talin, paxillin and α-actinin. Each point corresponds to individual focal adhesion measurements. d, Schematic model of focal adhesion molecular architecture, depicting experimentally determined protein positions. Note that the model does not depict protein stoichiometry.
To determine the vertical position of actin filaments which link to focal adhesions at stress fibre termini, we performed iPALM with an actin PA-FP probe and analysed focal adhesion regions near the cell edge. In contrast to the integrin localizations, this revealed a broader (average σvert=31.0±8.7 nm) and significantly higher vertical distribution for actin, peaking at average zcentre of 96.9±15.2 nm (Figs 1f, g and 4a, b and Supplementary Table 1), and which was separated from the plasma membrane by a ~40-nm region containing low actin density. The observed lack of integrin–actin physical overlap in focal adhesions is consistent with the absence of their binding interactions in vitro1,2, and stresses the importance of a ‘focal adhesion core’ domain bridging this gap.
We next sought to determine the nanoscale protein organization within the focal adhesion core domain. We imaged PA-FP fusions of key focal adhesion proteins representing three functional categories: integrin-mediated signalling (focal adhesion kinase (FAK), paxillin); cytoskeletal adaptors (vinculin, zyxin); and actin-regulatory proteins (vasodilator-stimulated phosphoprotein (VASP), α-actinin)1–4. Remarkably, we observed that each protein occupied a distinct and characteristic vertical position within focal adhesions, apparent from their different colours in iPALM images (Fig. 2) and statistics of their vertical localizations (Fig. 4a, b and Supplementary Table 1).
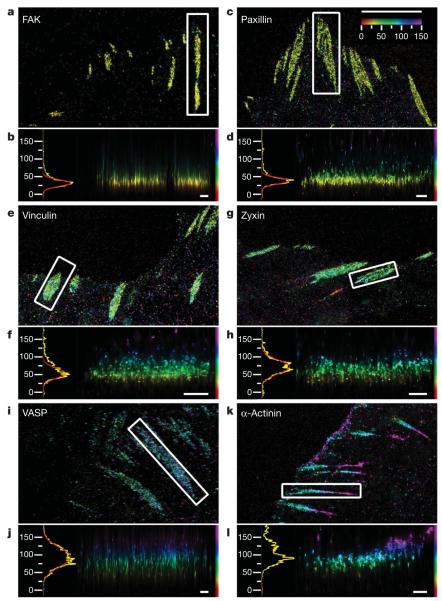
Figure 2. Protein stratification of the focal adhesion core.
Top view and side view iPALM images of focal adhesions (white boxes, top-view panels) and corresponding z histograms and fits. a, b, FAK–tdEos; c, d, paxillin–tdEos; e, f, vinculin–tdEos; g, h, zyxin–mEos2; i, j, VASP–mEos2; k, l, α-actinin–mEos2. The vertical distribution of α-actinin is non-Gaussian, so the focal adhesion peak fit is not shown. Paxillin and α-actinin shown are C-terminal PA-FP-tagged (N-terminal fusions in Supplementary Figs 21 and 22). Colours: vertical (z) coordinate relative to the substrate (z=0nm, red). Scale bars: 5 μm (a, c, e, g, i, k) and 500 nm (b, d, f, h, j, l).
We found that FAK and paxillin were both confined to a narrow plane at average zcentre of 36.0±4.7 nm and 43.1±6.1 nm, with average σvert of 10.0±2.5 nm and 8.6±2.8 nm, respectively (Figs 2a–d and 4a, b). In response to integrin engagement, FAK phosphorylates tyrosine residues on several focal adhesion proteins21 including paxillin, a key focal adhesion adaptor protein22. On the basis of their membrane proximity and signalling-related functions, these proteins may comprise a signalling/adaptor subcompartment of the focal adhesion core, organized by clustered integrins.
In contrast to the membrane-apposed position of integrin signalling proteins, other proteins were localized to distinctly higher vertical positions. The peak of vinculin distribution (average zcentre=53.7±5.5 nm, σvert=13.1±3.9 nm) coincided with the lower boundary of actin density (Figs 1g and 2f and Supplementary Figs 7, 8 and 14). Vinculin is believed to have a role in reinforcing the connection between ECM and actin23. Our results indicate that roughly half the vinculin molecules in focal adhesions may overlap with actin whereas the remainder reside lower in the focal adhesion core, positioning vinculin at a key site for regulating force transmission within focal adhesions. On the other hand, zyxin and VASP localized to higher vertical positions, overlapping to a greater degree with actin (zyxin: average zcentre=73.2±8.8 nm, σvert=17.5±3.5 nm; VASP: average zcentre=80.5±11.6 nm, σvert=23.4±4.5 nm). Their similar vertical localizations and overlaps with the lower actin boundary are consistent with their cooperative role in actin assembly regulation24. Although α-actinin was present in lamellipodia (Fig. 2k and Supplementary Fig. 17), it was virtually excluded from the focal adhesion core (Fig. 2l; average zcentre=103.9±14.6 nm, σvert=22.8 ±5.1 nm) but overlapped fully with actin localizations, exhibiting a tapered cross-section profile similar to that of actin stress fibres (Fig. 1 f, g and Supplementary Figs 7 and 8). This supports a role for α-actinin in actin organization at focal adhesions. The lack of overlap between α-actinin and integrin indicates that their interaction1,25 may only be transient or regulatory in vivo. Taken together, our results reveal the presence of protein-specific strata making up the focal adhesion core, bridging the ~40-nm gap between the integrin cytoplasmic tails and the actin cytoskeleton.
Importantly, we found that the vertical distributions for each focal adhesion component were highly consistent across focal adhesions of diverse size and shape that arise from the continual and asynchronous focal adhesion assembly and maturation occurring in the cell population (Fig. 4a–c and Supplementary Fig. 9). The vertical positions for each focal adhesion component were uncorrelated with the area and morphology (aspect ratio) of focal adhesions, and were also similar between U2OS and MEF cells (Supplementary Figs 9 and 10). This suggests that the observed stratification of focal adhesion proteins represents a cell-type-independent organizing principle that persists throughout focal adhesion maturation stages.
To address the origin of the protein-specific stratified architecture of focal adhesions, we explored the localization and orientation of talin by iPALM. Talin is a large (270-kDa) protein implicated in the initiation of integrin-mediated adhesion and transmission of force between integrin and actin, and which possesses multiple binding sites for focal adhesion proteins including integrin, FAK, paxillin, vinculin and actin26. We thus hypothesized that talin could form tethers that span the integrin–actin gap, thereby serving as a vertically oriented scaffold for the stratified focal adhesion core. To test this, we compared iPALM analyses of talin tagged with PA-FP probes at different sites (Fig. 3a). Both N- and C-terminally tagged talin PA-FP fusions dimerized with endogenous talin and localized to focal adhesions (Supplementary Figs 5 and 20). Imaging talin with the PA-FP probe at the N terminus (talin-N, Fig. 3a) revealed a narrow distribution close to the plasma membrane and similar in position to FAK, paxillin and integrin αv (Fig. 3b, c; average zcentre=42.8±3.8 nm; average σvert=9.8±2.4 nm). We next probed talin tail position using a C-terminal PA-FP fusion, talin-C, and observed a distinctly different distribution from talin-N (average zcentre=76.7±10.6 nm; average σvert=15.7±3.8 nm; Fig. 3d–f), indicating a highly polarized orientation of talin, with the tail vertically displaced by at least 30 nm from the head. The talin tail position also substantially overlapped with those of zyxin, VASP, α-actinin and actin. Although integrin and actin binding sites have been identified throughout the length of talin26, our results indicate that the integrin binding site in the N-terminal head and the C-terminal THATCH domain actin-binding site are the structurally relevant sites in focal adhesions. This is supported by iPALM analyses of talin fragments, which revealed membrane-proximal and upper localizations for the PA-FP-tagged head and THATCH domains, respectively (Supplementary Fig. 11). In contrast to the polarized talin orientation, we were unable to detect vertical polarizations for paxillin or α-actinin PA-FP tagged at either the N or C termini (Fig. 4c and Supplementary Figs 21 and 22). Together with the ~50–60 nm in vitro dimension of talin26, our results indicate that talins are organized into arrays of elongated molecular tethers that diagonally span the stratified focal adhesion core.
Our results demonstrate that focal adhesions possess a surprisingly well-organized molecular architecture in which integrins and actin are separated by a ~40-nm focal adhesion core region that contains multiple partially overlapping protein-specific strata. The stratification probably arises from spatial constraints in protein–protein interactions, but once formed may also impose spatial constraints on protein dynamics within focal adhesions. For example, distribution overlaps between given proteins in a focal adhesion should increase the frequency and duration of their interactions, whereas the lack of overlap indicates that the interactions may be transient or have no direct structural role. Partial overlaps between proteins as well as the width of the protein distributions in focal adhesions may also reflect heterogeneity in protein–protein binding interactions. The focal adhesion protein organization indicates a composite multilaminar architecture made up of at least three spatial and functional compartments that mediate the interdependent functions of focal adhesions: an integrin signalling layer, a force transduction layer, and an actin regulatory layer (Fig. 4d). FAK and paxillin represent a membrane-proximal integrin signalling layer of the focal adhesion core that probably relays integrin–ECM engagement into signalling cascades that control adhesion dynamics and gene transcription21,22. Talin and vinculin are observed in the broader central zone, with talin organized into arrays of diagonally oriented tethers that probably link integrin to actin directly. The distribution of vinculin is consistent with its binding to sites along talin rod domain and actin, which may serve to buttress the integrin–talin–actin linkages. Talin and vinculin have been implicated as regulatable force transmission links between actin and integrins23,27–29. Their positions together thus define the force-transduction layer, signifying a structural basis for the ‘molecular clutch’7,27,28 machinery. Finally, the similar vertical localizations of VASP, zyxin and actin filament termini in the uppermost region indicate that a VASP–zyxin complex may comprise an actin regulatory layer involved in focal adhesion strengthening via actin-barbed-end assembly and stress-fibre enlargement24. α-Actinin appears to localize predominantly along the actin stress fibres where it may mediate their formation through actin filament cross-linking activity25.
The observed molecular architecture also indicates how mechanical force may be essential for focal adhesion formation and maintenance1–3. The diagonal talin orientation could arise from actomyosin pulling of the talin tails relative to the integrin-bound talin heads, with the resulting intramolecular tension straightening or stretching the talin. Subsequently, distinct sites along the length of talin may serve as spatial templates giving rise to the observed protein-specific stratification in focal adhesions. Further stretching of talin is also indicated in iPALM images of some focal adhesions (Fig. 3d, f), where a fraction of talin-C localizations extend significantly upward in the proximal end of focal adhesions, implying a head-to-tail length greater than the nominal talin length of 50–60 nm. Stretching of talin rod fragment has been shown to unmask cryptic vinculin binding sites30, consistent with the observed talin and vinculin positions. Thus, via stretch-induced recruitment, talin may effectively serve as a molecular ruler that specifies focal adhesion molecular architecture.
METHODS SUMMARY
iPALM imaging
The principle and instrumentation for iPALM were described previously9 (see also Supplementary Fig. 1 and Supplementary Note 1). Gold nanoparticles (80–100 nm) immobilized to the coverglass were used as fiducials for calibration and drift correction. The vertical coordinate calibration was performed before each cell was imaged. For each cell, 25,000–75,000 image triplets were acquired, with 50 ms per frame exposure time, yielding ~106 localizations. Vertical coordinates relative to the coverglass surface are indicated by a colour scale from red to purple (z=0–150 nm). All side-view panels are shown with similar vertical scale and oriented with the nearest cell edge to the left.
Cell culture and fluorescent protein constructs
PA-FP protein fusions were constructed with green-to-red photoconvertible fluorescent protein, tandemdimer Eos (tdEos)19 or monomeric Eos2 (mEos2)20 fused to focal adhesion proteins via short linkers (Supplementary Table 3 and Supplementary Note 3). Fusion proteins were expressed in U2OS or MEF cells sparsely plated on fibronectin-coated, fiducialed coverglasses, and fixed for imaging ~18 h after re-plating.
Analysis of protein positions
The histogram of vertical localization coordinates was calculated for each focal adhesion region. The local z=0 nm level was defined by nonspecific fluorescence from the media that adsorbed to the coverglass, and was used to account for sample tilt. The centre positions (zcentre) and width parameter (σvert) were calculated from Gaussian fits or from the first and second moment of the distributions for non-Gaussian cases such as actin and α-actinin. For more detailed information see Methods and Supplementary Information.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Preparation of fiducialed coverglasses for cell culture
Fluorescent fiducials are critical for iPALM because they provide a constant internal reference for calibration, tracking and spatial drift correction. As described previously9, we use the plasmonic emission31 from 80 to 100 nm gold (Au) nanoparticles sparsely adsorbed (~2,000 permm2) to the coverglass surface and immobilized by 30–50 nm of sputtered SiO2. Although gold nanoparticles can be added after cell attachment, their rigid immobilization is critical for high localization accuracy, and additionally allows optimization of proper fiducial density before cell culture. Fiducialed coverglasses were ultraviolet-sterilized (15 min), rinsed with Dulbecco’s phosphate buffered saline (DPBS, Invitrogen), incubated at 4 °C overnight with 10 μgml−1 (U2OS) or 1 μg ml−1 (MEF) human plasma fibronectin (FC010-5MG, Chemicon International), and incubated with 1% heat-inactivated bovine serum albumin (A3059, Sigma) (1 h, 37 °C) before a final rinse with DPBS.
Cell culture and imaging sample preparation
U2OS (human osteosarcoma) cells were cultured in supplemented McCoy5A media (10% fetal bovine serum (FBS), 2 mM glutamine, and 100 units ml−1 of penicillin/streptomycin, Invitrogen). Mouse embryonic fibroblast (MEF) cells were cultured in supplemented DMEM media (10% FBS, 2 mM glutamine and 100 units ml−1 of penicillin/streptomycin). Cells were transfected by nucleofection with endotoxin-free expression vector DNA (U2OS, ~0.2–1 μg per ~1×106 cells; MEF, 5–6 μg per ~2–3×106 cells) per the manufacturer’s protocol (Lonza). Transfected cells were cultured overnight, replated onto fiducialed coverglasses, and incubated at 37 °C, 5% CO2. Cells were replated at a sparse density of ~60–100 cells mm−2 and fixed for imaging ~18 h after replating. At 18 h, most focal adhesions have not transformed into fibrillar adhesions, which are associated with fibronectin bundles that would affect the measurement of protein position relative to the substrate. Phenol-red free media was used to minimize background fluorescence. Cells were fixed with 2% paraformaldehyde and imaged in PHEM buffer (PIPES 60 mM, HEPES 25 mM, EGTA 10 mM, MgCl2 2 mM, pH 6.9). Imaging chambers (thickness ~10 μm) were assembled from the 18-mm cell-containing fiducialed coverglass and a 25-mm coverglass and sealed with 5-min epoxy (ITW Performance Polymers) and vaseline (Unilever). We imaged focal-adhesion-containing lamella areas, typically no greater than 15–20 μm from the cell edge, that also contained several fiducials for calibration and drift correction.
iPALM data acquisition and image processing
Imaging samples prepared as described above were mounted onto a piezo-electric-equipped sample holder. The optical configuration is described in Supplementary Note 1. Both top and bottom objectives were brought into focus and aligned using the images of the gold fiducials immobilized on the coverglass. The z-positions of the gold fiducials were determined and optimized for proper focus and interference modulation. This was carried out by piezo-based fine tuning of beamsplitter and mirror positions, while monitoring the z-calibration curve as described in detail below. Once the sample was in good initial alignment, the sample was translated laterally to find suitable cells for imaging. We imaged low-level expressing cells to avoid biological over-expression artefacts and to minimize background that can contribute to lower localization accuracy.
Once cells were located, the setup was fine-tuned until a z-calibration curve with an optimal modulation was attained and recorded. Key steps for the calibration are illustrated in Supplementary Fig. 1, with the fiducial positions shown in Supplementary Fig. 1c (inset: summed raw intensity data for calibration sets; main: iPALM image, note that fiducials are not prominent because the render program treats each fiducial as a single particle). The z-calibration data set was measured at 8-nm intervals as the sample was translated along the z-axis using piezo-electric translation stages (Physik Instrumente). This resulted in an intensity modulation between the three cameras due to interferometric effect9, as shown in Supplementary Fig. 1d for fiducial F1. To align the three-camera triplet of images for analysis, the coordinates of multiple fiducials were determined for each camera, using one camera image as reference. The similarity transformations for two other cameras with respect to the reference camera were determined using linear regression, and applied to the respective camera images to align them to the reference.
To extract calibration parameters, each of the molecule images in each frame of the triplet series was fit to a two-dimensional Gaussian, yielding Gaussian amplitudes Ik(z), where k=1, 2, 3, as plotted in Supplementary Fig. 1d for fiducial F1. Then a least square fit was used to determine the dependence of these amplitudes on z-coordinate according to the equation:
| (1) |
This yielded a set of z-calibration coefficients: ω,D,Ak,φk,Bk, k=1, 2, 3. The variable D in the denominator accounts for focal envelope function. A calibration curve and coefficients extracted for fiducial F1 are shown in Supplementary Fig. 1d, e.
Subsequently, we applied Newton’s method32 to extract the z-coordinate of each fluorescent molecule or fiducial. This method finds a value of molecular z-coordinate by minimizing the difference between the intensities calculated by equation (1) using the calibration parameters (for example, Supplementary Fig. 1e) and the measured two-dimensional Gaussian amplitudes from the data triplet. Performing this procedure on the calibration set also provides a check for calibration quality and consistency, as shown in Supplementary Fig. 1f, where the extracted z position (diamonds) for fiducials F1–4 are compared to the actual sample z position (solid lines).
During the main acquisition sequences, each frame triplet contained images of a few individual fluorescent proteins that emitted during each frame imaging interval. The activation power (405 nm) was adjusted so that these individual fluorescent proteins were sparsely distributed and their images did not overlap. The localization procedure consisted of the following steps: (1) the individual camera images were aligned by applying the similarity transformations determined from previously recorded images of multiple fiducials in each camera, as described above; (2) these three aligned images of each fluorescent particle from a given frame triplet were added together to form a sum image; (3) the fluorescent particle in each sum image was fit to a two-dimensional Gaussian by nonlinear leastsquare fitting to obtain x and y coordinates; (4) each individual camera image in the triplet was also fit to a two-dimensional Gaussian, yielding amplitudes Ik, k=1, 2, 3, which were used to extract the z-coordinate of each fluorescent molecule from equation (1) and the calibration parameters, using Newton’s method as described above. The refractive index difference between the calibration data (when the sample was translated, the varying path-length difference between the two arms is in immersion oil, n ~1.52) and main acquisition (sample was stationary, n ~1.40 for cell) was accounted for by applying appropriate linear scaling. (5) Sample drift was corrected using the fiducial localizations. Lateral sample drift during all measurements was significantly lower than the lateral localization accuracy, which was typically ~20 nm full-width at half-maximum. The vertical (z-coordinate) sample drift in all measurements varied between 10 nm and 50 nm over the course of the measurements. The vertical drift was traced by determining in each frame the z-position of the same gold fiducials as were used for z-calibration. This drift was then subtracted from z-coordinates of all fluorescent particles. The residual z-coordinate uncertainty was typically less than 5nm.
Typical measurements consisted of 25,000–75,000 image triplets (Supplementary Table 4) with the exposure time of 50 ms per frame; the 5–30-ms activation pulses were transmitted between the excitation pulses. Imaging parameters for iPALM data sets are summarized in Supplementary Table 4. Data acquisition was carried out using software written in LABVIEW (National Instruments). Data analysis and image processing was performed using software written in IDL (ITT Visual Information Solutions) and run on a Linux computational cluster at HHMI Janelia Farm Research Campus.
As described previously9, iPALM images were rendered from the processed list of three-dimensional molecular coordinates: the position and localization uncertainty of each localization is represented by a normalized two-dimensional Gaussian, whose width is proportional to localization uncertainty. Note that the lists of molecular coordinates were used for quantitative analysis, rather than the rendered images. For top-view (x, y) images, each molecule was represented by a normalized two-dimensional Gaussian. The width of the Gaussian is the positional uncertainties (σx, σy) of the calculated x, y position as described previously33. The z-coordinate is encoded by colour. For side-view images, the molecule was also rendered by a normalized two-dimensional Gaussian, but with the vertical width corresponding to σz, the vertical uncertainty. Because the vertical (z) resolution is ~2 times better than xy, σz is also ~2 times smaller. A gamma of 0.5 (top view) and 0.75 (side view) was used to compress the tonal range of iPALM images to within the dynamic range of print and computer monitor.
A single colour scheme was used from red to purple, covering the range from z=0 nm to z=150nm, where features within focal adhesions are seen. The same colour scheme was also used for side-view (xz) or (yz) image. Typically, the raw processed coordinates exhibited a minor tilt of <30–50 nm over the image field of ~50 μm across, due to sample tilt or optical alignments. These were corrected by simple coordinate rotation to achieve a flat vertical substrate level, which was set to z=0 nm for the z-colour-coded image rendering. For quantification of focal adhesion areas, local background level was used to control for long-range variations as described below.
Analysis of protein distributions in focal adhesions: zcentre and σvert calculation
iPALM localization data records both the fluorescent molecules localized within the focal adhesions as well as molecules in the cytoplasmic fraction and autofluorescent molecules inside and outside the cells. To quantify the spatial distribution of the proteins specifically residing within individual focal adhesion regions, we created binary region masks from top-view iPALM images, as depicted in Supplementary Fig. 3. These areas covered the focal adhesion and immediately surrounding space, primarily for the quantification of the local substrate background level. A program written in Java as an ImageJ (NIH) plug-in was used to export the three-dimensional molecular coordinates for each region into separate files, which were subsequently analysed using a program written in LABVIEW. A histogram of vertical positions was calculated with 1-nm bins. The centre vertical positions (zcentre) and width parameter (standard deviation of the vertical coordinate distributions, σvert, which relates to full-width at half-maximum (FWHM) by: FWHM=2.35σvert) were determined from a Gaussian fit to the focal adhesion molecule peak. In addition to the main peak, we also typically observed a smaller peak of substrate surface autofluorescent molecules. The observed background molecules probably originate from autofluorescence present in cell culture media, such as from fetal bovine serum and other trace contaminants34 and were primarily localized to the coverglass surface because non-surface-adsorbed background molecules diffuse too rapidly to be visualized as single molecules. We defined the zcentre of the local substrate distribution as the z=0 nm for each adhesion region. Because the substrate fluorescence density is low, typically the regional mask included the focal adhesion as well as a small surrounding region of a few square micrometres for good statistics. This local background provided an internal z=0 nm reference that controls for long-range vertical variation due to sample tilt or optical field curvature. Gaussian fitting used the least-absolute-residuals minimizing algorithm in LABVIEW. Most focal adhesion proteins exhibited Gaussian-like peaks, except for actin and α-actinin which extend into the stress fibre, instead of localizing as well-defined layers. Thus, to quantify their positions for comparison with other proteins, we calculated the zcentre and the width parameter (σvert) as the first moment and second moment , respectively, where N denotes the total number of molecules, zi denotes the z values for each histogram bin, i indexes the histogram z-bins, and f(zi) denotes the histogram of z position. The statistics for the average zcentre and σvert are shown in Fig. 4a, b and Supplementary Table 1. Fit parameters to individual focal adhesion regions in Figs 1–3 are shown in Supplementary Table 2.
Analysis of zcentre and focal adhesion morphologies
To determine whether a relationship existed between focal adhesion protein vertical position and focal adhesion morphology, we quantified morphometric properties of focal adhesions. We first calculated image maps of the number-density and average z position from the molecule coordinates measured by iPALM, using a 33×33 nm2 bin size. Programs written in MATLAB (Mathworks) were used to segment the areas corresponding to focal adhesions, calculate the areas, and measure the major and minor axes of the best-fit ellipse. The plots of protein position (zcentre) as a function of focal adhesion size (area in μm2) or aspect ratio (major divided by minor axes), as well as correlation coefficients, are shown in Fig. 4c and Supplementary Figs 9, 21c and 22c. These plots also indicate the range of focal adhesion size and shape (aspect ratio of round focal adhesion=1, and >1 for elongated focal adhesions) that were observed. As seen by the size distribution and as noted earlier, most regions analysed correspond to focal adhesion by previously defined morphometric criteria35 (area <8 μm2, aspect ratio <7). Note that small nascent adhesions were omitted if there was not sufficient background localization to allow accurate determination of local substratum level.
Supplementary Material
Acknowledgements
We thank J. Lippincott-Schwartz, G. Patterson and M. Parsons for sharing DNA; S. Xie for help with automation software; K. Jaqaman for MATLAB code; and HHMI Janelia Farm Scientific Computing and NIH Helix systems for computing resources. Funding: Division of Intramural Research, NHLBI (P.K., A.M.P. and C.M.W.); Howard Hughes Medical Institute (G.S. and H.F.H.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu. Rev. Cell Dev. Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 2.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix-cytoskeleton. Nature Rev. Mol. Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 3.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 4.Zaidel-Bar R, et al. Functional atlas of the integrin adhesome. Nature Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franz CM, Muller DJ. Analyzing focal adhesion structure by atomic force microscopy. J. Cell Sci. 2005;118:5315–5323. doi: 10.1242/jcs.02653. [DOI] [PubMed] [Google Scholar]
- 6.Chen WT, Singer SJ. Immunoelectron microscopic studies of the sites of cell-substratum and cell-cell contacts in cultured fibroblasts. J. Cell Biol. 1982;95:205–222. doi: 10.1083/jcb.95.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YL. Flux at focal adhesions: slippage clutch, mechanical gauge, or signal depot. Sci. STKE. 2007;2007:e10. doi: 10.1126/stke.3772007pe10. [DOI] [PubMed] [Google Scholar]
- 8.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 9.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl Acad. Sci. USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palade GE, Porter KR. Studies on the endoplasmic reticulum. I. Its identification in cells in situ. J. Exp. Med. 1954;100:641–656. doi: 10.1084/jem.100.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ledbetter MC, Porter KR. A “microtubule” in plant cell fine structure. J. Cell Biol. 1963;19:239–250. doi: 10.1083/jcb.19.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson GH, et al. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell. 2008;133:1055–1067. doi: 10.1016/j.cell.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Kaksonen M, Drubin DG, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc. Natl Acad. Sci. USA. 2006;103:10277–10282. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keren K, et al. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard TD, Berro J. Mathematical models and simulations of cellular processes based on actin filaments. J. Biol. Chem. 2009;284:5433–5437. doi: 10.1074/jbc.R800043200. [DOI] [PubMed] [Google Scholar]
- 16.Shroff H, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc. Natl Acad. Sci. USA. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hell SW, Schmidt R, Egner A. Diffraction-unlimited three-dimensional optical nanoscopy with opposing lenses. Nature Photon. 2009;3:381–387. [Google Scholar]
- 18.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 19.Wiedenmann J, et al. a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl Acad. Sci. USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinney SA, et al. A bright and photostable photoconvertible fluorescent protein. Nature Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nature Rev. Mol. Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 22.Brown MC, Turner CE. Paxillin: adapting to change. Physiol. Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- 23.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshigi M, et al. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 2005;171:209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil. Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 26.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 27.Hu K, et al. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 28.Brown CM, et al. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J. Cell Sci. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 29.Jiang G, et al. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424:334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 30.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dulkeith E, et al. Plasmon emission in photoexcited gold nanoparticles. Phys. Rev. B. 2004;70:205424. [Google Scholar]
- 32.Press WH, Flannery PP, Teukolsky SA, Vetterling WT. Numerical Recipes. Cambridge Univ. Press; 1986. [Google Scholar]
- 33.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubin JE. Autofluorescence of viable cultured mammalian cells. J. Histochem. Cytochem. 1979;27:36–43. doi: 10.1177/27.1.220325. [DOI] [PubMed] [Google Scholar]
- 35.Zamir E, et al. Molecular diversity of cell-matrix adhesions. J. Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.