Summary
C/EBP family proteins are important regulators of liver functions. Here we show the critical role of C/EBPα-mediated chromatin remodeling in the age-associated dysfunctions of the liver and in the maintenance of physiological homeostasis. Because ph-S193 isoform of C/EBPα is increased in livers of old mice, we have generated C/EBPα-S193D knockin mice which mimic the ph-S193 isoform of C/EBPα. Analyses of these mice showed that the S193D mutation causes chromatin remodeling leading to histological appearance of “foci-like” nodules, which are also observed in livers of old mice. These “foci–like” structures contain K9 thrimethylated histone H3, a marker of heterochromatin. The increase of heterochromatin regions in S193D mice correlates with the elevation of S193D-C/EBPα-HDAC1 complexes and with dys-regulation of gene expression including epigenetic silencing of cyclin D1 and D2 promoters and the inhibition of liver proliferation. The elimination of C/EBPα-HDAC1 complexes in S193D mice by inhibition of HDAC1 corrects chromatin structure and normalizes expression of cyclin D1 and D2. We found that epigenetic dys-regulation is also associated with the elevation of C/EBPβ and with the increase of C/EBPα/β hetero-dimers in S193D mice. The C/EBPα/β hetero-dimers activate transcription of Glut4 and increase levels of Glut4. As the result, S193D livers have accelerated uptake of glucose and accumulation of glycogen in the liver. Thus, this study demonstrates that phosphorylation of C/EBPα at S193 leads to the appearance of heterochromatin regions which correlates with the development of age-related dysfunctions of the liver.
Keywords: epigenetic control, liver, body homeostasis, C/EBPα, proliferation, aging
Introduction
Senescent liver is characterized by many alterations in biological processes leading to the decline of liver functions and to the alterations in body homeostasis (Schmucker, 2005; Timchenko, 2009). The most dramatic alteration in liver functions is the impaired liver regeneration after partial hepatectomy (PH) and after surgical resections (Bucher et al., 1964; Fry et al., 1984, Iakova et al., 2003, Conboy et al., 2005; Geilchinsky et al., 2010). The decline of the regenerative capacity of old livers is mediated by the appearance of the C/EBPα-Brm-HDAC1 complex (Iakova et al., 2003, Conboy et al., 2005; Wang et al., 2006) and following epigenetic silencing of E2F-dependent promoters (Wang et al., 2007; 2008a; Timchenko 2009). Recent reports have shown that the reduction of GSK3β in aged livers causes the age-associated elevation of the C/EBPα-HDAC1 complexes (Jin et al., 2009a; 2009b; 2009c). In agreement with these observations, Seo et al have found that the inhibition of GSK3β leads to development of senescence phenotype in human liver-derived Chang cells (Seo et al., 2008). The aging liver is also characterized by changes in the size of hepatocytes. Chipchase et al have found that livers of two years old mice have increased number of hepatocytes containing enlarged nuclei with increased ploidy; while livers of 3 months old mice do not contain enlarged hepatocytes (Chipchase et al., 2003). The premature liver polyploidy has been also described in DNA repair (Ercc1) deficient mice (Chipchase et al., 2003). One of the characteristics of the old livers is a lipid accumulation which leads to the development of steatosis (Kuk et al., 2009). It has been also shown that the activities of alanin transaminase (ALT) and aspartate transaminase (AST) and concentration of triglycerides are increased in the blood of old animals as the result of liver dysfunctions (Schmucker, 2005). Molecular basis for these age-associated dysfunctions of the liver has been not elucidated.
The liver is able to regenerate itself after partial hepatectomy (Fausto et al., 2006). The regeneration of the liver after PH is a very complex process which includes reorganizations of the networks of several signal transduction pathways (Michalopoulos, 2007). Despite the global re-organization of these pathways, the remaining portion of the liver proliferates and it is able completely support its differentiation functions and body homeostasis. The molecular basis for these enormous capabilities of the small remaining portion of the liver is not known. Two members of C/EBP family, C/EBPα and C/EBPβ, are expressed in the liver and play critical role in the regulation of liver biology (Timchenko, 2009; Johnson, 2005). Transcription factor C/EBPα is an important component of network since C/EBPα knockout mice die shortly after birth due to impaired energy homeostasis (Wang et al., 1995). C/EBPα displays many of its functions through direct interactions with proteins. C/EBPα interacts with the SWI/SNF chromatin remodeling complex and activates expression of genes involved in adipogenesis (Pedersen et al., 2001). The interaction of C/EBPα with Brm is involved in the inhibition of cell proliferation (Iakova et al., 2003, Conboy et al., 2005; Muller et al., 2004). In aging liver, C/EBPα is hyperphosphorylated at S193 by cyclin D3-cdk4 and is associated with histone deacetylase 1, HDAC1, and with heterochromatin protein 1α, HP1α (Wang et al., 2008a; 2008b). In agreement with these observations, Demarco et al have identified a direct interaction of C/EBPα with HP1α in the regions of centromeric heterochromatin (Demacro et al., 2006).
We have recently generated S193D-C/EBPα knockin mice and have shown that liver proliferation after PH is completely inhibited in these mice (Wang et al., 2010). Here we examined the role of phosphorylation of C/EBPα at S193 in the age-associated decline of liver functions. We found that the age-related mutation of C/EBPα leads to the significant increase of heterochromatin regions and that these alterations are similar to those observed in old mice which have abundant ph-S193 isoform of C/EBPα. The elevation of heterochromatin regions correlates with the development of age-like dys-functions such as inhibition of liver proliferation, accumulation of glycogen in the liver and the elevation of ALT/AST and triglycerides in the blood.
RESULTS
The C/EBPα–S193D increases size of hepatocytes and alters chromatin structure
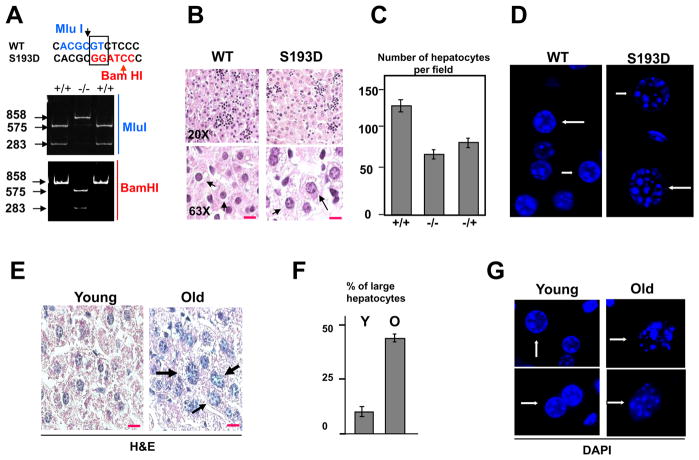
In these studies, we have examined liver functions in C/EBPα-S193D knockin mice, which have been recently generated (Wang et al., 2010). The Fig 1A shows an example of genotyping of mice used for these studies. We have mutated serine to aspartate by TC-GA substitution which also created new BamHI site and destroyed MluI site (Fig 1A). In the course of examination of S193D mice, we found that S193D isoform significantly changes liver morphology. The H&E staining showed that the size of hepatocytes and size of nuclei are significantly increased in livers of C/EBPα-S193D mice compared to WT mice (Fig 1B). The size of nuclei in S193D mice is around 2-fold bigger than that in WT mice (see bottom part of the Fig 1B). Since the liver weight/body weight ratio is not changed in S193D mice, the increase of the size of hepatocytes is consistent with a reduction of the number of hepatocytes in the liver down to 50% (Fig 1C). Since further studies showed that liver proliferation is inhibited in S193D mice during postnatal liver development (Fig 6), these observations suggested that normal size of the S193D livers is supported by increased cell growth leading to the increased size of hepatocytes. To determine if the alterations of the size of nuclei in S193D mice change chromatin structure, we have performed DAPI staining of livers of 2 mo old mice. Consistent with H&E staining, the DAPI staining showed that hepatocytes in C/EBPα-S193D livers have a larger size of nucleus. In addition to that, we found that hepatocytes in S193D livers have a significant increase of “foci-like” nodules (Fig 1D). Since livers of old mice are characterized by increased size of nuclei (Chipchase et al., 2003), we have next performed H&E and DAPI staining of livers of WT young (2 mo) and old (24 mo) mice (Fig. 1E–G). We found that up to 35–40% of hepatocytes are enlarged in livers of old mice (Fig 1F). Examination of chromatin structure by DAPI staining revealed the abundance of “foci-like” nodules in nuclei of livers from old mice, especially in cells with enlarged nuclei (Fig 1G and see additional images in Supplemental Figure 1).
Figure 1. The S193D isoform of C/EBPα causes alterations in liver morphology. A. An example of genotyping of the C/EBPα-S193D mice.
Upper image shows the sequence surrounding S193. TC nucleotides were mutated to GA leading to the disruption of the restriction site for MluI and creation of restriction site for BamHI. PCR products of 858 bp were generated using DNA from littermates and examined by restriction with MluI and BamHI (Bottom image). B. Livers of C/EBPα-S193D mice contain larger hepatocytes with enlarged nuclei. Livers of WT and S193D mice were stained with H&E. Images taken under 63X magnification are shown on the bottom. The hepatocytes are shown by arrows. Red lines show scale bars: 10 μm. C. Bar graphs show number of hepatocytes per field under 20X magnification. D. Chromatin structure is altered in hepatocytes of C/EBPα-S193D mice. WT and C/EBPα-S193D livers were stained with DAPI. A typical picture of DAPI staining is shown. Arrows show nuclei of hepatocytes. Original magnification, x63. E. A significant portion of hepatocytes in old mice has an increased size. A typical picture of H&E staining of livers of young and old mice is shown. Arrows show enlarged hepatocytes. Red lines show scale bars: 10 μm. F. Bar graphs show percentage of enlarged hepatocytes in livers of young and old mice. G. DAPI staining of livers from young and old mice. Two images of each age group are shown.
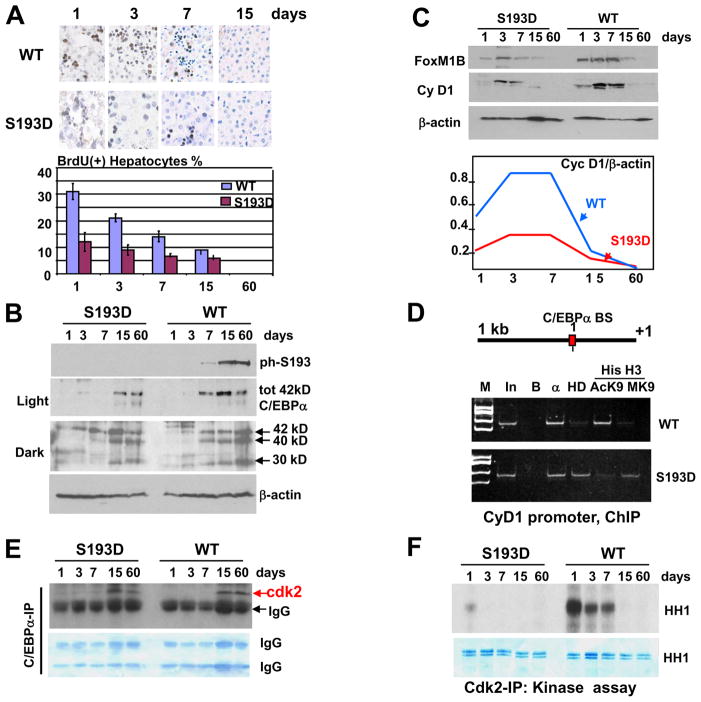
Figure 6. C/EBPα-S193D inhibits liver proliferation during post-natal development via repression the cyclin D1 promoter and inhibition of cdk2. A. C/EBPα-S193D mutant inhibits hepatocyte DNA synthesis during postnatal liver development.
BrdU uptake was examined in the livers of 1, 3, 7, 15 and 60 days old mice. Bar graphs show a summary of three independent experiments. B. Expression of C/EBPα during postnatal development. Western blotting was performed with nuclear extracts isolated from WT and S193D livers at different age after birth (shown on the top) using antibodies to total C/EBPα and to ph-S193 isoform of C/EBPα. Middle image shows a dark exposure of C/EBPα filter. The membranes were reprobed with β-actin. C. Expression of FoxM1B and cyclin D1 was determined by Western blotting. The levels of cyclin D1 were calculated as ratios toβ-actin (bottom image). D. C/EBPα-HDAC1 complex represses D1 promoter. Upper diagram shows locations of C/EBPα sites on the cyclin D1 promoter. ChIP assay was performed with 7 day old animals. M; molecular weight markers; In; input, B; agarose beads. HD; IP with antibodies to HDAC1, AcK9 and MK9 are IPs with antibodies to AcK9 and Trimethyl-K9 histone H3. E. C/EBPα-S193D is associated with cdk2 at all stages of postnatal development. C/EBPα was immunoprecipitated from nuclear extracts; and cdk2 was examined in these IPs. Commassie stain of the membrane is shown below. F. C/EBPα-S193D inhibits kinase activity of cdk2 in postnatal livers. Cdk2 was immunoprecipitated from nuclear extracts and examined in kinase assay with histone H1 (HH1) substrate. The bottom image shows a coomassie stain of the membrane to verify loading of HH1.
“Foci-like” structures in young S193D mice and in old wild type mice are heterochromatin regions
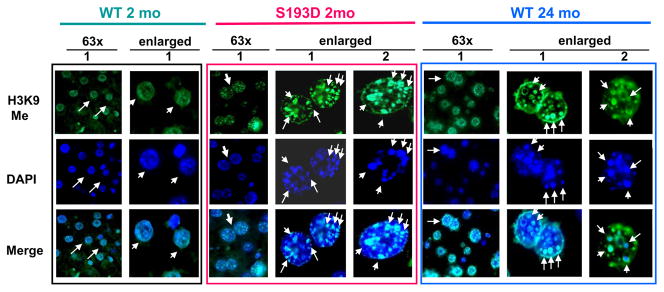
We have next examined if the “foci-like” structures in S193D and old WT hepatocytes might be heterochromatic regions. Livers of young WT, young S193D and old WT mice were stained with antibodies to the marker of heterochromatin, histone H3 trimethylated at K9 (H3K9Me). Figure 2 shows typical pictures of the staining. Nuclei of hepatocytes of young WT mice show mainly diffused staining of H3K9Me with a rare punctuated location of H3K9Me. On the contrary, nuclei in hepatocytes of young S193D mice and nuclei in large hepatocytes of old WT mice contain a much bigger number of H3K9Me located in “foci-like” structures. In S193D livers, around 80–90% of hepatocytes contain H3K9Me in “foci-like” structures. It is interesting to note that, in livers of old mice, “foci-like” structures determined by DAPI staining and H3K9me are mainly co-localized in enlarged hepatocytes. Thus, these studies show that the S193D mutation of C/EBPα leads to the elevation of heterochromatin regions. Similar alterations are observed in large hepatocytes of aged livers.
Figure 2. “Foci-like” structures in hepatocytes of young S193D and old WT mice contain a marker of heterochromatin; histone H3 trimethylated at K9 (H3K9me).
Livers of WT young (2mo), S193D young (2mo) and WT old (24 mo) mice were stained with antibodies to H3K9-trimethyl and with DAPI. Bottom panel shows the merge. 63x; shows bigger fields of the livers with multiple cells per field. These pictures were taken under original magnification 63X. Arrows mark nuclei of hepatocytes which are shown on the right side as enlarged images. Two-three typical pictures of enlarged nuclei are shown for each genotype on the right (enlarged). Arrows on enlarged images show big H3K9me foci which co-localize with DAPI foci.
Livers of S193D mice contain abundant C/EBPα-HDAC1 complexes and elevated levels of C/EBPβ
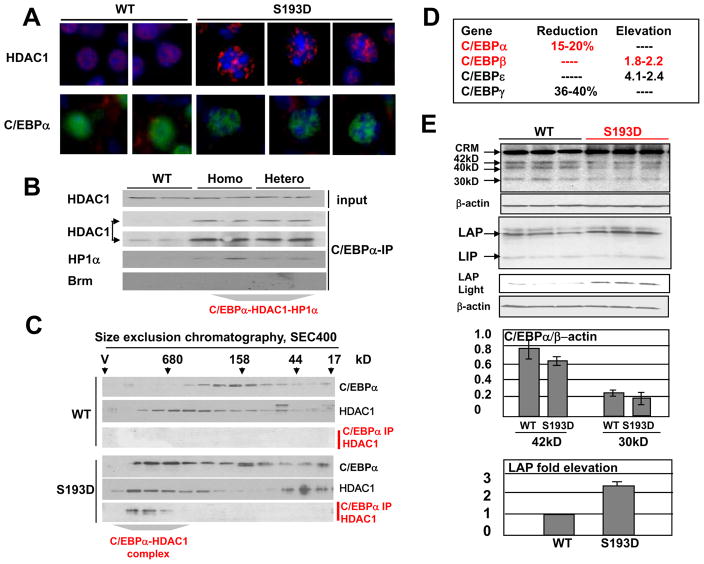
We have next examined pathways by which C/EBPα–S193D mutant causes alteration of chromatin structure. Our previous studies have shown that the S193-ph isoform of C/EBPα form complexes with a chromatin remodeling protein histone deacetylase 1, HDAC1, in livers of old mice (Wang et al., 2008a). Therefore, we determined if the C/EBPα-S193D might affect chromatin structure via interactions with HDAC1. Staining of livers of 2 mo old mice with Abs to HDAC1 and C/EBPα showed that livers of WT mice have a diffuse nuclear staining for both HDAC1 and C/EBPα; while HDAC1 and C/EBPα are observed in punctuate “foci-like” structures of the C/EBPα-S193D livers (Fig 3A). We have next examined interactions of C/EBPα with HDAC1 and with heterochromatin protein α, HP1α, using Co-IP approach and HPLC-based size exclusion chromatography. HP1α has been included in the studies since this protein cooperates with HDAC1 in the repression of promoters and because this protein interacts with C/EBPα (Wang et al., 2008a; Demacro et al., 2006). Co-IP approach showed that larger amounts of HDAC1 and HP1α are observed in the complexes with the mutant C/EBPα; while C/EBPα-HDAC1 complex is detected in WT mice only after longer exposure (Fig 3B). Previous studies have shown that the livers of old mice contain the C/EBPα-Brm complex (Iakova et al., 2003; Conboy et al., 2005) and that this complex is up-regulated by phosphorylation of C/EBPα at S193 (Wang et al., 2007). Therefore, we have examined if Brm might be in the complexes with C/EBPα in young S193D mice. Under sensitivity of our assay, we could not detect Brm in C/EBPα IPs (Fig 3B). The lack of Brm in C/EBPα complexes is perhaps associated with the very low levels of Brm in livers of 2 mo old mice. To confirm results of Co-IP and to determine the size of the C/EBPα-HDAC1 complex, we have utilized HPLC-based techniques. These studies showed that the complex is significantly larger (around 680kD) than the sum of masses of C/EBPα, HDAC1 and HP1α suggesting that it might contain additional components (Fig 3C).
Figure 3. Alterations of the chromatin structure in S193D mice are associated with an increase of C/EBPα-HDAC1 complexes. A. C/EBPα and HDAC1 are observed in foci-like structures in hepatocytes of S193D mice.
Livers were immunostained with antibodies to HDAC1 and C/EBPα. Two images for WT and three images for S193D mice are shown. Original magnification, x63. B. HDAC1 and HP1α form a complex with C/EBPα in C/EBPα-S193D mice. Proteins were immunoprecipitated with Abs to C/EBPα and examined by Western blotting with Abs to HDAC1, Brm1 and to HP1α. Light and dark exposures are shown for HDAC1. C. Size exclusion chromatography. Nuclear extracts of WT and S193D livers were fractionated by gel filtration and the fractions were examined by Western blotting with Abs to C/EBPα and HDAC1. C/EBPα-IP: HDAC1 Western: C/EBPα was immunoprecipitated from each fraction and examined by Western blotting with antibodies to HDAC1. D. Alterations of mRNA levels of C/EBP family proteins in livers of S193D mice. The table shows results of micro array studies which were further confirmed by Q-RT-PCR. E. Protein levels of C/EBPα and C/EBPβ in WT and in S193D livers. Three animals of each genotype were examined by Western blotting with Abs to C/EBPα and to C/EBPβ. Short exposure for the section with C/EBPβ-LAP isoform is shown. The filter was re-probed with β-actin. 42kD, 40kD and 30kD isoforms of C/EBPα and LAP and LIP isoforms of C/EBPβ are shown. CRM, cross reactive molecule. Middle image. Bar graphs show levels of C/EBPα 42kD and 30kD isoforms as ratios to β-actin. Bottom image. Bar graphs show levels of C/EBPβ-LAP as ratios to β-actin.
Given the elevation of heterochromatin regions in hepatocytes of S193D mice, we suggested that S193D mice might have changes in the transcription of many genes. Therefore, we have performed micro array studies with livers of 2 months old WT and S193D mice. These studies identified a number of genes with reduced or increased expression on livers of S193D mice (data not shown). In this paper, we have investigated in details a portion of these genes including C/EBP family proteins, genes which are involved in control of liver growth and in the control of glucose metabolism. The alteration in expression of these genes was confirmed by Q-RT-PCR and by Western blotting. These data are presented throughout the manuscript (see Figs 3, 5 and 8). C/EBP family is one of the groups of genes with altered mRNA levels in livers of S193D mice. Figure 3D shows differences in the levels of C/EBP family mRNAs which were further confirmed by Q-RT-PCR. We found that the levels of C/EBPβ and C/EBPε mRNAs are increased in livers of S193D mice, while levels of C/EBPα and C/EBPγ are slightly reduced. To examine if protein levels of C/EBP proteins are changed in S193D mice, we have performed Western blotting analyses. These studies have shown that, although C/EBPε and C/EBPγ mRNAs are detected by RT-PCR, these proteins are not detectable in livers of both WT and S193D mice by Western blotting (data not shown). Examination of C/EBPα showed that it is expressed as three isoform with MW 42kD, 40kD and 30kD. In agreement with a slight reduction of C/EBPα mRNA, the protein levels of these three isoforms of C/EBPα are slightly reduced in livers S193D mice (Fig 3E). However, this reduction is minor and represents less than 20% (Fig 3E, middle bar graphs). Quite different result was observed for C/EBPβ expression. C/EBPβ is also expressed in the liver as three isoforms: full-lengths protein (FL), C/EBPβ-LAP and C/EBPβ-LIP. We have found that protein levels of C/EBPβ-LAP are elevated in livers of S193D mice; while levels of a truncated isoform C/EBPβ-LIP are not changed. Calculation of C/EBPβ-LAP as ratios to β-actin showed around 2.5-fold increase in livers of S193D mice (Fig 3E, bottom). Taken together, these studies showed that the alterations of chromatin structure in S193D mice are associated with elevation of C/EBPα-HDAC1 complexes and with elevation of C/EBPβ.
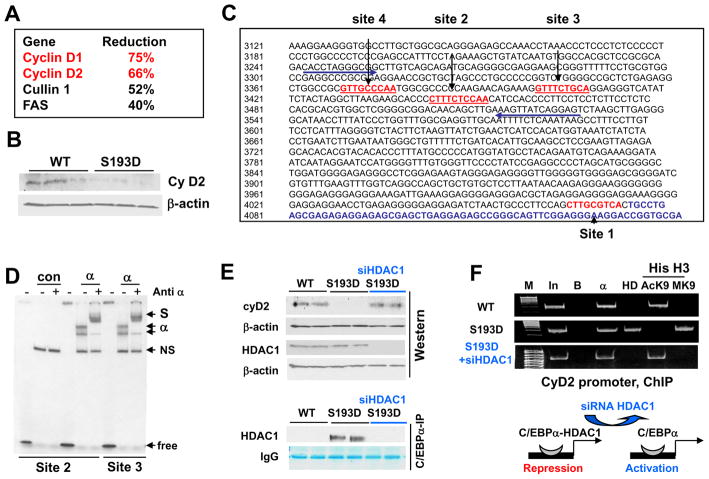
Figure 5. C/EBPα-HDAC1 represses the cyclin D2 promoter. A. List of cell cycle related mRNAs which are reduced in livers of C/EBPα-S193D mice.
The alterations of mRNA were found in micro-array studies and then confirmed by Q-RT-PCR. B. Protein levels of cyclin D2 are reduced C/EBPα-S193D mice. Western blotting was performed with antibodies to cyclin D2. The filter was re-probed with β-actin. C. Location of C/EBPα binding sites within the cyclin D2 promoter. Four C/EBPα sites are shown by red. Arrows show positions of primers used for ChIP experiments. D. C/EBPα binds to the cyclin D2 promoter. C/EBPα was overexpressed in cultured cells and incubated with DNA probes covering sites 2 and 3 of cyclin D2 promoter. Antibodies to C/EBPα were added to the binding reactions as shown on the top. Positions of C/EBPα shift, supershift (S) and free probe are shown on the right. NS, non-specific band. E. Elimination of C/EBPα-HDAC1 complexes in S193D mice increases expression of cyclin D2. Western blotting of cyclin D2 and HDAC1 was performed with nuclear extracts isolated from WT, S193D and S193D mice injected with siRNA to HDAC1. β-actin control is shown for the cyclin D2 filter. Bottom image shows C/EBPα-HDAC1 complexes determined by Co-IP using the same nuclear extracts. F. C/EBPα-HDAC1 complex represses the cyclin D2 promoter. ChIP assay was performed with chromatin solutions from livers of WT, S193D and S193D +siHDAC1 mice using primers covering C/EBPα sites on the promoter of cyclin D2. M; molecular weight markers; In; input, B; agarose beads. HD; IP with antibodies to HDAC1, AcK9 and MK9 are IPs with antibodies to AcK9 and trimethyl-K9 histone H3.
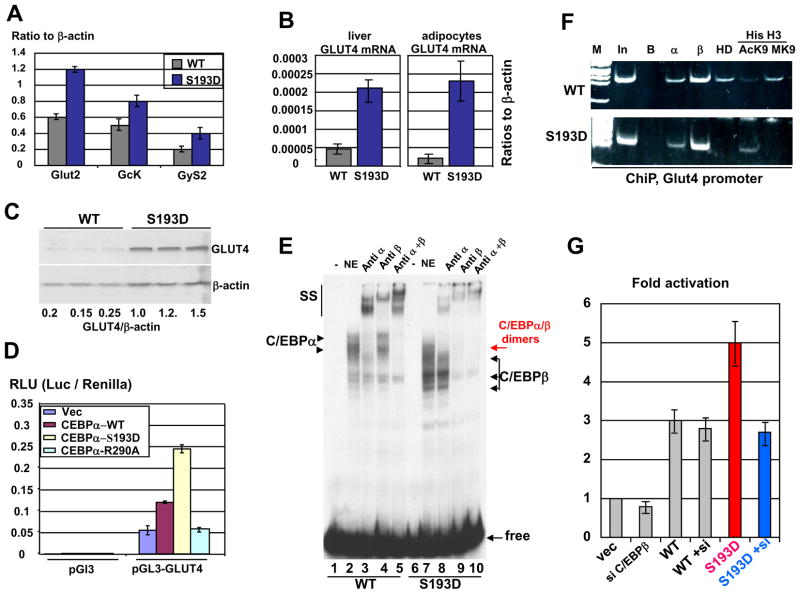
Figure 8. C/EBPα-S193D activates the Glut4 promoter in livers of S193D mice as a hetero-dimer with C/EBPβ. A. Expression of gluconeogenic genes in livers of C/EBPα-S193D mice.
Q-RT-PCR was performed with primers specific to Glut2, Gck and GyS2 mRNAs. The levels of mRNAs were calculated as ratios to β-actin. B. Levels of Glut4 mRNA are increased in livers and in adipose tissues. Levels of Glut4 mRNA were determined in the liver (left) and in adipocytes (right) by Q-RT-PCR. C. Levels of Glut4 protein are increased in livers C/EBPα-193D mice. Western blotting was performed with nuclear extracts from livers of WT and S193D mice using antibodies to Glut4. The membrane was re-probed with Abs to β-actin. D. S193D mutation enhances the ability of C/EBPα to activate the Glut4 promoter. The Glut4 promoter was transfected with C/EBPα proteins into HEK293 cells and the activity of the promoter was calculated and normalized to activity of Renilla. E. Binding of C/EBPβ and C/EBPα:C/EBPβ hetero-dimers to the Glut4 promoter is increased in livers of S193D mice. EMSA was performed with DNA probe covering C/EBP site within the Glut4 promoter and with nuclear extracts isolated from WT and S193D livers. Antibodies to C/EBPα and C/EBPβ were added to the reactions before the probe addition. Positions of C/EBPα and C/EBPβ are shown by arrows. SS; supershift. F. The Glut4 promoter is activated in S193D livers. ChIP assay was performed as described in the text. In; 1/100 input. B; beads. α and β; C/EBPα and C/EBPβ correspondingly. HD; HDAC1. AcK9 and MK9; acetylated and trimethylated histone H3. G. C/EBPβ is required for the increased ability of S193D-C/EBPα to activate the Glut4 promoter. The expression of C/EBPβ was inhibited by siRNA. WT C/EBPα and S193D-C/EBPα were co-transfected with the Glut4 promoter into cells with inhibited C/EBPβ. The activity of the Glut4 promoter was calculated as described above.
The inhibition of HDAC1 in S193D mice corrects size and chromatin structure of nuclei in hepatocytes
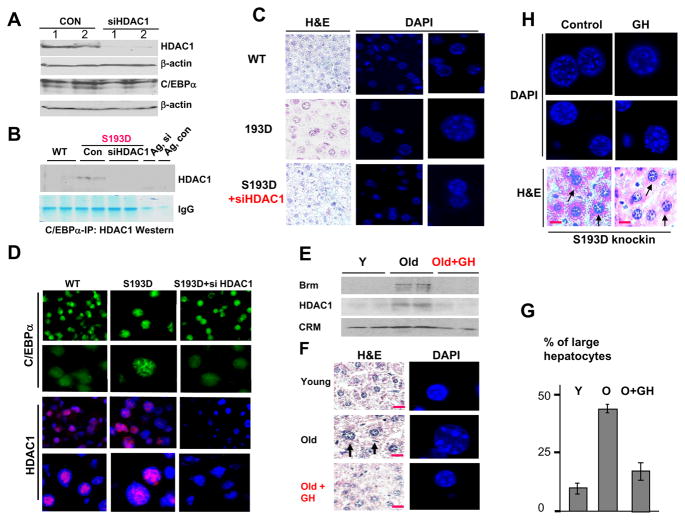
Given the abundance of the C/EBPα-HDAC1 complexes in livers of S193D mice, we suggested that these complexes might be involved in the alterations of chromatin structure. To test this hypothesis, we have inhibited expression of HDAC1 by siRNA in S193D mice and examined chromatin structure of the nuclei. We have previously established conditions for the efficient delivery of siRNA HDAC1 and for a strong inhibition of HDAC1 in the liver (Wang et al., 2008b). The animals were injected with siRNA to HDAC1 for three days and livers were harvested and analyzed. Using these conditions, we have inhibited expression of HDAC1 to less than 15% in livers of C/EBPα-S193D mice (Fig 4A). Examination of C/EBPα-HDAC1 complexes by Co-IP approach showed that siRNA-mediated inhibition of HDAC1 eliminates the C/EBPα-HDAC1 complexes in livers of S193D mice (Fig 4B). H&E staining and DAPI staining showed that the elimination of C/EBPα-HDAC1 complexes correlates with the correction of the size and chromatin structure of the nuclei in hepatocytes (Fig 4C). Staining of the livers with antibodies to C/EBPα and to HDAC1 showed that the reduction of HDAC1 by siRNA re-distributes C/EBPα from foci-like structures to the diffuse location within nucleus. In addition to the correction of chromatin structure, the inhibition of HDAC1 reduces the size of hepatocytes. We suggest that the enlarged size of hepatocytes in S193D mice might be the result of epigenetic alterations and that the elimination of foci-like structures also corrects the size of existing hepatocytes.
Figure 4. Inhibition of HDAC1 and the elimination of the C/EBPα-HDAC1 complexes correct size of hepatocytes and chromatin structure. A. Inhibition of HDAC1 by siRNA to HDAC1.
Western blotting was performed with nuclear extracts isolated from control S193D livers and from S193D livers of mice injected with siRNA to HDAC1. HDAC1 and C/EBPα levels were examined. Each membrane was re-probed with β-actin. B. The inhibition of HDAC1 eliminates C/EBPα-HDAC1 complexes in S193D livers. C/EBPα was immunoprecipitated from livers of WT, S193D and S193D siRNA injected mice and probed with Abs to HDAC1. Signals of heavy chains of IgGs are shown. C. H&E and DAPI staining of the livers of WT, S193D and S193D mice treated with siRNA to HDAC1. Right panel shows DAPI staining. Original magnification, x63. Enlarged images are shown on the right panels. D. Immunostaining of siHDAC1 treated livers with C/EBPα and HDAC1. Bottom images show pictures taken under 63X magnification. E–F. Elimination of C/EBPα-HDAC1 complexes in livers of old mice corrects size of the nuclei and chromatin structure. Old animals were treated with growth hormone (GH) for 3 days. E. C/EBPα was immunoprecipitated from the livers and probed with Abs to HDAC1 and to Brm. F. H&E and DAPI staining of young, old and old mice treated with GH. Representative pictures are shown. Red lines show scale bars: 10 μm. Single cells for DAPI staining are shown in this picture. Supplemental figure 1 contains more images of the DAPI staining. G. Percentage of enlarged hepatocytes in livers of young, old and old GH-treated mice. Bar graphs show a summary of experiments with three animals of each group. H. Treatment of young S193D mice with GH does not change size and chromatin structure of hepatocytes. S193D mice were treated with GH and with PBS (control) and stained with DAPI (upper) and with H&E (bottom) at 3 days of GH injections. Two images of DAPI staining are shown. Arrows show hepatocytes on the H&E staining. Red lines show scale bars: 10 μm.
Previous studies have shown that C/EBPα-HDAC1 complexes are also abundant in livers of old mice (Wang et al., 2008a) suggesting that these complexes might be involved in the age-associated alterations of chromatin structure in old animals. To determine the role of the C/EBPα-HDAC1 complexes in these age-associated alterations, we have initially tried to reduce the complexes by inhibiting HDAC1; however, the siRNA-mediated inhibition of HDAC1 in old mice caused a massive cell death shortly after injection of siRNA. Therefore, we have used an alternative approach to eliminate the complex, which is the treatment of mice with growth hormone (GH). It has been shown that GH activates pathways which de-phosphorylate C/EBPα at S193 and reduce amounts of C/EBPα-HDAC1-Brm complex (Wang et al., 2008a). In agreement with previous reports, the treatments of old mice with GH eliminated the C/EBPα-HDAC1-Brm complex (Fig 4E). H&E staining and examination of nuclei by DAPI staining revealed that the elimination of the C/EBPα-HDAC1-Brm complex also corrects the size and chromatin structure of the nuclei. The figure 4F shows typical pictures of H&E and DAPI staining. We found that the treatment of old mice with GH reduced the amounts of enlarged hepatocytes down to 10–15% (Fig 4G).
Previous studies showed that GH reduces the C/EBPα-HDAC1 complex by activation of pathways which de-phosphorylate C/EBPα at S193. Since S193D mutation prevents elimination of the negative charge on S193, we examined if S193D mutation will prevent GH-mediated correction of the chromatin structure in S193D mice. We found that the treatment of S193D mice with GH is not sufficient to alter chromatin structure and size of hepatocytes in S193D mice (Fig 4H). This result suggested that the S193D mutation took C/EBPα away from the GH-dependent control. Taken together, the studies of S193D mice and examinations of old mice showed that the elevation of the C/EBPα-HDAC1 complex correlates with alterations in chromatin structure and that elimination of these complexes in S193D and in old mice contributes to the correction of chromatin structure.
C/EBPα-HDAC1 complexes reduce levels of cyclin D2 in C/EBPα-S193D livers via repression of the cyclin D2 promoter
We have next examined biological consequences of the change of chromatin structure in S193D mice. The micro array analysis has determined a group of mRNAs which have reduced levels in livers of S193D mice. This group represents genes which are involved in regulation of liver growth including cyclins D1 and D2 (Fig 5A). Since D-type cyclins play critical role in the liver proliferation, we focused our further studies on the promoters of cyclin D1 and D2. Western blotting confirmed that the expression of cyclin D2 is reduced in C/EBPα-S193D livers (Fig 5B); while cyclin D1 protein was not detected in quiescent livers. Therefore, we first examined if C/EBPα-HDAC1 complexes down-regulate cyclin D2 promoter in quiescent livers. Examination of the cyclin D2 promoter identified four consensuses for C/EBPα (Fig 5C). To determine if C/EBPα binds to these sites, we have examined the interaction of C/EBPα with DNA probes covering C/EBPα sites within the cyclin D2 promoter using EMSA. These studies showed that C/EBPα binds to the cyclin D2 promoter in vitro (Figure 5D).
To test if C/EBPα-HDAC1 complexes down-regulate the cyclin D2 in S193D mice, we have examined expression of the cyclin D2 in livers of S193D mice treated with siRNA to HDAC1. Western blotting has shown that the levels of cyclin D2 are normalized in S193D mice by siRNA to HDAC1 and that this normalization correlates with the elimination of the C/EBPα-HDAC1 complexes (Fig 5E). To determine the causal role C/EBPα-HDAC1 complexes in the repression of the cyclin D2 promoter in the livers of S193D mice, we have performed chromatin immunoprecipitation (ChIP) analyses. ChIP studies showed that C/EBPα-HDAC1 complex is abundant on the cyclin D2 promoter in the C/EBPα-S193D livers, while only C/EBPα is observed on this promoter in WT mice (Fig 5F). The elimination of HDAC1-C/EBPα complexes leads to the removal of HDAC1 from the cyclin D2 promoter (Fig 5F). To determine if the cyclin D2 promoter is repressed in S193D livers, we examined modifications of histone H3 associated with the cyclin D2 promoter. K9 acetylated histone H3 occupies the cyclin D2 promoter in WT mice; while tri-methylated H3K9 is not detected. In contrast, the Ac-K9-histone H3 is dramatically reduced on the D2 promoter in livers of C/EBPα-S193D mice, but amounts of trimethylated K9H3 are increased (Fig 5F). The inhibition of C/EBPα-HDAC1 complexes reversed the patterns of modifications of histone H3 on the cyclin D2 promoter. These patterns of histone H3 modifications show that C/EBPα-HDAC1 complex inhibits the cyclin D2 promoter.
C/EBPα-HDAC1 complexes repress cyclin D1 promoter in the liver during post-natal development
Since expression of cyclin D1 protein is not detectable in quiescent livers of 2 months old mice, we have examined the role of C/EBPα-HDAC1 complexes in the regulation of cyclin D1 using mice during early post-natal development when liver proliferates. We have first asked if the C/EBPα-S193D inhibits liver proliferation after birth. Proliferation of the livers during postnatal development was examined by measuring BrdU uptake in 1, 3, 7, 15 and 60 days old mice. Figure 6A shows that the rate of liver proliferation in WT mice is relatively high at day 1 and then declines with age. We have found that the phosphomimetic C/EBPα-S193D reduces liver proliferation at all stages of postnatal liver development. We have next examined mechanisms by which C/EBPα-S193D mutant and ph-S193 isoform inhibit liver proliferation during postnatal liver development. Figure 6B shows that total protein levels of both WT and S193D-C/EBPα are increased with age and the phosphorylation of WT C/EBPα at S193 is detectable at days 7, 15 and 60 in WT mice. Since antibodies to S193-ph isoform of C/EBPα do not recognize S193D mutant (Wang et al., 2006; 2007), no specific signals were observed with protein extracts from the livers of C/EBPα-S193D mice. We have next examined expression of cyclin D1 during postnatal development. In addition to cyclin D1, expression of FoxM1 was examined since this protein is expressed only in proliferating livers and serves as an additional marker of liver proliferation. Figure 6C shows that the levels of cyclin D1 and FoxM1B are significantly lower in C/EBPα-S193D mutant livers during whole course of the studies. Calculations of cyclin D1 levels as ratios to β-actin revealed 2–3-fold higher levels of cyclin D1 protein at days 1–7 in WT mice compared to the levels in S193D mice. The expression of cyclin D1 is inhibited and is not detectable at days 15 and 60 in both WT and S193D mice.
We next asked if the inhibition of cyclin D1 in S193D livers is mediated by C/EBPα-HDAC1 complexes. Examination of the cyclin D1 promoter identified a consensus for C/EBPα to which C/EBPα binds (Supplemental Figure 2). To determine if the C/EBPα-HDAC1 complexes inhibit the cyclin D1 promoter through interactions with the identified site, we have performed ChIP assay with primers covering this site. For the ChIP studies, 7 day old animals were used since the differences in the expression of cyclin D1 between WT and S193D mice are significant at that time point. ChIP studies showed that the cyclin D1 promoter is occupied by C/EBPα in WT mice; while HDAC1 is not detectable on the promoter and histone H3 is acetylaed at K9 (Fig 6D). This pattern of histone H3 modifications shows that the promoter is active. On the contrary, the cyclin D1 promoter is occupied by C/EBPα-HDAC1 complex in S193D mice leading to partial de-acetylation of histone H3 and following trimethylation at K9. These alterations in the histone H3 modifications revealed that the cyclin D1 promoter is partially repressed in livers of S193D mice by C/EBPα-HDAC1 complex.
C/EBPα has several pathways of inhibition of cell proliferation. Previous studies showed that C/EBPα arrests liver proliferation in young mice through binding to and inhibition of cdk2 (Wang et al., 2001; 2004; Tan et al., 2005); therefore we have examined if this pathway might be also involved in the inhibition of liver proliferation during post-natal liver development. Figure 6E shows that cdk2 is associated with the mutant S193D-C/EBPα at all tested stages of postnatal liver development and that this association is proportional to amounts of C/EBPα. On the contrary, the association of cdk2 with C/EBPα in WT mice is observed only in livers of 15 and 60 days old mice. Examination of cdk2 IPs in in vitro kinase assay with histone H1 substrate shows that, in WT livers, cdk2 activity is high at days 1–7 and is reduced at later stages of liver development. However, cdk2 activity is inhibited in C/EBPα-S193D livers at all stages of postnatal development (Fig 6F). Taken together, these data demonstrate that the mutant S193D-C/EBPα and ph-S193 isoform of C/EBPα arrest liver proliferation during postnatal development through repression of the cyclin D1 promoter and via inhibition of cdk2.
Livers of C/EBPα-S193D mice have increased uptake of the glucose and accumulation of glycogen
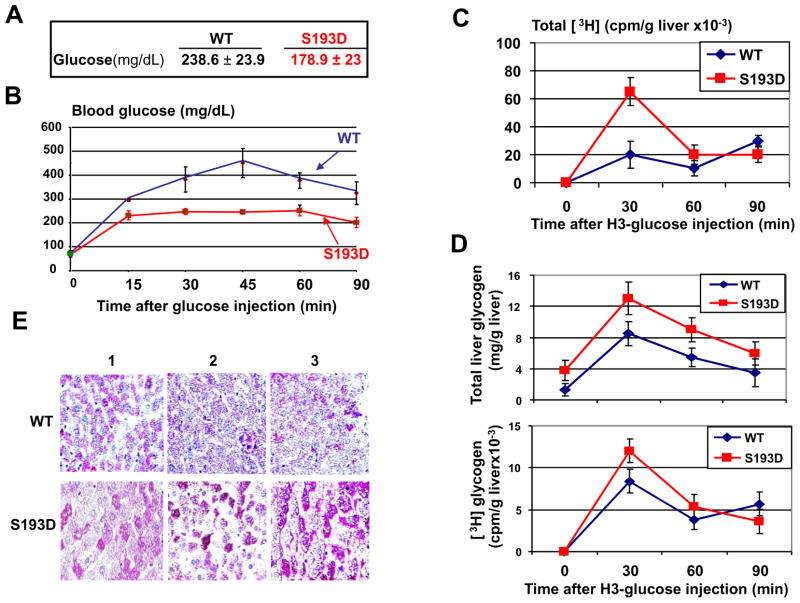
Examination of blood parameters showed that levels of glucose are reduced in S193D mice (Wang et al., 2010 and see Fig 7A). Therefore, we have investigated pathways by which S193D mutation affects glucose levels. The glucose tolerance test revealed that the C/EBPα-S193D animals remove glucose from the blood much faster than WT mice (Fig 7B). To determine if the liver is involved in the increased removing glucose from the blood, we have injected radioactive glucose and have examined the uptake of H3-glucose by livers in WT and in S193D mice. These studies revealed that livers of S193D mice utilize glucose much more efficiently than livers of WT mice (Fig 7C). These data suggest that the reduction of the glucose in the blood of C/EBPα-S193D mice is due to an increased uptake of glucose by the liver. Since one of the pathways of the utilization of the glucose in the liver is the conversion of glucose into glycogen, we have isolated glycogen from WT and S193D livers after injection with H3-glucose. Measurements of total and H3-glycogen showed that the utilized H3-glucose is converted into glycogen (Fig 7D). In agreement with these data, staining of WT and S193D livers for glycogen detected a significant increase of glycogen in livers of C/EBPα-S193D mice (Fig 7E).
Figure 7. C/EBPα-S193D mutant reduces the levels of glucose through activation of uptake of glucose by liver. A. Results of the examination of glucose in the blood of 2 months old WT and C/EBPα-S193D knockin mice.
The summary of analyses of eight animals of each genotype is shown. B. Glucose tolerance test. Mice were fasted for overnight and the glucose was injected. The concentration of glucose in the blood was examined at 15, 30, 45, 60 and 90 min after injection. C. Glucose up-take by the liver in WT and C/EBPα-S193D mice. H3-glucose was injected into mice and the level of H3 radioactivity was examined at different time points after injection. The picture shows amounts of H3 radioactivity per 1g of liver. D. Accumulation of glycogen is increased in livers of C/EBPα-S193D mice. Glycogen was isolated from the livers of mice injected by H3-glucose (see Fig 7C). Upper image shows amounts of total glycogen in livers of injected mice. Bottom image shows amounts of H3-glycogen in the livers of injected mice. E. Livers of S193D mice contain larger amounts of glycogen than livers of WT mice. Livers were stained with PAS. Livers of three animals of each genotype are shown.
The C/EBPα-S193D increases expression of genes involved in utilization of glucose
We have next examined molecular mechanisms by which the S193D mutant increases liver-mediated uptake of glucose in C/EBPα-S193D mice. Examination of expression of genes involved in glucose metabolism by Q-RT-PCR showed that levels of mRNAs coding for the glucose transporters Glut2 and Glut4, GcK and GyS2 are increased in livers of C/EBPα-S193D mice (Fig 8A and B). Since Glut4 is also expressed in adipose tissues, we have examined the levels of Glut4 mRNA in adipocytes and found around 15-fold elevation of Glut4 mRNA in adipose tissue of S193D mice. Because levels of Glut4 mRNA are dramatically activated in both liver and adipocytes, we determined mechanisms of regulation of Glut4 by C/EBPα-S193D. We have focused the studies on the livers of S193D mice, since the S193D liver has increased uptake of glucose. Western blotting confirmed that livers of C/EBPα-S193D mice contain higher levels of Glut4 protein (Fig 8C). We have next asked if there is a difference in the ability of WT C/EBPα and the mutant S193D-C/EBPα to activate the Glut4 promoter. The Glut4 promoter was cloned into pGL3 vector and co-transfected with WT C/EBPα and with S193D-C/EBPα into Hep3B2 cells. In addition to these constructs, C/EBPα-R290A mutant was used since it does not bind to DNA (Miller et al., 2003; Wang et al., 2004). Co-transfection studies showed that the S193D mutation significantly increases the ability of C/EBPα to activate the promoter (Fig 8D). Our further studies in tissue culture systems showed that the cyclin D3-cdk4-mediated phosphorylation of endogenous C/EBPα at S193 enhances its ability to activate the Glut4 promoter (Supplemental Figure 3). Taking together these data, we conclude that the S193D mutation and phosphorylation of C/EBPα at S193 enhance the ability of C/EBPα to activate the Glut4 promoter.
C/EBPβ forms hetero-dimers with C/EBPα-S193D and co-operates with C/EBPα-S193D in the activation of the Glut4 promoter
Because protein levels of C/EBPβ are increased in livers of S193D mice (Fig 3), we have examined if this elevation might be involved in the activation of the Glut4 promoter. We first performed EMSA with the probe covering C/EBP site within the Glut4 promoter. Specific antibodies were incorporated to distinguish C/EBPα and C/EBPβ isoforms. Consistent with elevation of protein levels of C/EBPβ, significantly higher amounts of C/EBPβ are bound to the Glut4 promoter in livers of S193D mice (Fig 8E). This is especially clear after supershift of the C/EBPα (Fig 8E, lane 8). Most important, the incorporation of antibodies to C/EBP proteins showed that the majority of S193D-C/EBPα binds to the Glut4 promoter as a heterodimer with C/EBPβ and that the binding of C/EBPα homodimers is not detectable after supershift with antibodies to C/EBPβ (Fig 8E, lane 9). It is important to note that antibodies to C/EBPβ specifically interact with C/EBPβ and do not cross-react and do not supershift C/EBPα in livers of WT mice (Fig 8E, lane 4). To determine if the elevation of C/EBPβ and C/EBPα/β heterodimers up-regulate Glut4 in livers of S193D mice, we have examined interactions of C/EBP proteins with the Glut4 promoter and modifications of histone H3 on the Glut4 promoter using ChIP analysis. HDAC1 protein was also included in these studies since it forms complexes with C/EBP proteins in the liver (Wang et al., 2008a; 2008b). These studies showed that the Glut4 promoter is occupied and partially repressed in WT mice by C/EBPβ-HDAC1 and C/EBPα-HDAC1 complexes because histone H3 is trimethylated at K9 on the Glut4 promoter (Fig 8F). On the contrary, HDAC1 is not detectable on the Glut4 promoter in S193D livers. Our data show that the Glut4 promoter is activated by S193D-C/EBPα and C/EBPβ because histone H3 is acetylated at K9 on this promoter.
C/EBPβ is required for the increased ability of C/EBPα-S193D to activate the Glut4 promoter
EMSA and ChIP assays suggested that C/EBPα-S193D activates the Glut4 promoter via formation of heterodimers with C/EBPβ. To directly examine the role of C/EBPα:C/EBPβ hetero-dimers, we have performed experiments in cultured HEK293 cells. These cells were chosen for these studies since they do not express endogenous C/EBPα; therefore, effects of the transfected S193D mutant will not be influenced by endogenous C/EBPα. The expression of C/EBPβ was inhibited in these cells by siRNA; and the WT and S193D mutant C/EBPα were co-transfected with the Glut4 promoter. As the control, the activity of Glut4 promoter was examined in cells transfected with C/EBPβ siRNA alone. Figure 8G shows that the inhibition of C/EBPβ in cells transfected with WT C/EBPα does not change significantly the activation of the Glut4 promoter. However, the inhibition of C/EBPβ in cells transfected with S193D mutant reduces the activity of the Glut4 promoter to the levels observed for activation by WT C/EBPα. These studies clearly demonstrated that the increased ability of C/EBPα-S193D to activate the Glut4 promoter is mediated via interactions with C/EBPβ, perhaps through the formation of hetero-dimers with C/EBPβ. Thus, these studies showed that the formation of C/EBPβ:C/EBPα-S193D heterodimers is involved in the activation of the Glut4 promoter and following increase of Glut4 protein in S193D mice.
DISCUSSION
Age-related dysfunctions of the liver are mediated by the ph-S193 isoform of C/EBPα
Senescent liver is characterized by decline of several functions and alterations in liver morphology (Schmucker, 2005; Timchenko, 2009). In this paper, we have examined the role of age-specific S193-ph isoform of C/EBPα in the age-related alterations using recently generated C/EBPα-S193D mice. In our previous paper, we have shown that C/EBPα-S193D mutant inhibits liver proliferation after partial hepatectomy (Wang et al., 2010). The impaired liver proliferation in S193D mice is similar to that observed in livers of old WT mice (Bucher et al, 1964; Timchenko, 2009). Examination of liver morphology and liver functions in young C/EBPα-S193D mice showed that hepatocytes of S193D mice have enlarged size and have abundant heterochromatin regions which are similar to those observed in livers of old WT mice. These data are in agreement with previous reports showing that mammalian aging is associated with remodeling of chromatin structure (Sedivy et al., 2008; Bandyopadhyay et al., 2007; Narita et al., 2003). Our data suggest that alterations of chromatin structure might be the cause of the increase of size of hepatocytes of old mice; however, it is also possible that the increased ploidy in old hepatocytes (Chipchase et al., 2003) explains the appearance of DAPI-bright heterochromatic foci, because the increased number of foci could reflect the heterochromatic centromeres of additional chromosomes.
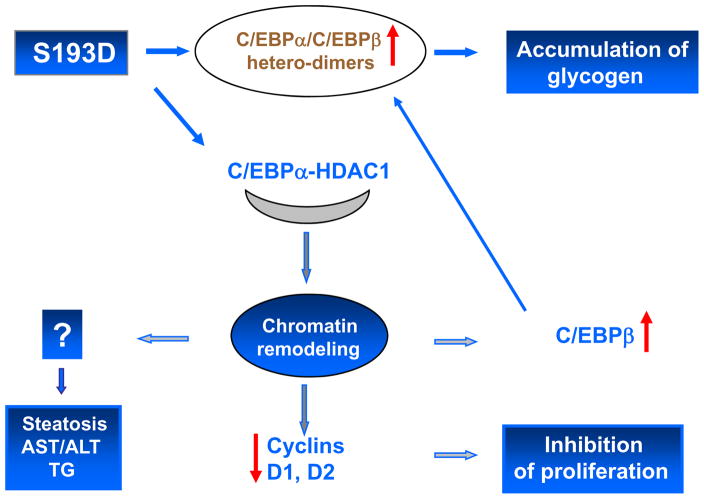
We have performed a detailed investigation of pathways by which S193D-C/EBPα and ph-S193 isoform of C/EBPα might change chromatin structure. Our data show that alterations in chromatin structure of hepatocytes correlate with the elevation of C/EBPα-HDAC1-HP1α complexes. These complexes are abundant in livers of old mice, but contain some additional proteins such as Brm and E2F4 (Iakova et al., 2003, Conboy et al., 2005; Wang et al., 2008a). The elevation of C/EBPα-HDAC1 complex seems to be responsible for the alterations of chromatin structure because the inhibition of the complex by siRNA to HDAC1 or by treatment of old mice with GH corrects chromatin structure in livers of S193D mice and in livers of old mice respectively. In this paper, we have shown that alterations of chromatin structure are involved in the inhibition of liver proliferation and in dys-regulation of gluconeogenesis. It is interesting to note that our previous paper has identified an additional age-associated and S193-ph- dependent alteration which is a high frequency of development of liver cancer in old mice. We have shown that the development of liver cancer in old mice is associated with the specific elimination of ph-S193 isoform of C/EBPα by gankyrin-mediated degradation through ubiquitin proteasome system (Wang et al., 2010). Taken together, the investigations of S193D mice show that the expression of the age-specific C/EBPα-S193D isoform in the liver of young mice leads to alterations which mimic liver dysfunctions observed in old animals (Schmucker, 2005, Timchenko, 2009). Alterations in S193D mice include enlarged hepatocytes, inhibition of liver proliferation, steatosis, accumulation of AST/ALT and TG in the blood and accumulation of glycogen in the liver (Fig 9).
Figure 9.
A hypothetical model for the pathways by which S193D isoform of C/EBPα causes the age-associated dysfunctions of the liver.
It is interesting to note that Seo at al have shown that the accumulation of glycogen itself led to senescence (Seo et al., 2008). Therefore, the accumulation of glycogen in livers of S193D mice might contribute to the age-associated changes in S193D mice. Our findings also suggest that the correction of the chromatin structure might be sufficient to improve liver functions in elderly. One of the possible tools for the correction of chromatin structure is a treatment of mice with growth hormone. Consistent with this hypothesis, Krupczak-Hollis et al have shown that the treatment of old mice with GH corrects liver regeneration via stimulation of proliferation of hepatocytes (Krupczak-Hollis et al., 2003). A recent paper has shown another pathway which might correct liver regeneration in aged mice. Gielchinsky et al found that activation of Akt/mTORC1 pathway in aged livers restores liver regeneration by activation of the process of cell growth (Gielchinsky et al., 2010).
Examination of the S193D mice has provided an answer to the critical question of liver biology. Liver supports body homeostasis and, during liver regeneration, the small portion of remaining liver completely maintains its functions of the differentiated tissue. The majority of studies of liver regeneration were performed with 70% model of partial hepatectomy; however, there are many reports of successful liver regeneration after 90% partial hepatectomy (Moser et al., 2001; Gorbin et al., 2002; Benko et al., 2010; Eipel et al., 2010). How does the remaining, very small portion of the liver control the multiple functions? Data in this paper and our published observations (Wang et al., 2010) present an example of pathways which liver uses to control proliferation and differentiation functions at the same time. Our data suggest that the enormous capabilities of the liver to control multiple functions are associated with a proper regulation of C/EBPα activities by timely phosphorylation/de-phosphorylation at a single amino acid residue and that disruption of this regulation causes pathological alterations in the liver and in blood.
Multiple pathways of C/EBPα-mediated arrest of cell proliferation
C/EBPα inhibits cell proliferation mainly through the interactions with cell cycle proteins (Wang et al., 2001, Tan et al., 2005; Timchenko, 2009). The analysis of S193D mice allowed us to identify additional pathways by which C/EBPα inhibits liver proliferation and additional pathways which regulate the biological activities of C/EBPα. The additional pathway of growth arrest involves silencing the cyclin D1 and D2 promoters by C/EBPα-HDAC1 complexes through direct interactions with these promoters. Although this pathway requires protein-protein interactions with HDAC1, it differs from previously described pathways and also requires DNA binding activity of C/EBPα. This is a first observation showing that direct interaction of C/EBPα with the promoters of D-type cyclin genes is involved in the arrest of proliferation. Examination of the S193D mice has also identified additional levels of complexity of the regulation of C/EBPα activities. Data in our paper show that S193D-C/EBPα activates the Glut4 promoter, but it inhibits cyclin D2 and cyclin D1 promoters. How are these opposite activities of C/EBPα regulated? One possible pathway is supported by the findings that S193D mice have the increased amounts of S193D-C/EBPα/β heterodimers. Our data show that the C/EBPα/β heterodimers bind to the Glut4 promoter and activate the promoter stronger than homo-dimers of each of these proteins. We have also shown that the interactions of C/EBPα with chromatin remodeling proteins might be one of the mechanisms which control these opposite transcriptional functions of C/EBPα. We have found that C/EBPα is observed on the cyclin D1 and cyclin D2 promoters in the complexes with HDAC1; while C/EBPα binds to the Glut4 promoter independently on HDAC1, mainly in the complexes with C/EBPβ. Thus, our data show that the transcriptional activities of C/EBPα are also controlled, at least in part, by phosphortylation at S193.
Experimental Procedures
Antibodies and Reagents
Antibodies to cyclin D3 (C-16), C/EBPα (14AA and N19), Glut4, C/EBPβ (C19), cyclin D1, cyclin D2 HDAC1, HP1α, cdk4 and cdc2 are from Santa Cruz Biotechnology. Monoclonal anti-β-actin antibodies were from Sigma.
C/EBPα-S193D knockin mice
The generation of C/EBPα-S193D knockin mice was described in our previous paper (Wang et al., 2010). Briefly, the C/EBPα-S193D mice were generated by replacement of the endogenous C/EBPα gene with knockin construct which contained a substitution of TC to GA in the position of Ser193 (See Fig 1A). This substitution leads to the mutation of Ser to Asp and to alterations in restriction sites for MluI and BamH1. Therefore, the genotyping of these mice included the examination of 858 bp PCR product by restriction enzymes MluI and BamHI. Our previous paper contains more details for the generation and genotyping of these mice (Wang et al., 2010). For all experiments presented in this paper, we have bred heterozygous mice and have examined WT, hetero and homozygous mice from the same littermates. Animal experiments were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine (protocol AN-1439).
Experiments with growth hormone (GH) and with siRNA to HDAC1
For GH treatment of mice, recombinant mouse growth hormone (rmGH) provided by the National Hormone and Peptide Program was injected into mice (2 mg/kg) subcutaneously for 3 days. The physiological saline (PS, 0.9% NaCl) was used as vehicle for rmGH. The control animals were injected with PS. Liver tissues were collected and used for the staining with H&E and DAPI and for the examination of protein expression as described below. The inhibition of HDAC1 by siRNA was performed as described in our previous paper (Wang et al, 2008b). The control animals were injected with a control siRNA containing random composition of nucleotides (Wang et al., 2008b).
Blood test, H&E and DAPI staining
Examination of the blood parameters, H&E staining and DAPI staining were performed in Baylor College of Medicine Facilities.
Immunostaining of livers with antibodies to C/EBPα, HDAC1 and K9 trimethylated histone H3
The immunostaining of the WT, S193D and WT old livers was performed as described in our previous papers (Wang et al., 2008a; 2008b). The antibodies to C/EBPα (14AA) and HDAC1 were from Santa Cruz Biotechnology. Antibodies to H3K9-trimethyl were from Abcam.
Examination of the Glut4 promoter
The proximal region of the mouse Glut4 promoter (containing C/EBPα site) was cloned into pGl3 vector. The reporter construct was co-transfected with WT C/EBPα and with C/EBPα mutants (S193D and R290A) into HEK293 cells. The activity of the promoter was calculated as a ratio to Renilla luc control.
Size exclusion chromatography
Nuclear extracts were isolated from livers as described earlier (Timchenko et al., 1999) and fractionated by size-exclusion column SEC400 (HPLC, BioLogic HR, BioRad). The detailed procedure for the analysis of C/EBPα complexes is described in our previous papers (Wang et al., 2001; 2004). Briefly. Gel filtration fractions were loaded on denaturing gradient (4–20%) PAAG, blotted onto membrane and probed with antibodies to C/EBPα (14AA), HDAC1, HP1α and Brm (Santa Cruz Biotechnology). To detect C/EBPα complexes, C/EBPα was immunoprecipitated from each fraction, and IPs were probed with antibodies to HDAC1.
Protein isolation and Western blotting
Nuclear extracts were isolated from cultured cells and from livers as described in previous papers (Wang et al., 2001; 2004). Briefly, tissues were homogenized with buffer A (20mM Tris-HCL pH 7.5, 30mM KCl, 10% glycerol and inhibitors of phosphatases) and spun down at 12,000rpm for 10 min at 4°C. Supernatant (cytoplasm) was frozen and the pellet was treated with high salt buffer (20mM Tris-HCl pH7.5, 0.42M NaCl, 25% sucrose, 5mM DTT and inhibitors of phosphatases). After centrifugation, the supernatant (NE) was used for the Western blotting and for Co-IP or frozen in −80°C freezer. Nuclear extracts were isolated from mouse livers of WT and S193D mice and examined by Western blotting as described below.
Co-Immunoprecipitation
C/EBPα was immunoprecipitated from nuclear extracts or from gel filtration fractions with polyclonal antibodies (14AA, Santa Cruz Biotechnology), and the presence of HDAC1 and HP1α in C/EBPα IPs was examined by Western blotting with monoclonal antibodies to the mentioned proteins.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed with the Glut4 promoter and with promoters of cyclin D1 and cyclin D2. The sequence of the Glut4 probe is as follows. 5′-CTGCACTCAATTCTTTCAGAAATTTCGCAGT-3′ (C/EBP site is shown in bold). The sequences of the probes with C/EBP sites within cyclin D1 and cyclin D2 promoters are shown in Fig 5C and Supplemental Fig 2. The double stranded probes were gel-purified and labeled by Klenow “fill in” reaction with radioactive P32-dCTP. The DNA probes were incubated with nuclear extracts isolated from WT and from S193D livers. The binding buffer contained 20mM Tris-HCl pH 7.5, 100mM KCl, 5mM MgCl2, 5mM DDT, 1μg/10μl poly (dI-dC) and 10% glycerol. Antibodies were added to the binding reactions before probe addition. The reaction mixtures were separated by 5% native gel electrophoresis; the gel was dried and exposed with X-ray film.
Chromatin immunoprecipitation assay (ChIP)
ChIP assay was performed using Chip-It kit according to the instruction. Briefly, chromatin solutions were prepared from livers of WT and S193D mice. C/EBPα, C/EBPβ, HDAC1, histone H3K9 and histone H3-trimethyl K9 were immunoprecipitated from the solutions. DNA was isolated and used for the PCR reactions with primers covering C/EBPα sites within the Glut4, cyclin D1 and cyclin D2 promoters (see Figure 5 and Supplemental Figure 2). The sequences of these primers for the Glut4 promoter are as follows, 5′-ACACACACACACACACACACAC-3′ (forward) and 5′-TAAGGTTCCCGCCTGCTTCTGAGTT-3′ (reverse). PCR mixtures were amplified for one cycle of 95°C (5 min), 60°C (5 min) and 72°C (2 min). Then PCR mixtures were amplified for 31 cycles of 95°C (1 min), 60°C (2 min) 72°C (1.5 min). PCR products were separated by 8 % PAGE.
Micro array studies
Total RNA was isolated from three animals of each genotype and used for micro array assay. The micro array was performed as described in our previous paper (Wang et al., 2008c). Briefly, two hundred nanograms of total RNA were amplified using Illumina TotalPrep RNA Amplification Kit (Ambion, Cat# IL1791) following kit instructions. In vitro transcription was performed and biotinylated cRNA was synthesized by 14-hour amplification with dNTP mix containing biotin-dUTP and T7 RNA polymerase. Amplified cRNA was subsequently purified and concentration was measured by NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, DE). The amplified products were used for analysis of Illumina Sentrix Beadchip Array Mouse-6 arrays. The arrays were scanned with BeadArray Reader (Illumina, CA). Data were analyzed using BeadStudio software (Illumina, CA). Clustering and pathway analysis were performed with BeadStudio and Ingenuity Pathway Analysis (Ingenuity Systems, Inc.) softwares respectively.
Supplementary Material
Supplemental figure 1. GH-mediated elimination of C/EBPα-HDAC1 complex reduces size of the nuclei and alters chromatin structure of the nuclei in hepatocytes of old mice. DAPI staining of livers of three young, old and old GH treated mice. Arrows show nuclei in old livers which are enlarged and have changed chromatin structure.
Supplemental figure 2. The cyclin D1 promoter contains binding site for C/EBPα. Left part shows the sequence of the cyclin D1 promoter. The start of transcription is shown in blue. C/EBPα site is shown by red bold letters. Arrows show the positions of primers used in ChIP analysis. Right part shows EMSA analysis with DNA probe from the cyclin D1 promoter (underlined on the left part). Antibodies to C/EBPα were added before the probe addition.
Supplemental figure 3. A. Cyclin D3-cdk4-mediated phosphorylation of C/EBPα at S193 enhances the ability of C/EBPα to activate the Glut4 promoter. The Glut4 promoter was co-transfected with WT, C/EBPα-S193D and C/EBPα-R290A with or without cyclin D3. B. Phosphorylation of endogenous C/EBPα in Hep3B2 activates the Glut4 promoter. Glut4 promoter was co-transfected with empty vector, with cyclin D3 and with Cyclin D3 + siRNA to C/EBPα.
Acknowledgments
We thank Estela Medrano and Gretchen Darlington for the discussion of this work and for useful suggestions. This work is supported by NIH grants GM55188, CA100070 and AG20752 (NAT).
Footnotes
Author contributions
JJ has performed experiments with the examination of heterochromatin regions in S193D and old mice, ChIP assay, treatments of S193D and old mice with growth hormone, examination of glucose metabolism in S193D mice and genotyping. GW has performed Q-RT-PCR, immunostaining and a portion of Western blotting studies. PI has performed immunostaining, ChIP and Western blotting experiments. XS was responsible for genotyping and maintenance of mice. SH and MF have provided their expertise for the analyses of blood parameters and for examination of liver tissues. NT performed Co-IP and HPLC-based examination of protein-protein complexes, was overall supervisor of the research and contributed to the writing and editing the manuscript.
Additional supporting information is included in on-line version of the manuscript.
References
- Bandyopadhyay B, Curry JL, Lin Q, Richards HW, Chen D, Hornsby P, Timchenko NA, Medrano EE. Dynamic Assembly of Chromatin during Cellular Senescence: Implications for the Growth Arrest of Human Melonocytic Nevi. Aging Cell. 2007;6:577–591. doi: 10.1111/j.1474-9726.2007.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko T, Frede S, Gu Y, Best J, Baba HA, Schlaak JF, de Groot H, Fandrey J, Rauen U. Glycine pretreatment ameliorates liver injury after partial hepatectomy in the rat. J Invest Surg. 2010;1:12–20. doi: 10.3109/08941930903469466. [DOI] [PubMed] [Google Scholar]
- Bucher NLR, Glinos MN, Di Troi F. The influence of age upon the incorporation of thymidine-2C14 into the DNA of regenerating rat liver. Cancer Res. 1964;24:509–512. [PubMed] [Google Scholar]
- Chipchase MD, O’Neill M, Melton DW. Characterization of premature liver polyploidy in DNA repair (Ercc1)-deficient mice. Hepatology. 2003;38:968–966. doi: 10.1053/jhep.2003.50421. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma IR, Weisman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;43:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Demarco IA, Periasamy A, Broker CF, Day RN. Monitoring dynamic protein interactions with photoquencing FRET. Nature Methods. 2006;7:519–524. doi: 10.1038/nmeth889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipel C, Abshagen K, Ritter J, Cantré D, Menger MD, Vollmar B. Splenectomy improves survival by increasing arterial blood supply in a rat model of reduced-size liver. Transplant International. 2010 doi: 10. 1111/j.1432–2277.2010.01079.x. [DOI] [PubMed] [Google Scholar]
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S43–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- Fry M, Silber J, Loeb LA, Martin GM. Delayed and reduced cell replication and diminishing levels of DNA-polymerase alpha in regenerating liver of aging mice. J Cell Physiol. 1984;118:225–232. doi: 10.1002/jcp.1041180302. [DOI] [PubMed] [Google Scholar]
- Gielchinsky V, Laufer N, Weitman E, Abramovitch R, Granot Z, Bergman Y, Pikarsky Pregnancy restores the regenerative capacity of the aged liver via activation of an mTORC1-controlled hyperplasia/hypertrophy switch. Genes & Dev. 2010;24:543–548. doi: 10.1101/gad.563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin IR, Buist R, Volotovskyy V, Peeling J, Zhang M, Minuk GY. Regenerative activity and liver function following partial hepatectomy in the rat using (31)P-MR spectroscopy. Hepatology. 2002;36:345–353. doi: 10.1053/jhep.2002.34742. [DOI] [PubMed] [Google Scholar]
- Jin J, Wang G-L, Shi X, Darlington GJ, Timchenko NA. The age-associated decline of GSK3β plays a critical role in the inhibition of liver regeneration. Mol Cell Biol. 2009;29:3867–3880. doi: 10.1128/MCB.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wang G-L, Salisbury E, Timchenko LT, Timchenko NA. GSK3β-cyclin D3-CUGBP1-eIF2 pathway in aging and in Myotonic Dystrophy. Cell Cycle. 2009;15:2356–2359. doi: 10.4161/cc.8.15.9248. [DOI] [PubMed] [Google Scholar]
- Jin J, Wang G-L, Timchenko LT, Timchenko NA. GSK3β and aging liver. Aging. 2009;6:582–585. doi: 10.18632/aging.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Science. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- Iakova P, Awad SS, Timchenko NA. Aging reduces proliferative capacities of liver by switching pathways of C/EBPα growth arrest. Cell. 2003;113:495–506. doi: 10.1016/s0092-8674(03)00318-0. [DOI] [PubMed] [Google Scholar]
- Krupczak-Hollis K, Wang X, Dennewitz MB, Costa RH. Growth hormone stimulates proliferation of old-aged regenerating liver through forkhead box m1b. Hepatology. 2003;38:1552–1562. doi: 10.1016/j.hep.2003.08.052. [DOI] [PubMed] [Google Scholar]
- Kuk JL, Saunders TJ, Davidson LE, Toss R. Age-related changes in total and regional fat distribution. Ageing Res Rev. 2009;4:339–348. doi: 10.1016/j.arr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Shuman JD, Sebastian T, Dauter Z, Johnson PF. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/Enhancer Binding Protein alpha. J Biol Chem. 2003;278:15178–15184. doi: 10.1074/jbc.M300417200. [DOI] [PubMed] [Google Scholar]
- Moser MJ, Gong Y, Zhang MN, Johnston J, Lipschitz J, Minuk GY. Immediate-early protooncogene expression and liver function following various extents of partial hepatectomy in the rat. Dig Dis Sci. 2001;46:907–914. doi: 10.1023/a:1010791915733. [DOI] [PubMed] [Google Scholar]
- Muller K, Calkhoven CF, Sha X, Leutz A. The CCAAT/Enhancer binding protein α (C/EBPα) requires a SWI/SNF complex for proliferation arrest. J Biol Chem. 2004;297:7353–7358. doi: 10.1074/jbc.M312709200. [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular cenescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPα, TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes & Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker DL. Age-related changes in liver structure and functions: Implications for disease? Exp Gerontol. 2005;40:650–659. doi: 10.1016/j.exger.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Sedivy JM, Banumathy G, Adams PD. Aging by epigenetics – A consequence of chromatin change? Exp Cell Res. 2008;314:1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y-H, Jung H-L, Shin H-T, Kim Y-M, Yim H, Chung H-Y, Lim IK, Yoon G. Enhanced glycogenesis is involved in cellular senescence via GSK/GS modulation. Aging Cell. 2008;7:894–907. doi: 10.1111/j.1474-9726.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- Tan EH, Hooi SC, Laban M, Wong E, Ponniah AW, Wang N-D. CCAAT/Enhancer Binding Protein α Knock-in Mice Exhibit Early Liver Glycogen Storage and Reduced Susceptibility to Hepatocellular Carcinoma. Cancer Res. 2005;65:10330–10337. doi: 10.1158/0008-5472.CAN-04-4486. [DOI] [PubMed] [Google Scholar]
- Timchenko NA. Aging and liver regeneration. Trends Endocr Metab. 2009;20:171–176. doi: 10.1016/j.tem.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Harris TE, Wilde M, Bilyeu TA, Burgess-Beusse BL, Finegold MJ, Darlington GJ. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde M, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- Wang H, Iakova P, Wilde M, Welm A, Goode T, Roesler WJ, Timchenko NA. C/EBPα arrests cell proliferation through direct inhibition of cdk2 and cdk4. Mol Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- Wang G-L, Iakova P, Wilde M, Awad SS, Timchenko NA. Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBPα growth inhibitory activity. Genes & Dev. 2004;18:912–925. doi: 10.1101/gad.1183304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Salisbury E, Shi X, Timchenko LT, Medrano EE, Timchenko NA. HDAC1 cooperates with C/EBPα in the inhibition of liver proliferation in old mice. J Biol Chem. 2008a;283:26196–26178. doi: 10.1074/jbc.M803544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Salisbury E, Shi X, Timchenko LT, Medrano EE, Timchenko NA. HDAC1 promotes liver proliferation in young mice via interaction with C/EBPα. J Biol Chem. 2008b;283:26179–26187. doi: 10.1074/jbc.M803545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Shi X, Salisbury E, Sun Y, Albrecht JH, Smith RG, Timchenko NA. Cyclin D3 maintains growth inhibitory activity of C/EBPα by stabilizing C/EBPα-cdk2 and C/EBPα-Brm complexes. Mol Cell Biol. 2006;26:2570–2582. doi: 10.1128/MCB.26.7.2570-2582.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Shi X, Salisbury E, Sun Y, Albrecht JH, Smith RG, Timchenko NA. Growth Hormone Corrects Proliferation and Transcription of PEPCK in Livers of Old Mice via Elimination of C/EBPα-Brm Complex. J Biol Chem. 2007;28:1468–1478. doi: 10.1074/jbc.M608226200. [DOI] [PubMed] [Google Scholar]
- Wang G-L, Shi X, Salisbury E, Timchenko NA. Regulation of apoptotic and growth inhibitory activities of C/EBPα _in different cell lines. Exp Cell Research. 2008c;314:1626–1639. doi: 10.1016/j.yexcr.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Shi X, Haefliger S, Jin J, Major A, Iakova P, Finegold M, Timchenko NA. UPS-mediated elimination of C/EBPα is required for the development of liver cancer. J Clin Invest. 2010;120:2549–2562. doi: 10.1172/JCI41933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Timchenko NA. Dephosphorylated C/EBPα accelerates cell proliferation through sequestering retinoblastoma protein. Mol Cell Biol. 2005;25:1325–1338. doi: 10.1128/MCB.25.4.1325-1338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1. GH-mediated elimination of C/EBPα-HDAC1 complex reduces size of the nuclei and alters chromatin structure of the nuclei in hepatocytes of old mice. DAPI staining of livers of three young, old and old GH treated mice. Arrows show nuclei in old livers which are enlarged and have changed chromatin structure.
Supplemental figure 2. The cyclin D1 promoter contains binding site for C/EBPα. Left part shows the sequence of the cyclin D1 promoter. The start of transcription is shown in blue. C/EBPα site is shown by red bold letters. Arrows show the positions of primers used in ChIP analysis. Right part shows EMSA analysis with DNA probe from the cyclin D1 promoter (underlined on the left part). Antibodies to C/EBPα were added before the probe addition.
Supplemental figure 3. A. Cyclin D3-cdk4-mediated phosphorylation of C/EBPα at S193 enhances the ability of C/EBPα to activate the Glut4 promoter. The Glut4 promoter was co-transfected with WT, C/EBPα-S193D and C/EBPα-R290A with or without cyclin D3. B. Phosphorylation of endogenous C/EBPα in Hep3B2 activates the Glut4 promoter. Glut4 promoter was co-transfected with empty vector, with cyclin D3 and with Cyclin D3 + siRNA to C/EBPα.