Abstract
Recruitment of a dynamin-like GTPase (Drp1/Dlp1/Dnm1) to membranes requires the mitochondrial dynamics protein Fis1. Mdv1 has been proposed to act as an adaptor between Fis1 and Dnm1 in Saccharomyces cerevisiae. We show that S. cerevisiae Fis1 binds directly to Dnm1 and to Mdv1. Two Fis1 regions have been previously implicated in Mdv1 recruitment: an N-terminal “arm” and a concave surface formed by evolutionarily conserved residues in the tetratricopeptide repeat domain. Perturbing either Fis1 region does not affect Mdv1 binding, but both regions influence Dnm1 binding. Fis1 lacking its N-terminal arm binds tightly to Dnm1, and binding is abolished by mutations to the Fis1 concave surface. The Fis1-Dnm1 interaction decreases more than 100-fold in the presence of the Fis1 arm, suggesting that the arm acts in an autoinhibitory manner to restrict access to the Dnm1 binding site on Fis1. Our data indicate that the concave surface of the Fis1 tetratricopeptide repeat-like domain is evolutionarily conserved to bind the dynamin-like GTPase Dnm1 and not Mdv1 as previously predicted.
Mitochondria are dynamic organelles that often divide or fuse independently of cellular division (1). The purpose of mitochondrial fission and fusion is not firmly established, but mitochondrial morphology appears important for organelle function (2–8). Supporting this idea, mitochondrial morphological changes occur during apoptosis in humans and other organisms (9–14). Three autosomal dominant neuropathies in humans are caused by defects in mitochondrial fusion (8, 15, 16), and a defect in mitochondrial fission was recently reported as being lethal (17).
Mitochondrial morphology is proposed to be maintained by a balance of mitochondrial fusion and fission involving at least four dynamin-like GTPases: three in fusion (in humans Mfn1, Mfn2, and Opa1) and one in fission (in humans Drp1, also known as Dlp1). In mitochondrial fission, the dynamin-related GTPase is thought to polymerize at points of constriction on the mitochondria, a fraction of which result in membrane scission in a manner that may be similar to dynamin in endocytosis (3, 18–20). The mitochondrial fission GTPase (Dnm1/Drp1) is also implicated in fission of peroxisomes (21–23). Localization of this molecule to either mitochondria or peroxisomes appears to require the evolutionarily conserved mitochondrial fission protein 1, or Fis1 (24–31). Fis1 is localized to the surface of these organelles by a C-terminal transmembrane domain (26, 31) and is proposed to mediate the assembly of a functional fission complex that includes Dnm1/Drp1 (27–29).
In addition to Dnm1 and Fis1, mitochondrial fission in Saccharomyces cerevisiae involves at least two other proteins: Mdv1 and Caf4 (24–26, 32–36). Mdv1 is proposed to act as a molecular adaptor and is composed of three domains: an N-terminal extension (NTE)4 domain, a coiled-coil (CC) domain, and a C-terminal WD repeat domain (3) (Fig. 1). Caf4 shares a similar domain architecture and may be redundant with Mdv1 (36) but is also thought to be important in establishing Dnm1 polarity at sites of membrane scission (37). To date, Mdv1 or Caf4 orthologs have been identified only in fungi.
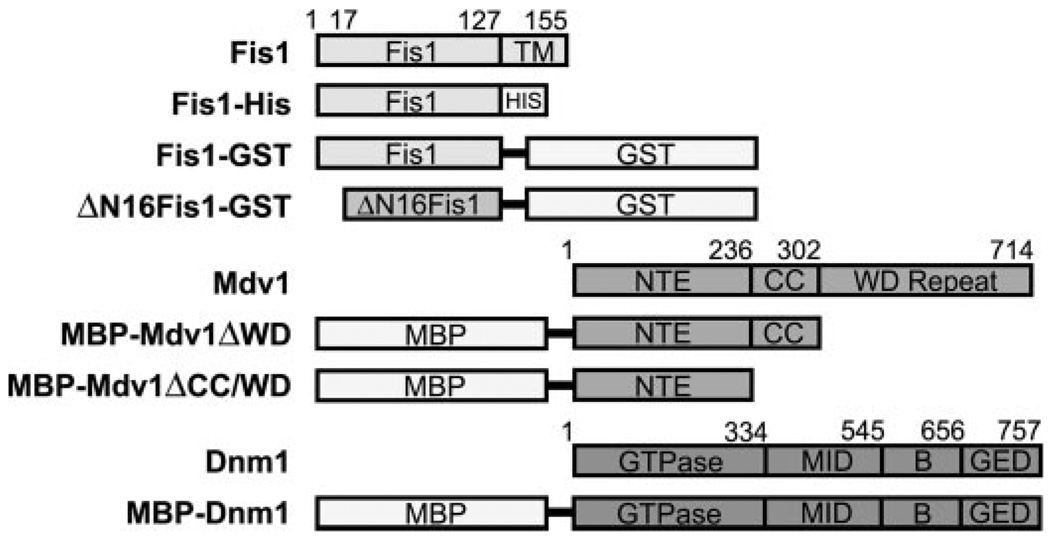
FIGURE 1. Domain architecture of the Fis1, Mdv1, and Dnm1 constructs used in this study.
For Fis1, constructs were created by replacing the C-terminal transmembrane domain with either His6 sequence (HIS) or GST. For Mdv1 and Dnm1, constructs were created by placing MBP before the Mdv1 domains.
Based on cytological, yeast two-hybrid, and co-immunoprecipitation data, a model has emerged for the assembly of these proteins. Fis1 binds to Mdv1, which subsequently recruits Dnm1 to sites of scission (24–26, 32–36, 38–42). Thus, in S. cerevisiae, Mdv1 is proposed to act as a bridge between Fis1 and Dnm1. Competing models exist for the temporal order of a Fis1-Mdv1-Dnm1 assembly, but in these models Fis1 plays a central role in recruiting proteins to sites of fission (3, 39). In humans, where no Mdv1 ortholog has yet been identified (40, 41), Drp1 recruitment is also Fis1-dependent (31, 43). However, the recombinant proteins do not form a stable complex (30), suggesting that other factors may be required in mammals.
High resolution structures of Fis1 from human, mouse, and budding yeast reveal a tetratricopeptide repeat (TPR)-like fold (40, 45, 46),5 a common protein-protein interaction domain (47). These structures share a concave surface formed by evolutionarily conserved residues in the TPR-like domain that probably bind other proteins important in mitochondrial dynamics. However, these structures differ in the orientation of the N-terminal region of Fis1, which we refer to as the Fis1 arm. The arm is not part of the TPR-like domain and, in S. cerevisiae, contains an additional 8 residues not found in mammalian Fis1 sequences (40) (supplemental Fig. 1). In the structure of S. cerevisiae Fis1 (40), this arm is ~16 residues and blocks access to the conserved, concave surface (Fig. 3, A–D). However, hydrogen-deuterium exchange experiments show that the Fis1 arm is less structured than the TPR-like domain, suggesting that it could move aside to regulate access to a putative binding surface (40).6 Here we show, using pull-down experiments with recombinantly expressed proteins, that the concave surface of the Fis1 TPR-like domain binds the dynamin-like GTPase Dnm1 and not Mdv1. Our results suggest a new model for the assembly of these proteins in which the conserved concave surface of the Fis1 TPR-like domain binds the dynamin-like GTPase and the Fis1 arm regulates access to this GTPase binding pocket.
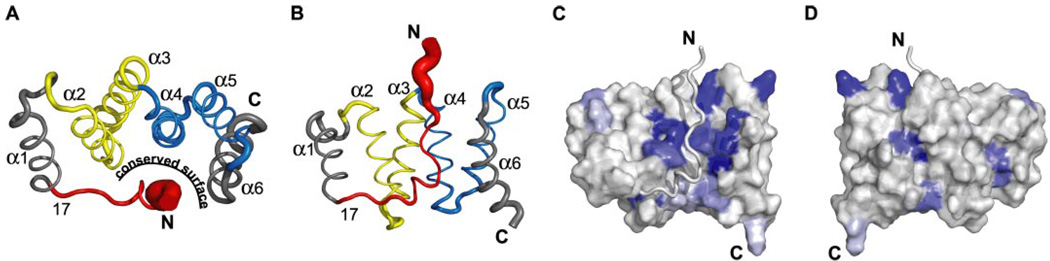
FIGURE 3. The Fis1 arm (residues 1–16) prevents access to a conserved concave surface of the TPR-like domain.
A, the ensemble of NMR structures (Protein Data Bank code 1Y8M) (40) represented as a single tubular structure. The tube diameter is scaled to the Cα root mean square deviations of each residue with respect to the average structure calculated with a PyMOL script (Mura, C., and McCammon, J. A.; available on the World Wide Web). The molecule is colored to highlight the arm (red), the first (yellow), and second (blue) TPR motifs. The concave surface is formed by α-helices 2, 4, and 6. B, rotation of the structure in A by+65° about the x axis. C, Fis1 amino acid sequence conservation from 41 species (supplemental Fig. 1) displayed onto a surface representation of the yeast Fis1 structure oriented as in B. The most conserved residues are dark blue, and least conserved are white. D, rotation of the surface representation in C by 180° about the y axis. The figures were made using PyMOL (W. L. DeLano; available on the World Wide Web).
EXPERIMENTAL PROCEDURES
Mutagenesis, Protein Expression, and Purification
DNA encoding S. cerevisiae Fis1 lacking the C-terminal 27 residues was subcloned into pET29b (EMD Biosciences) to give a gene product of the cytosolic domain of Fis1 with a C-terminal His6 tag (Fis1ΔTM-His).A similar Fis1 construct except with GST at the C terminus was created by subcloning DNA encoding GST from pGEX vector (GE Healthcare) into a pET29b vector containing Fis1ΔTM. A ΔN16Fis1-GST construct lacking the first 16 residues and replacing Leu17 with Met was created by cohesive end ligation and PCR (49). Mutations in ΔN16Fis1-GST were created using the QuikChange kit (Stratagene). Fusion and mutant plasmids were transformed into Escherichia coli BL21(DE3) cells and grown at 37 °C in Luria broth with kanamycin (30 µg/ml) to A600 of ~0.7. Protein expression was with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 15–18 h. Cells harvested by centrifugation were resuspended in column buffer (20 mm NaH2PO4, 150 mm NaCl, 2 mm dithiothreitol, pH 7.4) containing protease inhibitors (Roche Applied Science). Buffers for mutant and wild-type Fis1ΔTM-GST fusion constructs also contained 5 mm EDTA. Cells were lysed by French press or by sonication, DNase was added to 1 µg/ml with 10mm MgCl2, and lysates were clarified by centrifugation. Proteins were isolated from the resulting supernatant by affinity chromatography using either glutathione or Ni2+ affinity beads (GE Healthcare). Sample purity was checked by Coomassie-stained SDS-PAGE and was typically greater than 90%.
DNA encoding Mdv1 residues 1–302 lacking the WD repeat domain (Mdv1ΔWD) or residues 1–236 lacking both the WD repeat and coiled-coil domains (Mdv1ΔCC/WD) was sub-cloned into pMALc2x expression vectors (New England Bio-Labs) (Fig. 1A). For overexpression, E. coli Rosetta cells (EMD Biosciences) were transformed with the Mdv1-containing vector and grown at 18 °C in autoinducible Studier ZYP-5052 defined media (50) containing carbenicillin (50 µ/ml) and chloramphenicol (34 µg/ml) to A600 of ~8.0. Harvesting and lysis was similar to the Fis1 constructs. Cell lysate was applied to an amylose affinity resin and eluted with either 1 m α-methylglucoside or 10 mm maltose. For both constructs, the purity was assessed by Coomassie-stained SDS-PAGE and found to be typically greater than 90% homogeneous. However, the Mdv1ΔCC/WD construct was less homogeneous with degradation products that ranged from 20 to 40% of the parent protein based on quantifying immunoaffinity Western blots with anti-maltose-binding protein (MBP) antibodies.
DNA encoding the full-length Dnm1 protein was cloned into pMALc2x expression vectors (New England BioLabs). For overexpression, E. coli Rosetta cells (EMD Biosciences) were transformed and grown at 37 °C in Luria broth containing kanamycin (34µg/ml) and chloramphenicol (34µg/ml) to A600 of ~0.7 and induced with 1 mm isopropyl 1-thio-β-d-galacto-pyranoside at 18 °C for 16 h. The harvested cells were resuspended in 20 mm Tris-HCl, 250 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol, pH 7.4, and frozen until purification, which was similar to the MBP-Mdv1 construct using amylose affinity chromatography with elution by 1 m α-methylglucoside or 10 mm maltose. Purity of MBP-Dnm1 was assessed by Coomassie-stained SDS-PAGE and found to be typically greater than 90% homogeneous. Recombinant Dnm1 appears to be properly folded, since its GTPase activity was similar to Dnm1 after cleavage of the MBP fusion tag (data not shown).
All constructs were verified by DNA sequencing, and the resulting isolated proteins were found to be at the expected molecular weight by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry.
Protein-Protein Interaction Assays
For MBP-Mdv1 pull-downs, 2 µm protein was incubated with Fis1 constructs at the indicated concentrations at 23 °C for 2 h in 150 µl of 20 mm Tris, 150 mm NaCl, 5 mm EDTA, 2 mm dithiothreitol, pH 7.4, buffer while rotating. For MBP-Dnm1 pull-downs, 1 µm protein was incubated with Fis1 constructs at the indicated concentrations at 23 °C for 2 h in 150 µl of 20 mm Tris, 250 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol, pH 7.4, buffer while rotating. For the pull-downs, 20µl of slurry mixture containing glutathione-affinity beads (diluted 3:5 with buffer) were added to the mixture and rotated for an additional 30–60 min. The affinity beads were washed five times with the same buffer to remove nonspecific binding. Protein was eluted with 30 µl of buffer containing 50 mm glutathione, diluted into 4× SDS-PAGE loading buffer, and a fraction was analyzed by a 10% SDS-PAGE with Western blot detection using commercially available anti-MBP (New England BioLabs), anti-GST (GE Healthcare), or anti-His (AbCam) tag antibodies. The Western blots were visualized by either the CN/DAB colorimetric assay (Bio-Rad) or with enhanced chemiluminescence (Pierce) and quantified by densitometry using ImageQuant (Amersham Biosciences) software or a Typhoon 9410 variable mode imager (GE Healthcare). Protein-protein interactions were also confirmed by reverse pull-downs using amylose affinity beads (New England BioLabs) and elution with 50mm maltose or with pull-down using Ni2+ affinity beads (GE Healthcare) and elution with 1 m imidazole. Control reactions were performed concurrently to ensure specificity of Fis1-Mdv1 interactions. No binding between MBP-Mdv1 constructs and GST, glutathione beads, or Ni2+ affinity beads was detected. No binding between Fis1-GST and MBP or amylose beads was detected.
RESULTS
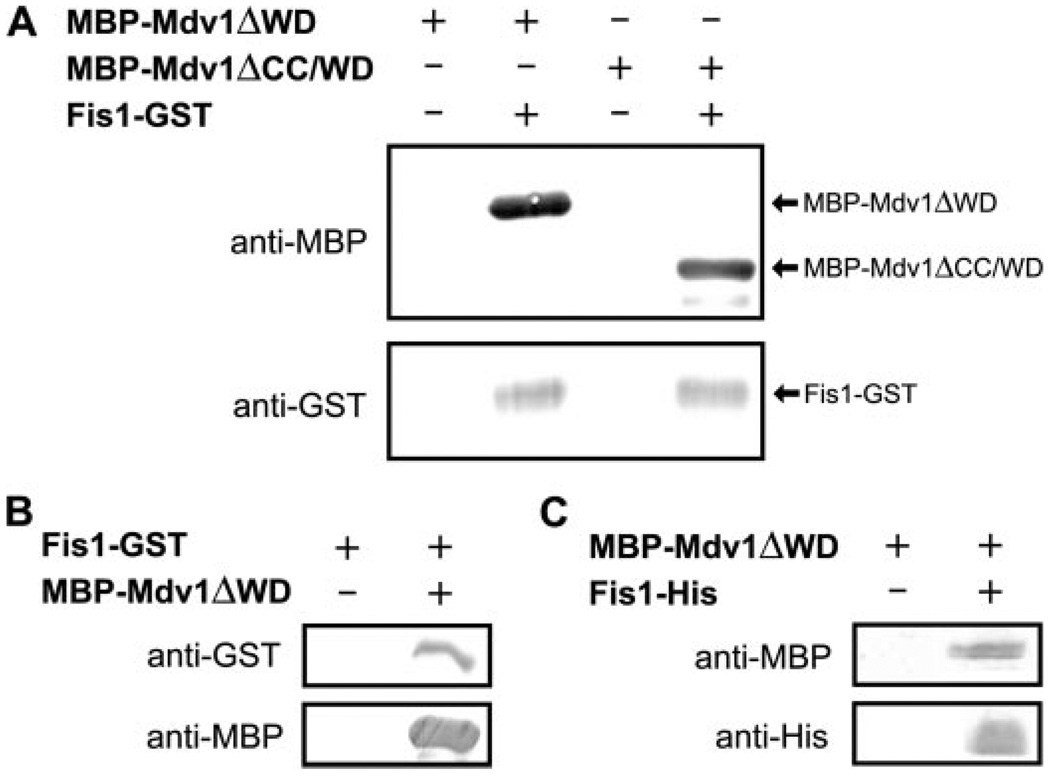
Fis1 appears to localize Mdv1 to sites of future mitochondrial fission that may arise from a Fis1-Mdv1 interaction, which has been observed in two-hybrid and co-immunoprecipitation studies suggesting a direct interaction (20, 38–41). To test for a direct interaction, we attempted to recombinantly express and purify Mdv1 in E. coli for pull-down experiments with the cytosolic domain of Fis1. Various expression constructs of full-length Mdv1 did not yield soluble, folded protein (data not shown), but we isolated two constructs that had MBP fused to the N terminus of Mdv1 (Fig. 1). One construct lacked the WD repeat domain (MBP-Mdv1ΔWD), and another shorter construct lacked both the WD repeat and CC domains (MBP-Mdv1ΔCC/WD). Each of these Mdv1 constructs bound to Fis1-GST in pull-down experiments using GST affinity beads (Fig. 2A). In the reverse experiment using MBP affinity beads, MBP-Mdv1ΔCC/WD was able to pull down Fis1-GST (Fig. 2B). This interaction appeared to be the result of direct binding to Fis1, since control experiments showed that GST did not bind MBP or Mdv1 constructs, and Fis1 did not bind MBP (supplemental Fig. 2). A direct Fis1-Mdv1 interaction was also observed in a pull-down experiment using a Fis1 construct with a His6 tag and Ni2+ affinity beads (Fig. 2C). Thus, the NTE domain of Mdv1 binds directly to the cytosolic domain of Fis1 in agreement with several studies (20, 38–41).
FIGURE 2. Fis1 binds directly to Mdv1 independent of the CC and WD repeat domains.
A, Western blot analysis of GST pull-down experiments with Fis1-GST and MBP-Mdv1. Control experiments confirm the specificity of the interaction (supplemental Fig. 2). B, Western blot analysis of the inverse experiment using amylose beads to pull down MBP-Mdv1ΔWD. C, Western blot analysis of a similar pull-down experiment using Ni2+ affinity beads to pull down a His6-tagged construct of Fis1, Fis1-His.
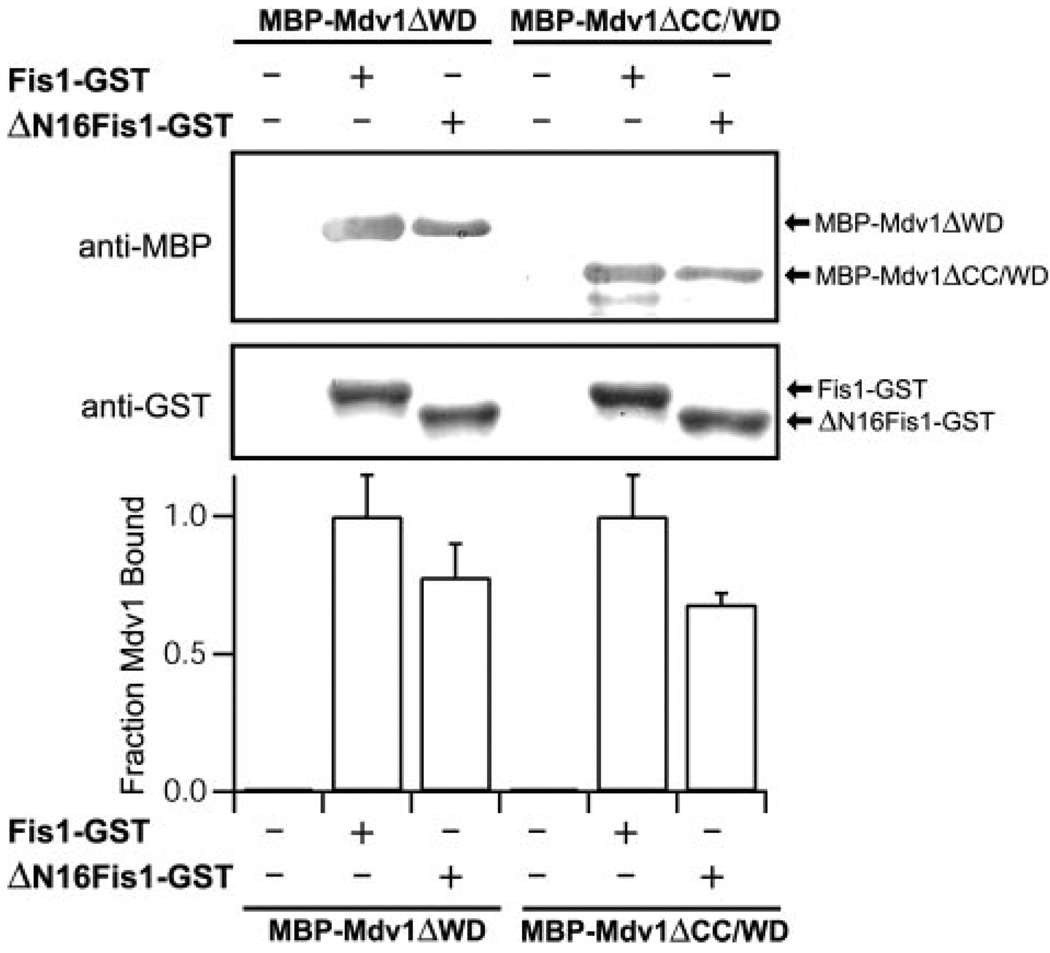
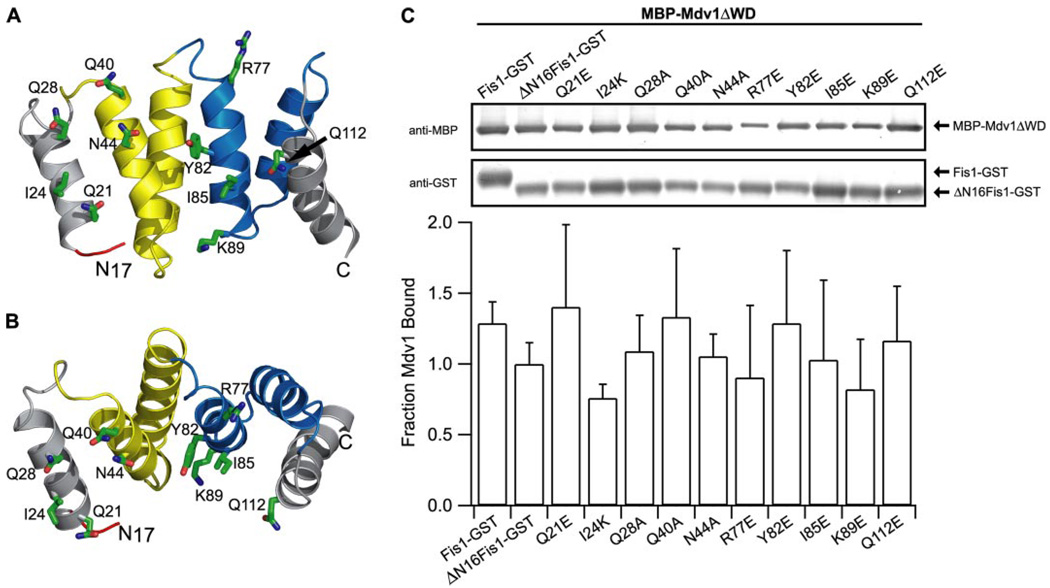
The structure of the cytosolic domain of Fis1 from S. cerevisiae reveals a concave surface formed by α-helices 2, 4, and 6 (Fig. 3, A and B). This surface is lined with evolutionarily conserved residues (Fig. 3, C and D), but access to it is blocked by the Fis1 arm. In vivo mutagenesis experiments have implicated both the Fis1 arm and the conserved surface of the TPR-like domain in Mdv1 recruitment to mitochondria (40, 41). To define the relative contributions of these regions for Mdv1 binding, we designed a mutant of Fis1 lacking the N-terminal 16 residues (ΔN16Fis1) and tested its binding to the Mdv1 domains with a GST pull-down assay. We reasoned that if Mdv1 bound into the concave surface of the Fis1 TPR-like domain as proposed (41), then removal of the Fis1 arm would enhance Mdv1 binding, because the Fis1 arm occludes access to this conserved surface (Fig. 3, A–C). However, Fis1 binding to either Mdv1 construct decreased with the deletion of the Fis1 arm (Fig. 4). This decrease was small but reproducible and did not appear to arise from a decrease in folding of ΔN16Fis1, which was as folded as Fis1 by circular dichroism spectropolarimetry (supplemental Fig. 3).
FIGURE 4. Removal of the Fis1 arm does not enhance Mdv1 binding.
GST pull-down experiments with Fis1-GST and ΔN16Fis1-GST indicate that binding to Mdv1 is reduced, not enhanced, upon deletion of the Fis1 arm (the first 16 residues). The binding is reported as fraction of Mdv1 bound, which is measured as the ratio of anti-MBP to anti-GST relative to Fis1-GST binding of MBP-Mdv1ΔWD (lane 2). The error bars represent the S.D. from at least three independent experiments. The experiments were performed as described in the legend to Fig. 2A.
This decreased Mdv1 binding to Fis1 upon deletion of the Fis1 arm suggested that the arm may stabilize Mdv1 binding to an adjacent region. To test this idea, we attempted to disrupt the Fis1-Mdv1 interaction by site-directed mutagenesis of ΔN16Fis1. Point mutants of Fis1 residues that lie in the concave surface of the TPR-like domain (R77E, Y82E, I85E, and K89E) did not disrupt Mdv1 binding; nor did point mutants adjacent to the Fis1 arm (Q21E, Q28A, Q40A, N44A, Q112E) with the exception of I24K, which showed a small but reproducible decrease in Mdv1 binding (Fig. 5). Poor solubility of E32K, W47A, and L116E prevented testing these conserved residues. Although these data do not define the Mdv1 binding site on Fis1, they indicate that the NTE and CC domains do not bind tightly into the concave surface of the TPR-like domain of Fis1 and that the Fis1 arm is not essential for this Mdv1 interaction.
FIGURE 5. Mdv1 does not appear to bind into the concave surface of the Fis1 TPR-like domain.
A, evolutionarily conserved residues of Fis1 that were selected for site-directed mutagenesis (Gln21, Ile24, Gln28, Gln40, Asn44, Arg77, Tyr82, Ile85, Lys89, and Gln112) are depicted on a ribbon model of ΔN16Fis1 derived from the structure of the Fis1, Protein Data Bank code 1y8m (40). B, rotation of the structure in A by −65° about the x axis. C, GST pull-down experiments between point mutants of ΔN16Fis1 and Mdv1. The binding is reported as fraction of Mdv1 bound, which is measured as the ratio of anti-MBP to anti-GST relative to ΔN16Fis1-GST binding of MBP-Mdv1ΔWD(lane 2). The error bars represent the S.D. from at least three independent experiments. Experiments were performed as in Fig. 2A.
Many Fis1 residues that constitute the concave surface of the TPR-like domain are evolutionarily conserved and therefore might be expected to bind a more highly conserved protein than Mdv1, which appears to be conserved only in fungi (40, 41). In addition to Fis1, the only known mitochondrial outer membrane fission protein that is highly conserved is the dynamin-related GTPase (Dnm1 in budding yeast, Drp1 in humans). Fis1 and the dynamin-related GTPase are reported to co-immunoprecipitate in budding yeast (42) and in humans (27, 31, 43), but this interaction has not been observed by yeast two-hybrid experiments (3, 39) and also was not observed when tested with purified human proteins (30). These considerations, in light of the dynamic nature of the Fis1 arm,5 led us to hypothesize that the Fis1 arm might regulate binding of the dynamin-like GTPase into the concave surface of the Fis1 TPR-like domain.
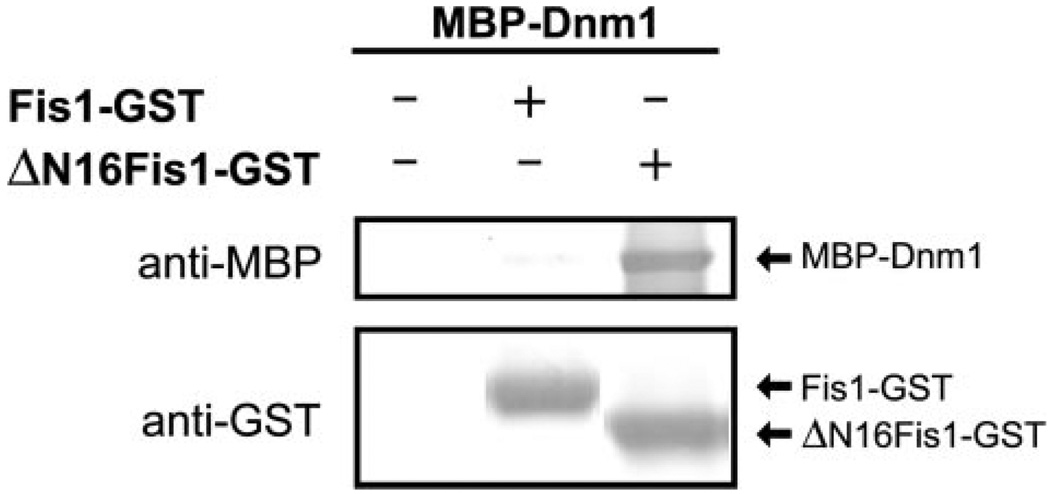
To test this idea, we expressed and purified the budding yeast GTPase, Dnm1, in E. coli with MBP fused to the N terminus for pull-down experiments with our Fis1 constructs. Dnm1 did not appear to bind Fis1 with a native N terminus, consistent with previously published results (Fig. 6). However, Dnm1 bound robustly to ΔN16Fis1 lacking the N-terminal arm. MBP-Dnm1 appears to be properly folded, since this construct hydrolyzes GTP similarly to Dnm1 without the MBP affinity tag (data not shown). These data show that the Fis1 arm inhibits Dnm1 binding and suggest that binding may occur directly into the concave surface of the Fis1 TPR-like domain.
FIGURE 6. Dnm1 binds directly to Fis1 lacking the N-terminal arm.
GST pull-down experiments with Fis1-GST and ΔN16Fis1-GST indicate that binding to Dnm1 is enhanced upon deletion of the Fis1 arm (the first 16 residues). Experiments were performed as in Fig. 2A.
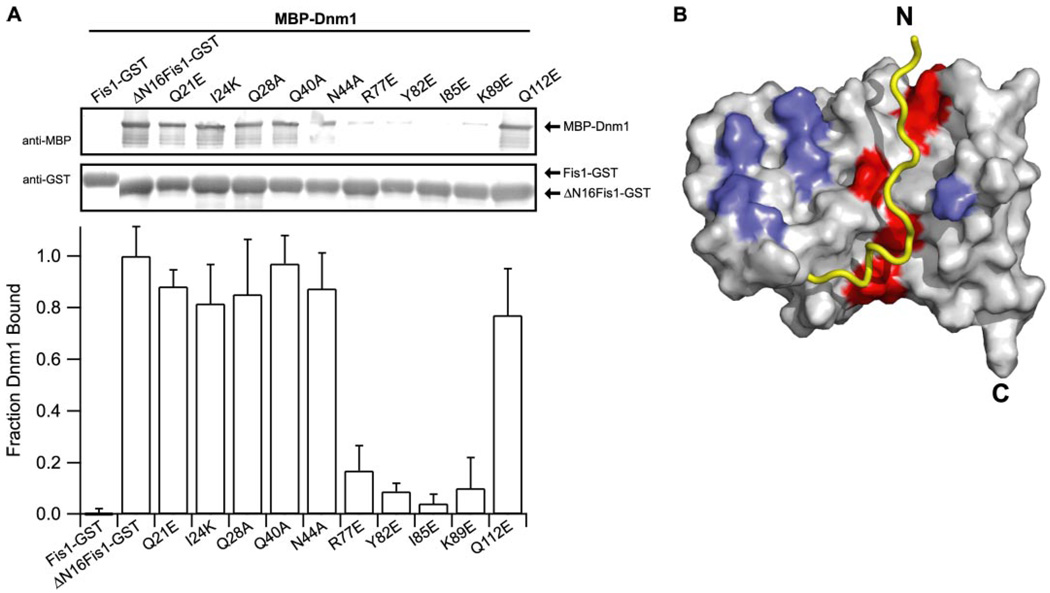
To further define the Dnm1 binding surface on Fis1, we attempted to disrupt this interaction by site-directed mutagenesis of ΔN16Fis1 followed by pull-down experiments with Dnm1. Point mutants of residues proximal to the Fis1 arm (Q21E, I24K, Q28A, Q40A, N44A, and Q112E) did not substantially affect the Fis1-Dnm1 interaction (Fig. 7A), but point mutants of residues that line the concave surface of the Fis1 TPR-like domain (R77E, Y82E, I85E, and K89E) dramatically decreased the Fis1-Dnm1 interaction (Fig. 7A). This result did not arise from misfolding of the Fis1 mutants, since they were competent for Mdv1 binding (Fig. 5C). The disruptive mutants all lie on α-helix 4, which is the central helix of the concave surface created by α-helices 2, 4, and 6 (Fig. 5, A and B). The residues on this surface of α-helix 4 are highly conserved and shielded from solvent by interactions with the Fis1 arm (Fig. 7B). These results suggest that Dnm1 binds to the concave surface on the Fis1 TPR-like domain, and the Fis1 arm regulates access to this binding surface.
FIGURE 7. The Fis1 arm regulates access to a Dnm1 binding pocket.
A, GST pull-down experiments between point mutants of ΔN16Fis1 and MBP-Dnm1. The binding is reported as fraction of Dnm1 bound, which is measured as the ratio of anti-MBP to anti-GST relative to ΔN16Fis1-GST binding of MBP-Dnm1 (lane 2). The error bars represent the S.D. from at least three independent experiments. Experiments were performed by GST pull-down as in Fig. 2A. B, structural representation of Dnm1 binding data showing Fis1 mutants that did (red) or did not (blue) affect Dnm1 binding. Residues that were not tested are colored light gray. The Fis1 arm was not present in these experiments but is shown in yellow for illustrative purposes. The figure was made using PyMOL (W. L. DeLano; available on the World Wide Web).
DISCUSSION
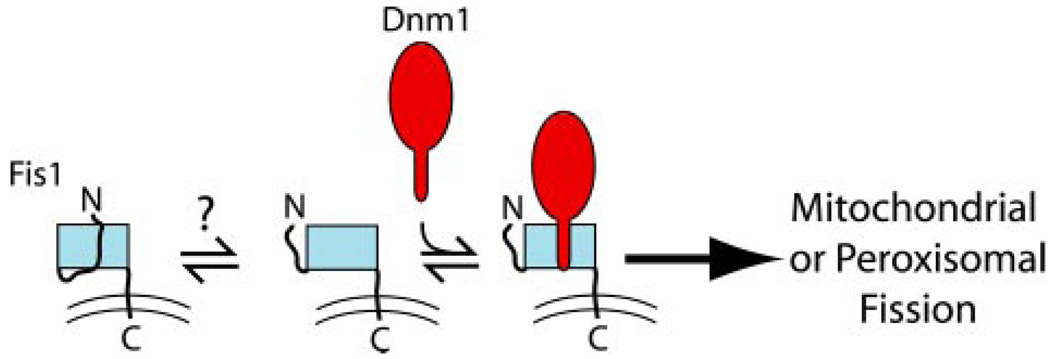
In this report, we use in vitro pull-down assays to test protein-protein interactions thought to be essential for fission of the mitochondrial outer membrane and peroxisomes of S. cerevisiae. We find that Dnm1 and Mdv1 bind directly to Fis1; Mdv1 binds via its NTE domain and does so independently of its coiled-coil domain, its WD repeat domain, or other proteins. Dnm1 binds directly into the concave surface of the Fis1 TPR-like domain and does so independently of Mdv1. The Fis1 arm appears to regulate access to the Dnm1 binding site (Fig. 8).
FIGURE 8. Autoinhibition model for Fis1 binding of Dnm1.
The Fis1 arm blocks access to the Dnm1 binding pocket. Upon an unknown stimulus (protein binding, phosphorylation, membrane) the Fis1 arm autoinhibition may be relieved to allow Dnm1 binding.
Our Fis1 mutagenesis data on Dnm1 binding suggests an interaction between these two proteins that does not share a large buried surface area. The disruptive mutants of Fis1 lie in a narrow groove of the concave surface that is primarily hydrophobic (Fig. 7B). Point mutants in either α-helix 2 (Q40A and N44A) or α-helix 6 (Q112E) that lie adjacent to α-helix 4 did not affect Dnm1 binding (Fig. 5, A and B). This finding is consistent with binding by a part of Dnm1 in extended conformation, such as the N or C terminus or an unstructured region between domains. Similar interactions are observed in other TPR domain proteins. The TPR1 domain of Hop specifically recognizes the C-terminal heptapeptide of Hsp70, whereas the TPR2A domain of Hop binds the C-terminal pentapeptide of Hsp90 (51). Also, the TPR domain of Pex5, a protein involved in peroxisomal targeting, recognizes a C-terminal tripeptide motif that is in extended conformation (52). Identifying the Dnm1 residues important for binding to Fis1 will be the focus of future experiments.
Our results indicate that Mdv1 does not bind into the conserved surface of the Fis1 TPR-like domain. However, we cannot rule out that this surface mediates interactions with the C-terminal, WD repeat domain of Mdv1, which is not present in our constructs. Yeast two-hybrid data conflict on whether this interaction is possible (38, 39). Mdv1 is found only in fungi (40, 41), so we tested residues Gln28, Gln40, and Asn44 because they are more conserved in fungi (supplemental Fig. 1). However, point mutants of these residues did not affect Mdv1 binding (Fig. 5). Based on sequence considerations, Fis1 residues on the convex side of the TPR-like domain between α1 and α2 are more conserved in fungi than mammals. Future studies will test the role of these Fis1 residues (Arg42 and Tyr45) in Mdv1 binding.
Given the new information presented in this study, previous in vivo observations may be better understood. Several laboratories have reported interactions between Fis1 and Mdv1 by cytological or yeast two-hybrid experiments (25, 26, 35, 36, 38–42). However, many attempts to observe an interaction between Fis1 and Dnm1 by yeast two-hybrid analyses have not revealed this interaction, although a recent report that budding yeast Fis1 and Dnm1 co-immunoprecipitate suggests that a direct interaction is possible (42). This discrepancy may now be resolved in light of the present work, which suggests that a likely explanation for the lack of the yeast two-hybrid interaction may arise from the autoinhibition of Fis1 by its N-terminal arm.
Fis1 autoinhibition of Dnm1 binding may be relieved in the presence of Mdv1. We attempted to test this possibility with our constructs in a competition assay but did not observe an increase in Fis1-Dnm1 interaction upon increasing concentrations of Mdv1ΔWD (supplemental Fig. 4). These data suggest that the NTE and CC domains are insufficient to enhance Dnm1 binding to Fis1. Perhaps full-length Mdv1 or another protein is necessary for relieving the autoinhibition of Fis1. Alternatively, it may be the presence of the mitochondrial outer membrane or a covalent modification, such as phosphorylation. Intriguingly, the Dbl homology domain of the proto-oncogene Vav also contains an N-terminal arm that inhibits Rho GTPase activation, and this inhibition is relieved by tyrosine phosphorylation that exposes the GTPase interaction surface (48). Fis1 contains many putative phosphorylation sites, but whether this is important in regulating Fis1 activity in mitochondrial dynamics has not been reported to our knowledge.
The loss of the Fis1 arm in vivo alters mitochondrial morphology (40), which appeared to arise from failed recruitment of Mdv1 to mitochondria, and subsequently has been shown to be rescued by Mdv1 overexpression (41). Our data are consistent with these physiological results, since they suggest that the Fis1 arm enhances the Mdv1 interaction. In the absence of this arm, we observe a decrease in binding to both Mdv1 constructs (Fig. 4). Thus, we think the reported loss of Mdv1 recruitment with ΔN-Fis1 in vivo arises from decreased binding between Mdv1 and ΔN-Fis1 that we detect in vitro. This decrease in affinity appears small enough to be rescued by Mdv1 overexpression in vivo as reported (41).
Our results may also help to explain the aberrant mitochondrial morphology of a temperature-sensitive allele of Fis1 that arises from mutations in three residues (E78D/I85T/Y88H) that lie in α-helix 4 of the TPR-like domain (41). This temperature-sensitive phenotype is suppressed by either Mdv1 overexpression or a point mutant in the CC domain of Mdv1, implying a direct interaction between the conserved surface of Fis1 and the CC domain of Mdv1 (41). Our mutagenesis data suggests another possible interpretation; perhaps this Fis1 triple mutant affects Dnm1 binding that results in aberrant morphology. In this situation, overexpression of Mdv1 might then be expected to stabilize Dnm1 on the Fis1 surface due to the reported interaction between the WD repeat domain of Mdv1 and Dnm1 (25, 35, 38, 39). Additionally, the point mutation in the CC domain may serve to relax an autoinhibitory ability of the Mdv1 protein. Consistent with this possibility, overexpression of an Mdv1 construct containing both NTE and CC domains inhibits mitochondrial fission (39).
A direct interaction between yeast Fis1 and Dnm1 has implications for the mammalian orthologs, hFis1 and hDrp1. Previous studies show that hFis1 and hDrp1 are in close proximity by fluorescence resonance energy transfer and that these two proteins co-immunoprecipitate together (27, 43). We predict here that these results arise from a direct interaction between these proteins in a manner similar to the yeast molecules. Whereas recombinant human Fis1 and Drp1 were tested for a stable interaction that was not found (30), perhaps human Fis1 is also autoinhibited by its own N-terminal arm in a manner similar to the yeast molecule. Two lines of evidence support this speculation. First, an hFis1 construct lacking its first 20 residues co-immunoprecipitates with hDrp1 to a much greater extent than Fis1 with a native N terminus (43). Second, structural studies on the human and mouse Fis1 molecules suggest that such autoinhibition may be possible, although the mammal Fis1 arm is eight residues shorter than the yeast arm. The mouse Fis1 solution structure reveals the Fis1 arm blocking access to the TPR-like domain,5 whereas the human Fis1 solution structure allows access (45). These differences speak to the dynamic nature of the Fis1 arm, since the amino acid sequences only differ by eight mostly conserved substitutions that are not proximal to either the Fis1 arm or the conserved surface. Thus, human Fis1 may also be autoinhibited in a manner similar to the yeast molecule. Alternatively, the Fis1 arm may mediate self-association to restrict access to the TPR-like domain, as observed in the human Fis1 crystal structure (46).
In conclusion, we present data indicating that the S. cerevisiae Fis1 arm acts in an autoinhibitory manner to regulate access to a binding pocket that is evolutionarily conserved for binding the dynamin-like GTPase involved in fission of the mitochondrial outer membrane and peroxisomes (Fig. 8). We anticipate that this mechanism is evolutionarily conserved.
Supplementary Material
Acknowledgments
We thank David M. Merrick for technical expertise, Kevin R. MacKenzie for careful reading of the manuscript, and Kevin R. MacKenzie and John M. Flanagan for helpful discussions during the course of this work. We also thank members of the Hill laboratory.
Footnotes
This work was supported by National Institutes of Health Grant RO1GM067180 and Training Grant 2T32GM007231 (to R. C. W., L. K. P., and F. J. T.) and American Cancer Society Award IRG-58-005-41.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Supported in part by the Howard Hughes Medical Institute summer under-graduate research fellowship program.
Supported in part by a Department of Defense NDSEG fellowship, and a Millipore Foundation Dimitri V. d’Arbeloff award.
The abbreviations used are: NTE, N-terminal extension of Mdv1; CC, coiled-coil; TM, transmembrane domain; GST, glutathione S-transferase; MBP, maltose-binding protein; TPR, tetratricopeptide repeat.
W. Ohashi, H. Hirota, T. Yamazaki, S. Koshiba, T. Hamada, M. Yoshida, and S. Yokoyama, submitted for publication.
S. A. Casares, L. K. Picton, A. C. Monahan, A. Majumdar, and R. B. Hill, submitted for publication.
Supplemental Material can be found at: http://www.jbc.org/cgi/content/full/M700807200/DC1
REFERENCES
- 1.Bereiter-Hahn J, Voth M. Microsc. Res. Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 2.Johnson LV, Walsh ML, Chen LB. Proc. Natl. Acad. Sci. U. S. A. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw JM, Nunnari J. Trends Cell Biol. 2002;12:178–184. doi: 10.1016/s0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott SV, Cassidy-Stone A, Meeusen SL, Nunnari J. Curr. Opin. Cell Biol. 2003;15:482–488. doi: 10.1016/s0955-0674(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 5.Gorsich SW, Shaw JM. Mol. Biol. Cell. 2004;15:4369–4381. doi: 10.1091/mbc.E03-12-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youle RJ, Karbowski M. Nat. Rev. Mol. Cell. Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 7.Jendrach M, Pohl S, Voth M, Kowald A, Hammerstein P, Bereiter-Hahn J. Mech. Ageing Dev. 2005;126:813–821. doi: 10.1016/j.mad.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Chan DC. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 10.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. J. Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basanez G, Hardwick JM. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jagasia R, Grote P, Westermann B, Conradt B. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 13.Delivani P, Adrain C, Taylor RC, Duriez PJ, Martin SJ. Mol. Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 14.Parone PA, James DI, Da Cruz S, Mattenberger Y, Donze O, Barja F, Martinou JC. Mol. Cell. Biol. 2006;26:7397–7408. doi: 10.1128/MCB.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olichon A, Guillou E, Delettre C, Landes T, Arnauné-Pelloquin L, Emorine LJ, Millo V, Daloyau M, Hamel C, Amati-Bonneau P, Bonneau D, Reynier P, Lenaers G, Belenguer P. Biochim. Biophys. Acta. 2006;1763:500–509. doi: 10.1016/j.bbamcr.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Leinninger GM, Edwards JL, Lipshaw MJ, Feldman EL. Nat. Clin. Pract. Neurol. 2006;2:620–628. doi: 10.1038/ncpneuro0320. [DOI] [PubMed] [Google Scholar]
- 17.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. N. Engl. J. Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 18.Legesse-Miller A, Massol RH, Kirchhausen T. Mol. Biol. Cell. 2003;14:1953–1963. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. J. Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. J. Biol. Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Gould SJ. J. Biol. Chem. 2003;278:17012–17020. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 22.Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M. J. Biol. Chem. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 23.Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. J. Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 24.Fekkes P, Shepard KA, Yaffe MP. J. Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tieu Q, Nunnari J. J. Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mozdy AD, McCaffery JM, Shaw JM. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobs S, Martini N, Schauss AC, Egner A, Westermann B, Hell SW. J. Cell Sci. 2003;116:2005–2014. doi: 10.1242/jcs.00423. [DOI] [PubMed] [Google Scholar]
- 29.James DI, Parone PA, Mattenberger Y, Martinou JC. J. Biol. Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 30.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. J. Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 31.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. Mol. Biol. Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sesaki H, Jensen RE. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerveny KL, McCaffery JM, Jensen RE. Mol. Biol. Cell. 2001;12:309–321. doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffin EE, Graumann J, Chan DC. J. Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schauss AC, Bewersdorf J, Jakobs S. J. Cell Sci. 2006;119:3098–3106. doi: 10.1242/jcs.03026. [DOI] [PubMed] [Google Scholar]
- 38.Tieu Q, Okreglak V, Naylor K, Nunnari J. J. Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerveny KL, Jensen RE. Mol. Biol. Cell. 2003;14:4126–4139. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki M, Neutzner A, Tjandra N, Youle RJ. J. Biol. Chem. 2005;280:21444–21452. doi: 10.1074/jbc.M414092200. [DOI] [PubMed] [Google Scholar]
- 41.Karren MA, Coonrod EM, Anderson TK, Shaw JM. J. Cell Biol. 2005;171:291–301. doi: 10.1083/jcb.200506158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhar D, Karren MA, Babst M, Shaw JM. J. Biol. Chem. 2006;281:17312–17320. doi: 10.1074/jbc.M513530200. [DOI] [PubMed] [Google Scholar]
- 43.Yu T, Fox RJ, Burwell LS, Yoon Y. J. Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- 44.Deleted in proof
- 45.Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N. J. Mol. Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. [DOI] [PubMed] [Google Scholar]
- 46.Dohm JA, Lee SJ, Hardwick JM, Hill RB, Gittis AG. Proteins. 2004;54:153–156. doi: 10.1002/prot.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blatch GL, Lassle M. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 48.Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 50.Studier FW. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 52.Gatto GJ, Jr, Geisbrecht BV, Gould SJ, Berg JM. Nat. Struct. Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.