Abstract
Introduction
Current attempts to identify genetic modifiers of BRCA1 and BRCA2 associated risk have focused on a candidate gene approach, based on knowledge of gene functions, or the development of large genome-wide association studies. In this study, we evaluated 24 SNPs tagged to 14 candidate genes derived through a novel approach that analysed gene expression differences to prioritise candidate modifier genes for association studies.
Methods
We successfully genotyped 24 SNPs in a cohort of up to 4,724 BRCA1 and 2,693 BRCA2 female mutation carriers from 15 study groups and assessed whether these variants were associated with risk of breast cancer in BRCA1 and BRCA2 mutation carriers.
Results
SNPs in five of the 14 candidate genes showed evidence of association with breast cancer risk for BRCA1 or BRCA2 carriers (P < 0.05). Notably, the minor alleles of two SNPs (rs7166081 and rs3825977) in high linkage disequilibrium (r2 = 0.77), located at the SMAD3 locus (15q22), were each associated with increased breast cancer risk for BRCA2 mutation carriers (relative risk = 1.25, 95% confidence interval = 1.07 to 1.45, Ptrend = 0.004; and relative risk = 1.20, 95% confidence interval = 1.03 to 1.40, Ptrend = 0.018).
Conclusions
This study provides evidence that the SMAD3 gene, which encodes a key regulatory protein in the transforming growth factor beta signalling pathway and is known to interact directly with BRCA2, may contribute to increased risk of breast cancer in BRCA2 mutation carriers. This finding suggests that genes with expression associated with BRCA1 and BRCA2 mutation status are enriched for the presence of common genetic modifiers of breast cancer risk in these populations.
Introduction
BRCA1 and BRCA2 mutation carriers are at increased risk for developing breast cancer and/or ovarian cancer. Estimates of the cumulative risk of breast cancer by age 70 years range from 46% to 87% for BRCA1 mutation carriers and from 43% to 84% for BRCA2 mutation carriers [1-6]. Evidence from these studies suggests that breast cancer risks in mutation carriers are modified by environmental or genetic factors. A number of large studies, facilitated through the Consortium of Investigators of Modifiers of BRCA1/BRCA2 (CIMBA), have evaluated associations between genetic polymorphisms and breast cancer risk in BRCA1 and BRCA2 mutation carriers [7-15].
The candidate gene (or candidate SNP) approach for identifying potential risk modifiers has been successfully used to identify a SNP in the 5' untranslated region of RAD51. Until recently, this finding has provided the most reliable evidence for a genetic modifier in BRCA2 mutation carriers [7]. A major disadvantage of using this approach to identify common genetic modifiers of breast cancer, however, is the limited understanding of mechanisms and pathways that underlie breast cancer development in families carrying mutations in BRCA1 or BRCA2. An alternative and powerful approach that can overcome such issues is the use of genome-wide association (GWA) studies to identify candidate SNPs. Analysis of breast cancer risk-associated SNPs identified by a large population-based GWA study of breast cancer [16] has shown that several of these SNPs also appear to modify risk in BRCA1 and/or BRCA2 mutation carriers [8]. Not all of the breast cancer-associated SNPs assessed have been found to modify risk in carriers, however, and some of the risk associations are specific for BRCA2 mutation carriers only and not BRCA1 [8]. While GWA studies specifically addressing risk for BRCA1 and/or BRCA2 carriers are a more direct approach to identifying modifiers of these genes using an agnostic approach, GWA studies require large sample sizes to identify genetic modifiers with confidence. To address the problem of inadequate sample size, the CIMBA was established in 2005 to link clinical and epidemiological data from many groups from around the world [17]. The GWA approach is still limited, however, in that study designs involve predefined stringent selection criteria for which SNPs identified from the initial whole genome scan are going to be analysed in subsequent replication studies, a study design enforced by current genotyping costs. Moreover, GWA studies are often limited in information about exogenous risk factors, such as environmental exposures, which confounds any effort to explore the effect of environmental factors in modifying gene-disease associations. Global gene expression analysis as a means to agnostically identify candidate genetic modifiers has the potential to prioritise SNPs for candidate genes for association studies. This may be particularly valuable given recent observations that SNPs associated with risk of cancer in the general population appear to reside in noncoding regions that may modulate gene expression.
An alternative approach to prioritising SNPs and candidate genes for association studies in BRCA1 and BRCA2 mutation carriers could rely on the selection of genes displaying associations with BRCA1 or BRCA2 mutation status at the expression level in response to DNA damage. In a previous study, we used a novel combinatorial approach to identify a subset of 20 irradiation responsive genes as high-priority candidate BRCA1 and/or BRCA2 modifier genes [18]. The expression levels of these genes were shown to be associated with BRCA1 and/or BRCA2 mutation status in irradiated lymphoblastoid cell lines from female carriers when compared against irradiated lymphoblastoid cell lines from healthy controls. Furthermore, each of the genes were tagged with one or more SNPs shown to be associated with breast cancer risk from the Cancer Genetic Markers of Susceptibility (CGEMS) Phase 1 Breast Cancer Whole Genome Association Scan [19,20]. In the present study we investigated the association of these polymorphisms, tagged to genes demonstrated in vitro to be involved in irradiation response, with risk of breast cancer for BRCA1 and BRCA2 mutation carriers.
Materials and methods
Study participants
Eligibility of study participants was restricted to female BRCA1 or BRCA2 pathogenic mutation carriers who were aged 18 years or older. Fifteen clinic and population-based research studies from the USA, Canada, Australia, the UK and Europe submitted data to the present study (Table 1). Information collected included year of birth, age at diagnosis of breast cancer or ovarian cancer, age at last observation, family membership, ethnicity and information on bilateral prophylactic mastectomy and oophorectomy. All centres have obtained informed consent from study participants and the institutional review board approved protocols. In total, this study included up to 4,724 BRCA1 and 2,693 BRCA2 eligible female mutation carriers. Of the 2,193 and 1,189 unaffected BRCA1 and BRCA2 carriers, respectively, 972 (44.3%) and 589 (49.5%) had a relative that was in the affected group.
Table 1.
Distribution of BRCA1 and BRCA2 mutation carriers by study site
| Study | Countrya | BRCA1 | BRCA2 | Genotyping platform |
|---|---|---|---|---|
| HEBON | The Netherlands | 807 | 308 | iPLEXb; Golden Gatec |
| EMBRACE | UK | 841 | 656 | iPLEXb; Golden Gatec |
| FCCC | USA | 82 | 53 | iPLEXb; Golden Gatec |
| GC-HBOC | Germany | 398 | 163 | Golden Gatec |
| GEMO | France/USA | 408 | 226 | Golden Gatec |
| Georgetown | USA | 27 | 14 | iPLEXb; Golden Gatec |
| HEBCS | Finland | 103 | 104 | iPLEXb; Golden Gatec |
| ILUH | Iceland | 6 | 87 | iPLEXb; Golden Gatec |
| kConFab | Australia/New Zealand | 531 | 427 | iPLEXb; Golden Gatec |
| Mayo | USA | 227 | 123 | iPLEXb; Golden Gatec |
| ModSQuaD | USA | 158 | 91 | Golden Gatec |
| MUV | Austria | 298 | 126 | iPLEXb; Golden Gatec |
| PBCS | Italy | 76 | 43 | iPLEXb |
| SWE-BRCA | Sweden | 489 | 141 | iPLEXb; Golden Gatec |
| UPENN | USA | 273 | 131 | iPLEXb; Golden Gatec |
EMBRACE, Epidemiological Study of BRCA1 and BRCA2 Mutation Carriers; FCCC, Fox Chase Cancer Center; HEBON, Hereditary Breast and Ovarian Cancer Research Group Netherlands; ILUH, Iceland Landspitali - University Hospital Study; kConFab, Kathleen Cunningham Consortium for Research into Familial Breast Cancer; ModSQuaD, Modifier Study of Quantitative Effects on Disease; MUV, Medical University of Vienna; PBCS, Pisa Breast Cancer Study. aCoordinating centre. bSamples were genotyped at the Queensland Institute of Medical Research. cSamples were genotyped at the Mayo Clinic.
SNP selection and genotyping
In a previous report, we proposed 13 genes (ARHGEF2, HNRPDL, IL4R, JUND, LSM2, MAGED2, MLF2, MS4A1, SMAD3, STIP1, THEM2, TOMM40, VNN2) as candidate modifiers of breast cancer risk for BRCA1 mutation carriers, and 14 genes (ARHGEF2, JUND, MLF2, SMAD3, STIP1, THEM2, TOMM40, ABL1, ELMO1, EPM2AIP1, PER1, PLCG2, PLD3, SLC20A1) as candidate modifiers of breast cancer risk for BRCA2 mutation carriers (see Additional file 1) [18]. Thirty-seven SNPs denoted by CGEMS as being tagged to these genes were initially identified as showing some association with breast cancer risk (P < 0.05) (see Additional file 2). Of these 37 SNPs, a panel of 32 variants were selected after successful assay design and genotyped on two platforms, using the Illumina GoldenGate assay (Illumina Inc., San Diego, California, USA) and the Sequenom MassARRAY iPLEX platform (Sequenom, San Diego, CA, USA), as previously described [21,22]. The genotyping method used for each participating study is detailed in Table 1. Five SNPs tagged to five candidate genes (JUND, MAGED2, MLF2, MLH1, STIP1) had call rates <95% and were excluded from the analysis. The minor allele frequencies of three SNPs (rs2893535 - ELMO1, minor allele frequency = 0.033; rs2304911 - PER1, minor allele frequency = 0.043; and rs3802957 - MS4A1, minor allele frequency = 0.04) were considered too small for reliable analysis. The number of genes assessed for their associations with breast cancer risk for BRCA1 and BRCA2 mutation carriers was therefore eight and 10, respectively.
Statistical methods
Relative risks (RRs) and 95% confidence intervals were estimated using weighted Cox proportional hazards models. Each subject was followed from birth to the earliest of breast cancer, bilateral mastectomy, ovarian cancer, last follow-up, or age 80. The phenotype of interest was time to breast cancer. Mutation-specific weights were calculated using the age distribution of affected and unaffected individuals according to the methods previously outlined by Antoniou and colleagues [23]. Analyses were stratified by year of birth, ethnicity, country of residence, study site, and mutation status. A robust variance estimate was used to account for relatedness amongst individuals. Primary SNP analyses assumed a log-additive relationship between the number of minor alleles carried by each individual and time to breast cancer. Wald P values below 0.05 were declared of interest. Secondary analyses were carried out in which RR estimates were separately generated for those carrying one and two copies of the minor allele versus those with two copies of the major allele. Between-study heterogeneity was examined in each SNP by including an interaction term between the genotype and study centre.
Owing to the highly-selected nature of subjects, a number of sensitivity analyses were examined. To limit the effect of potential survival bias, subjects diagnosed more than 5 years prior to study enrolment were excluded (number affected analysed = 1,342 and 762 for BRCA1 and BRCA2 carriers, respectively). Other models were examined that excluded women with ovarian cancer (number excluded = 491 and 151 BRCA1 and BRCA2 carriers, respectively). Finally, as risk of breast cancer is reduced after bilateral oophorectomy [24,25], analyses were carried out treating oophorectomy as a time-dependent covariate in the Cox proportional hazards models. All P values are two sided and analyses were carried out using R software [26].
Results and Discussion
A cohort of up to 4,724 BRCA1 and 2,693 BRCA2 female mutation carriers was used for the present study. Of these, 4,035 mutation carriers were diagnosed with breast cancer or ovarian cancer at the end of follow-up and 3,382 were censored as unaffected at a mean age of 44 years. The patient characteristics of BRCA1 and BRCA2 mutation carriers are presented in Table 2.
Table 2.
Patient characteristics
| Characteristic | BRCA1 mutation carriers | BRCA2 mutation carriers | ||
|---|---|---|---|---|
| Unaffected | Breast cancer | Unaffected | Breast cancer | |
| Number of carriers | 2,193 | 2,531 | 1,189 | 1,504 |
| Length of follow-up (person-years) | 93,521 | 102,870 | 53,147 | 66,764 |
| Mean (SD) age at censure (years) | 43 (12.6) | 41 (9.4) | 45 (13.2) | 44 (9.7) |
| Age at censure, n (%) | ||||
| < 30 years | 344 (16%) | 252 (10%) | 144 (12%) | 52 (3%) |
| 30 to 39 years | 658 (30%) | 1060 (42%) | 343 (29%) | 474 (32%) |
| 40 to 49 years | 608 (28%) | 809 (32%) | 331 (28%) | 587 (39%) |
| 50 to 59 years | 374 (17%) | 310 (12%) | 215 (18%) | 273 (18%) |
| 60 to 69 years | 143 (6%) | 87 (3%) | 101 (8%) | 93 (6%) |
| 70+ years | 66 (3%) | 13 (1%) | 55 (5%) | 25 (2%) |
| Year of birth, n (%) | ||||
| Before 1949 | 523 (24%) | 840 (33%) | 281 (24%) | 602 (40%) |
| 1949 to 1959 | 508 (23%) | 816 (32%) | 307 (26%) | 518 (35%) |
| 1960 to 1968 | 594 (27%) | 602 (24%) | 302 (25%) | 303 (20%) |
| After 1968 | 568 (26%) | 273 (11%) | 299 (25%) | 81 (5%) |
| Oophorectomy | 260 (12%) | 77 (3%) | 126 (11%) | 47 (3%) |
| Ethnicity, n (%) | ||||
| Caucasian | 2127 (97%) | 2446 (97%) | 1159 (97%) | 1464 (97%) |
| Ashkenazi Jewish | 66 (3%) | 85 (3%) | 30 (3%) | 40 (3%) |
SD, standard deviation.
The RR estimates for the association between SNP genotypes and risk of breast cancer for BRCA1 and BRCA2 mutation carriers are presented in Table 3 and Table 4 respectively. Of the 24 SNPs that passed quality control, the minor alleles of two SNPs were found to be associated with increased risk for BRCA1 mutation carriers (rs10242920 - ELMO1, P = 0.043; and rs480092 - LSM2, P = 0.015) and the minor alleles of three SNPs to be associated with increased risk for BRCA2 mutation carriers (rs1559949 - HNRPDL, P = 0.021; rs3825977 - SMAD3, P = 0.018; and rs7166081 - SMAD3, P = 0.004). The minor alleles of two SNPs, rs1559949 (HNRPDL) and rs3808814 (ABL1), were associated with decreased risk for BRCA1 (P = 0.022) and BRCA2 (P = 0.030) mutation carriers, respectively.
Table 3.
Genotype distributions of 24 candidate modifier SNPs and hazard ratio estimates for BRCA1 mutation carriers
| SNP | Gene | Minor allele | MAF | Heterozygous | Homozygous | Per allele | P trend | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | |||||
| rs7026988 | ABL1 | A | 0.12 | 1.08 | 0.88 to 1.34 | 1.61 | 0.82 to 3.15 | 1.13 | 0.93 to 1.36 | 0.212 |
| rs3808814 | ABL1 | A | 0.09 | 0.90 | 0.72 to 1.14 | 0.65 | 0.22 to 1.91 | 0.89 | 0.72 to 1.10 | 0.284 |
| rs1889532 | ARHGEF2 | G | 0.25 | 0.99 | 0.83 to 1.18 | 1.12 | 0.80 to 1.56 | 1.03 | 0.90 to 1.18 | 0.708 |
| rs10242920 | ELMO1 | A | 0.24 | 1.06 | 0.89 to 1.27 | 1.61 | 1.12 to 2.32 | 1.16 | 1.00 to 1.33 | 0.043 |
| rs6964474 | ELMO1 | C | 0.22 | 1.08 | 0.90 to 1.29 | 0.71 | 0.49 to 1.02 | 0.96 | 0.84 to 1.10 | 0.568 |
| rs2541095 | ELMO1 | G | 0.12 | 1.03 | 0.84 to 1.27 | 1.12 | 0.48 to 2.63 | 1.04 | 0.86 to 1.26 | 0.683 |
| rs6956864 | ELMO1 | G | 0.41 | 1.04 | 0.76 to 1.42 | 1.35 | 0.18 to 10.03 | 1.05 | 0.78 to 1.42 | 0.755 |
| rs1559949 | HNRPDL | G | 0.14 | 0.78 | 0.65 to 0.94 | 0.91 | 0.51 to 1.64 | 0.82 | 0.70 to 0.97 | 0.022 |
| rs4285076 | HNRPDL | A | 0.29 | 0.95 | 0.80 to 1.12 | 1.00 | 0.73 to 1.37 | 0.98 | 0.86 to 1.11 | 0.746 |
| rs4787956 | IL4R | G | 0.34 | 0.99 | 0.83 to 1.18 | 1.11 | 0.84 to 1.46 | 1.03 | 0.91 to 1.17 | 0.611 |
| rs16976728 | IL4R | A | 0.38 | 0.92 | 0.77 to 1.10 | 1.07 | 0.82 to 1.39 | 1.00 | 0.88 to 1.13 | 0.978 |
| rs480092 | LSM2 | G | 0.16 | 1.25 | 1.04 to 1.51 | 1.30 | 0.81 to 2.08 | 1.21 | 1.04 to 1.42 | 0.015 |
| rs2253820 | PER1 | A | 0.17 | 1.02 | 0.9 to 1.16 | 0.72 | 0.52 to 1.00 | 0.96 | 0.87 to 1.06 | 0.412 |
| rs4888201 | PLCG2 | A | 0.16 | 1.10 | 0.91 to 1.34 | 1.39 | 0.77 to 2.51 | 1.13 | 0.95 to 1.33 | 0.168 |
| rs10514519 | PLCG2 | A | 0.18 | 1.06 | 0.88 to 1.28 | 1.59 | 0.92 to 2.75 | 1.11 | 0.95 to 1.31 | 0.195 |
| rs4997772 | PLCG2 | A | 0.39 | 1.13 | 0.95 to 1.35 | 1.07 | 0.84 to 1.37 | 1.05 | 0.94 to 1.18 | 0.377 |
| rs3936112 | PLCG2 | A | 0.39 | 0.98 | 0.87 to 1.10 | 0.97 | 0.82 to 1.16 | 0.98 | 0.91 to 1.07 | 0.700 |
| rs4254419 | PLD3 | A | 0.15 | 0.98 | 0.81 to 1.18 | 0.87 | 0.50 to 1.52 | 0.96 | 0.82 to 1.13 | 0.648 |
| rs10758 | SLC20A1 | G | 0.26 | 0.98 | 0.82 to 1.17 | 0.95 | 0.68 to 1.33 | 0.98 | 0.85 to 1.12 | 0.729 |
| rs3825977 | SMAD3 | A | 0.20 | 0.98 | 0.88 to 1.11 | 0.94 | 0.71 to 1.24 | 0.98 | 0.89 to 1.07 | 0.638 |
| rs7166081 | SMAD3 | G | 0.24 | 0.98 | 0.87 to 1.11 | 1.03 | 0.80 to 1.33 | 1.00 | 0.91 to 1.10 | 0.995 |
| rs3777663 | THEM2 | G | 0.24 | 1.02 | 0.86 to 1.22 | 1.17 | 0.84 to 1.63 | 1.05 | 0.92 to 1.20 | 0.453 |
| rs2075642 | TOMM40 | A | 0.20 | 0.97 | 0.81 to 1.16 | 1.06 | 0.69 to 1.63 | 0.99 | 0.86 to 1.15 | 0.931 |
| rs12211125 | VNN2/VNN3 | G | 0.09 | 0.96 | 0.83 to 1.11 | 1.19 | 0.73 to 1.94 | 0.99 | 0.87 to 1.12 | 0.882 |
MAF, minor allele frequency; HR, hazard ratio; CI, confidence interval.
Table 4.
Genotype distributions of 24 candidate modifier SNPs and hazard ratio estimates for BRCA2 mutation carriers
| SNP | Gene | Minor allele | MAF | Heterozygous | Homozygous | Per allele | P trend | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | |||||
| rs7026988 | ABL1 | A | 0.12 | 0.96 | 0.73 to 1.26 | 0.93 | 0.41 to 2.10 | 0.96 | 0.76 to 1.20 | 0.713 |
| rs3808814 | ABL1 | A | 0.09 | 0.72 | 0.51 to 1.02 | 0.50 | 0.17 to 1.42 | 0.71 | 0.53 to 0.97 | 0.030 |
| rs1889532 | ARHGEF2 | G | 0.25 | 1.32 | 1.05 to 1.67 | 0.99 | 0.64 to 1.54 | 1.13 | 0.95 to 1.33 | 0.172 |
| rs10242920 | ELMO1 | A | 0.24 | 1.01 | 0.79 to 1.30 | 0.86 | 0.52 to 1.43 | 0.97 | 0.80 to 1.17 | 0.747 |
| rs6964474 | ELMO1 | C | 0.22 | 1.09 | 0.86 to 1.40 | 1.47 | 0.89 to 2.45 | 1.15 | 0.95 to 1.39 | 0.153 |
| rs2541095 | ELMO1 | G | 0.12 | 1.04 | 0.8 to 1.35 | 0.72 | 0.27 to 1.94 | 1.00 | 0.78 to 1.26 | 0.971 |
| rs6956864 | ELMO1 | G | 0.41 | 0.76 | 0.48 to 1.18 | 2.20 | 0.16 to 29.7 | 0.78 | 0.50 to 1.22 | 0.279 |
| rs1559949 | HNRPDL | G | 0.14 | 1.29 | 0.96 to 1.72 | 2.06 | 0.99 to 4.28 | 1.33 | 1.04 to 1.70 | 0.021 |
| rs4285076 | HNRPDL | A | 0.29 | 0.86 | 0.67 to 1.09 | 1.47 | 0.95 to 2.26 | 1.03 | 0.85 to 1.25 | 0.737 |
| rs4787956 | IL4R | G | 0.34 | 1.10 | 0.87 to 1.41 | 1.31 | 0.90 to 1.91 | 1.13 | 0.95 to 1.35 | 0.167 |
| rs16976728 | IL4R | A | 0.38 | 1.16 | 0.91 to 1.48 | 1.38 | 0.95 to 1.99 | 1.17 | 0.98 to 1.39 | 0.075 |
| rs480092 | LSM2 | G | 0.16 | 0.92 | 0.72 to 1.18 | 1.09 | 0.55 to 2.16 | 0.96 | 0.78 to 1.19 | 0.735 |
| rs2253820 | PER1 | A | 0.17 | 0.85 | 0.70 to 1.02 | 1.13 | 0.68 to 1.87 | 0.90 | 0.77 to 1.06 | 0.209 |
| rs4888201 | PLCG2 | A | 0.16 | 0.98 | 0.75 to 1.27 | 1.16 | 0.54 to 2.52 | 1.01 | 0.80 to 1.27 | 0.964 |
| rs10514519 | PLCG2 | A | 0.18 | 0.85 | 0.66 to 1.09 | 1.92 | 0.95 to 3.88 | 0.99 | 0.79 to 1.24 | 0.933 |
| rs4997772 | PLCG2 | A | 0.39 | 1.21 | 0.95 to 1.55 | 1.26 | 0.89 to 1.78 | 1.14 | 0.97 to 1.34 | 0.107 |
| rs3936112 | PLCG2 | A | 0.39 | 0.91 | 0.76 to 1.09 | 0.94 | 0.73 to 1.22 | 0.96 | 0.85 to 1.08 | 0.483 |
| rs4254419 | PLD3 | A | 0.15 | 0.95 | 0.73 to 1.22 | 0.74 | 0.36 to 1.51 | 0.92 | 0.74 to 1.14 | 0.448 |
| rs10758 | SLC20A1 | G | 0.26 | 1.12 | 0.88 to 1.42 | 1.11 | 0.67 to 1.84 | 1.08 | 0.90 to 1.30 | 0.388 |
| rs3825977 | SMAD3 | A | 0.20 | 1.10 | 0.91 to 1.33 | 1.83 | 1.23 to 2.73 | 1.20 | 1.03 to 1.40 | 0.018 |
| rs7166081 | SMAD3 | G | 0.24 | 1.17 | 0.97 to 1.42 | 1.74 | 1.21 to 2.49 | 1.25 | 1.07 to 1.45 | 0.004 |
| rs3777663 | THEM2 | G | 0.24 | 0.95 | 0.75 to 1.21 | 1.11 | 0.69 to 1.79 | 0.99 | 0.82 to 1.20 | 0.945 |
| rs2075642 | TOMM40 | A | 0.20 | 1.14 | 0.89 to 1.46 | 1.19 | 0.68 to 2.09 | 1.12 | 0.92 to 1.37 | 0.267 |
| rs12211125 | VNN2 /VNN3 | G | 0.09 | 1.01 | 0.81 to 1.26 | 1.31 | 0.49 to 3.56 | 1.02 | 0.83 to 1.26 | 0.818 |
MAF, minor allele frequency; HR, hazard ratio; CI, confidence interval.
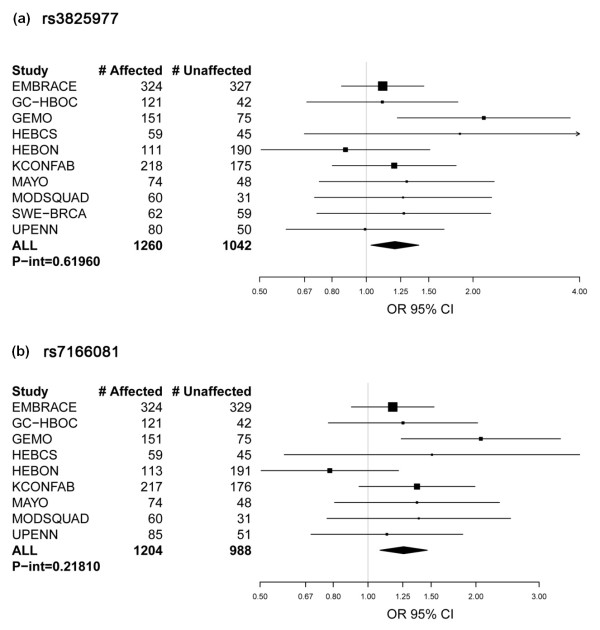
All SNPs selected for the present study (see Additional file 2) had previously been reported to be at least marginally associated (P < 0.05) with breast cancer risk through the CGEMS Phase 1 Breast Cancer Whole Genome Association Scan [18], and to be tagged to a gene whose expression level was associated with BRCA1 and/or BRCA2 mutation status in irradiated lymphoblastoid cell lines [18]. The minor allele of four out of six SNPs shown here to be associated with risk in BRCA1 mutation carriers (rs1559949 - HNRPDL; rs480092 - LSM2) or BRCA2 (rs3825977 and rs7166081 - SMAD3) had risk estimates for the homozygous genotype that were concordant with the odds ratio reported by the CGEMS study (Table 3 and 4, and Additional file 2). Furthermore, the expression of HNRPDL and LSM2 was associated with BRCA1 mutation status and the expression of SMAD3 was associated with BRCA2 mutation status [18]. The risk estimate of rs10242920 (ELMO1) was also concordant with the odds ratio determined by the CGEMS study; and although the expression of ELMO1 was not associated with BRCA1 mutation status at P < 0.001, there was an association with gene expression at P < 0.005 [18]. In contrast, the risk estimates of rs3808814 (ABL1) and rs1559949 (HNRPDL) in BRCA2 mutation carriers are not concordant with the odds ratio determined by the CGEMS study. Forest plots of study groups with 70 or more carriers and tests of heterogeneity are shown for two of the most significant SNPs (rs3825977, P-het = 0.619 and rs7166081, P-het = 0.218 at the SMAD3 locus), stratified by study site (Figure 1). The minor alleles of rs3825977 and rs7166081 are in high linkage disequilibrium (r2 = 0.77), which would be expected if their association with increased breast cancer risk is bona fide.
Figure 1.
BRCA2 plot of relative risk for rs3825977 and rs7166081 at the SMAD3 locus. BRCA2 plots of study group-specific relative risk (RR) for rs3825977 and rs7166081 at the SMAD3 locus. Study groups with 70 or more carriers and tests of heterogeneity are shown for (a) rs3825977 (overall RR (95% confidence interval (CI)) = 1.20 (1.03, 1.40), Ptrend = 0.018) and (b) rs7166081 (overall RR (95% CI) = 1.25 (1.07, 1.45), Ptrend = 0.004). OR, odds ratio.
Although further study is required to confirm whether genetic variation in SMAD3 plays a role in modifying risk of breast cancer, SMAD3 has been shown to interact with the BRCA2 protein - suggesting a possible mechanism through which SMAD3 may modify BRCA2 function [27]. Furthermore, SMAD3 is a critical regulatory factor of the transforming growth factor beta pathway, which is known to play a key role in the development of breast cancer as well as many other cancers [28,29]. In addition, a recent study comparing dense breast tissue (a known breast cancer risk factor) with nondense tissue identified reduced expression of SMAD3 to be associated with dense tissue, indirectly supporting a role of SMAD3 expression with breast cancer risk [29].
Choosing candidate BRCA1 and BRCA2 modifier genes from a novel combinatorial approach [18], we show that four SNPs tagged to three of the 14 candidate genes show an association with breast cancer risk for BRCA1 or BRCA2 mutation carriers. We initiated the present study, however, with the expectation that SNPs in eight of the 14 genes may be associated with altered expression in BRCA1 mutation carriers, and 10 of the 14 genes with altered expression in BRCA2 carriers. We can thus argue that three out of 18 (17%) valid comparisons showed an association with risk. For either interpretation, the rate of observed association is greater than the one in 20 (5%) expected by chance. In addition, post hoc mining of the expression dataset showed that another SNP (rs10242920 - ELMO1) association that was consistent with the effect reported in the CGEMS dataset was also actually associated with altered expression in carriers, albeit with less significance (P = 0.005) than originally used for gene and SNP selection. These findings suggest that the combinatorial approach may be a useful method to prioritise candidate modifier genes for polymorphism association studies. It is notable that CIMBA GWA studies of BRCA1 and BRCA2 mutation carriers are currently underway [30]. One might therefore anticipate that the combinatorial approach would provide even greater enrichment for prioritising SNPs from GWA studies that directly relate to the disease state under study. Further studies with larger cohort size are therefore warranted to assess the benefit of carrying out such an approach.
Conclusions
We have explored the value of using biological information embedded in gene expression data to prioritise candidate modifier genes for SNP association studies. Using this combinatorial approach we were able to demonstrate a threefold enrichment of genes that contain SNPs associated with breast cancer risk for BRCA1 or BRCA2 mutation carriers. Most notable was the evidence that the SMAD3 gene, which encodes a key regulatory protein in the transforming growth factor beta signalling pathway, may contribute to increased risk of breast cancer in BRCA2 mutation carriers. These results suggest that the combinatorial approach may be a useful method to prioritise candidate modifier genes for polymorphism association studies.
Abbreviations
CGEMS: Cancer Genetic Markers of Susceptibility; CIMBA: Consortium of Investigators of Modifiers of BRCA1 and BRCA2; GWA: genome-wide association; RR: relative risk; SNP: single nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LCW, ABS and FJC conceived and designed the study. LCW, GC-T, ABS and FJC coordinated the study, and LCW drafted the manuscript. ABS and FJC supervised the analysis and participated in manuscript writing. ZSF and VSP carried out the statistical analysis, and ZSF contributed to the manuscript writing. XW, RT, NML, JB, and XC processed samples and acquired data. DS-L, CT, SG, SM, DM, J-PF, CD, RKS, BW, CE, IS, HD, AM, FBH, MV, MJH, AMWvdO, MRN, MGEMA, CMA, CJvA, PD, MMG, QW, CIS, DFE, SP, MC, CTO, DF, PH, DGE, FL, RE, LI, CC, RD, DE, K-RO, JC, TR, KLN, SMD, CFS, DG-K, A-CD, GP, AKG, TH, HN, BAA, MAC, HO, UK, AL, BA, PK, BM, OMS, LM, ACA, GC-T and FJC provided samples and information on the BRCA1 and BRCA2 mutation carriers included in this study. SH and OMS provided assistance with mutation nomenclature and classifications. LM and ACA maintained the database of BRCA1 and BRCA2 mutation carriers. All authors read and approved the manuscript.
Supplementary Material
Supplementary Table S1. Genes predicted to modify risk. Genes predicted to modify risk in BRCA1 and/or BRCA2 mutation carriers by Walker and colleagues [18].
Supplementary Table S2. List of 37 candidate BRCA1/2 risk modifier SNPs. Each SNP listed was tagged to a gene and shown to be associated with breast cancer risk from the CGEMS Study version 1.
Contributor Information
Logan C Walker, Email: logan.walker@qimr.edu.au.
Zachary S Fredericksen, Email: Fredericksen.Zachary@mayo.edu.
Xianshu Wang, Email: wang.xianshu@mayo.edu.
Robert Tarrell, Email: Tarrell.robert@mayo.edu.
Vernon S Pankratz, Email: pankratz.vernon@mayo.edu.
Noralane M Lindor, Email: Lindor.noralane@mayo.edu.
Jonathan Beesley, Email: Jonathan.beesley@qimr.edu.au.
Sue Healey, Email: Sue.healey@qimr.edu.au.
Xiaoqing Chen, Email: XiaoQing.Chen@qimr.edu.au.
Dominique Stoppa-Lyonnet, Email: dominique.stoppa-lyonnet@curie.net.
Carole Tirapo, Email: carole.tirapo@curie.net.
Sophie Giraud, Email: sophie.giraud@chu-lyon.fr.
Sylvie Mazoyer, Email: MAZOYER@lyon.fnclcc.fr.
Danièle Muller, Email: dmuller@strasbourg.fnclcc.fr.
Jean-Pierre Fricker, Email: jfricker@strasbourg.fnclcc.fr.
Capucine Delnatte, Email: c-delnatte@nantes.fnclcc.fr.
Rita K Schmutzler, Email: Rita.Schmutzler@uk-koeln.de.
Barbara Wappenschmidt, Email: Barbara.Wappenschmidt@uk-koeln.de.
Christoph Engel, Email: Christoph.engel@imise.uni-leipzig.de.
Ines Schönbuchner, Email: Schoenbuchner@biozentrum.uni-wuerzburg.de.
Helmut Deissler, Email: helmut.deissler@uniklinik-ulm.de.
Alfons Meindl, Email: Alfons.Meindl@lrz.tu-muenchen.de.
Frans B Hogervorst, Email: f.hogervorst@nki.nl.
Martijn Verheus, Email: m.verheus@nki.nl.
Maartje J Hooning, Email: m.hooning@erasmusmc.nl.
Ans MW van den Ouweland, Email: a.vandenouweland@erasmusmc.nl.
Marcel R Nelen, Email: m.nelen@antrg.umcn.nl.
Margreet GEM Ausems, Email: m.g.e.m.ausems@umcutrecht.nl.
Cora M Aalfs, Email: c.m.aalfs@amc.uva.nl.
Christi J van Asperen, Email: C.J.van_Asperen@lumc.nl.
Peter Devilee, Email: P.Devilee@lumc.nl.
Monique M Gerrits, Email: Monique.gerrits@mumc.nl.
Quinten Waisfisz, Email: q.waisfisz@vumc.nl.
Csilla I Szabo, Email: Szabo.Csilla@mayo.edu.
Douglas F Easton, Email: douglas@srl.cam.ac.uk.
Susan Peock, Email: susan.peock@srl.cam.ac.uk.
Margaret Cook, Email: margaret.cook@srl.cam.ac.uk.
Clare T Oliver, Email: clare.oliver@srl.cam.ac.uk.
Debra Frost, Email: debra.frost@srl.cam.ac.uk.
Patricia Harrington, Email: patricia.harrington@srl.cam.ac.uk.
D Gareth Evans, Email: Gareth.Evans@cmft.nhs.uk.
Fiona Lalloo, Email: Fiona.Lalloo@cmft.nhs.uk.
Ros Eeles, Email: Rosalind.Eeles@icr.ac.uk.
Louise Izatt, Email: Louise.izatt@nhs.net.
Carol Chu, Email: carol.chu@leedsth.nhs.uk.
Rosemarie Davidson, Email: Rosemarie.Davidson@ggc.scot.nhs.uk.
Diana Eccles, Email: D.M.Eccles@soton.ac.uk.
Kai-Ren Ong, Email: Kai-ren.Ong@bwhct.nhs.uk.
Jackie Cook, Email: Jackie.Cook@sch.nhs.uk.
Tim Rebbeck, Email: rebbeck@mail.med.upenn.edu.
Katherine L Nathanson, Email: knathans@exchange.upenn.edu.
Susan M Domchek, Email: Susan.Domchek@uphs.upenn.edu.
Christian F Singer, Email: Christian.singer@meduniwien.ac.at.
Daphne Gschwantler-Kaulich, Email: Daphne.gschwantler-kaulich@meduniwien.ac.at.
Anne-Catharina Dressler, Email: Catharina.Dressler@meduniwien.ac.at.
Georg Pfeiler, Email: georg.pfeiler@meduniwien.ac.at.
Andrew K Godwin, Email: Andrew.Godwin@fccc.edu.
Tuomas Heikkinen, Email: tuomas.heikkinen@helsinki.fi.
Heli Nevanlinna, Email: heli.nevanlinna@hus.fi.
Bjarni A Agnarsson, Email: bjarniaa@landspitali.is.
Maria Adelaide Caligo, Email: m.caligo@med.unipi.it.
Håkan Olsson, Email: hakan.olsson@med.lu.se.
Ulf Kristoffersson, Email: ulf.kristoffersson@med.lu.se.
Annelie Liljegren, Email: annelie.liljegren@karolinska.se.
Brita Arver, Email: brita.wasteson-arver@karolinska.se.
Per Karlsson, Email: per.karlsson@oncology.gu.se.
Beatrice Melin, Email: beatrice.melin@onkologi.umu.se.
Olga M Sinilnikova, Email: sinilnik@lyon.fnclcc.fr.
Lesley McGuffog, Email: l.mcguffog@srl.cam.ac.uk.
Antonis C Antoniou, Email: antonis@srl.cam.ac.uk.
Georgia Chenevix-Trench, Email: Georgia.Trench@qimr.edu.au.
Amanda B Spurdle, Email: Amanda.spurdle@qimr.edu.au.
Fergus J Couch, Email: couch.fergus@mayo.edu.
Acknowledgements
For the Kathleen Cunningham Consortium for Research into Familial Breast Cancer (kConFab), the authors thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow-Up Study (funded by National Health and Medical Research Council (NHMRC) grants 145684, 288704 and 454508) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia.
The MAYO study is supported in part by National Institute of Health grants (CA116167, CA116167Z, CA128978), a Specialized Program of Research Excellence (SPORE) grant in Breast Cancer (P50 CA116201), a grant from the Breast Cancer Research Foundation, and a grant from the Komen Foundation for the Cure.
For the Modifier Study of Quantitative Effects on Disease (ModSQuaD), CIS is supported by the Mayo Rochester Early Career Development Award for Non-Clinician Scientists. The authors acknowledge the contributions of Petr Pohlreich and Zdenek Kleibl (Department of Biochemistry and Experimental Oncology, First Faculty of Medicine, Charles University, Prague, Czech Republic) and the support of the Research Project of the Ministry of Education, Youth, and Sports of the Czech Republic (grant MSM0021620808 to MZ, Zdenek Kleibl, and Petr Pohlreich). Lenka Foretova, Machackova Eva, and Lukesova Miroslava are supported through the Ministry of Health (grant CR-MZ0 MOU 2005). The authors acknowledge the contribution of Kim De Leeneer, Bruce Poppe and Anne De Paepe. This research was supported by grant 1.5.150.07 from the Fund for Scientific Research Flanders (FWO) to Kathleen Claes, and by grant 12051203 from the Ghent University to Anne De Paepe. Bruce Poppe is Senior Clinical Investigator of the Fund for Scientific Research of Flanders (FWO - Vlaanderen).
SWE-BRCA collaborators include: Per Karlsson, Margareta Nordling, Annika Bergman and Zakaria Einbeigi (Gothenburg, Sahlgrenska University Hospital); Marie Stenmark-Askmalm and Sigrun Liedgren (Linköping University Hospital); Åke Borg, Niklas Loman, Håkan Olsson, Ulf Kristoffersson, Helena Jernström Katja Harbst and Karin Henriksson (Lund University Hospital); Annika Lindblom, Brita Arver, Anna von Wachenfeldt, Annelie Liljegren, Gisela Barbany-Bustinza and Johanna Rantala (Stockholm, Karolinska University Hospital); Beatrice Melin, Henrik Grönberg, Eva-Lena Stattin and Monica Emanuelsson (Umeå University Hospital); and Hans Ehrencrona, Richard Rosenquist Brandell and Niklas Dahl (Uppsala University Hospital).
The UPENN study is supported by the Breast Cancer Research Foundation (to KLN), and by the Mariann and Robert MacDonald Foundation (to SMD).
The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) collaborating centers include: coordinating center - Netherlands Cancer Institute, Amsterdam (Frans BL Hogervorst, Senno Verhoef, Martijn Verheus, Laura J van 't Veer, Flora E van Leeuwen, Matti A Rookus); Erasmus Medical Center, Rotterdam (Margriet Collée, Ans MW van den Ouweland, Agnes Jager, Maartje J Hooning, Madeleine MA Tilanus-Linthorst, Caroline Seynaeve); Leiden University Medical Center, Leiden (Christi J van Asperen, Juul T Wijnen, Maaike P Vreeswijk, Rob A Tollenaar, Peter Devilee); Radboud University Nijmegen Medical Center, Nijmegen (Marjolijn J Ligtenberg, Nicoline Hoogerbrugge); University Medical Center Utrecht, Utrecht (Margreet G Ausems, Rob B van der Luijt); Amsterdam Medical Center (Cora M Aalfs, Theo A van Os); VU University Medical Center, Amsterdam (Johan JP Gille, Quinten Waisfisz, Hanne EJ Meijers-Heijboer); University Hospital Maastricht, Maastricht (Encarna B Gomez-Garcia, Cees E van Roozendaal, Marinus J Blok); University Medical Center Groningen University (Jan C Oosterwijk, Annemarie H van der Hout, Marian J Mourits); and The Netherlands Foundation for the Detection of Hereditary Tumours, Leiden, The Netherlands (Hans F. Vasen). The HEBON study is supported by the Dutch Cancer Society grants NKI 1998-1854, NKI 2004-3088 and NKI 2007-3756.
The Epidemiological Study of BRCA1 and BRCA2 Mutation Carriers (EMBRACE) is supported by Cancer Research UK Grants C1287/A10118 and C1287/A8874. PH is supported by Cancer Research UK Grant C8197/A10123. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. RE is also supported by Cancer Research UK Grant C5047/A8385. DGE and FL are supported by an NIHR grant to the Biomedical Research Centre, Manchester. DFE is the principal investigator of the study. EMBRACE collaborating centres include: Coordinating Centre, Cambridge (Susan Peock, Margaret Cook, Clare Oliver, Debra Frost); North of Scotland Regional Genetics Service, Aberdeen (Helen Gregory, Zosia Miedzybrodzka); Northern Ireland Regional Genetics Service, Belfast (Patrick Morrison, Lisa Jeffers); West Midlands Regional Clinical Genetics Service, Birmingham (Trevor Cole, Carole McKeown, Kai-Ren Ong, Laura Boyes); South West Regional Genetics Service, Bristol (Alan Donaldson); East Anglian Regional Genetics Service, Cambridge (Joan Paterson); Medical Genetics Services for Wales, Cardiff (Alexandra Murray, Mark T Rogers, Emma McCann); St James's Hospital, Dublin & National Centre for Medical Genetics, Dublin (M John Kennedy, David Barton); South East of Scotland Regional Genetics Service, Edinburgh (Mary Porteous); Peninsula Clinical Genetics Service, Exeter (Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman); West of Scotland Regional Genetics Service, Glasgow (Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt, Sarah Gibson); South East Thames Regional Genetics Service, Guys Hospital London (Louise Izatt, Chris Jacobs, Caroline Langman); North West Thames Regional Genetics Service, Kennedy-Galton Centre, Harrow (Huw Dorkins); Leicestershire Clinical Genetics Service, Leicester (Julian Barwell); Yorkshire Regional Genetics Service, Leeds (Carol Chu, Tim Bishop, Julie Miller); Merseyside & Cheshire Clinical Genetics Service, Liverpool (Ian Ellis, Catherine Houghton); Manchester Regional Genetics Service, Manchester (D Gareth Evans, Fiona Lalloo, Jane Taylor); North East Thames Regional Genetics Service, NE Thames (Alison Male, Lucy Side, Cheryl Berlin); Nottingham Centre for Medical Genetics, Nottingham (Jacqueline Eason, Rebecca Collier); Northern Clinical Genetics Service, Newcastle (Fiona Douglas, Oonagh Claber); Oxford Regional Genetics Service, Oxford (Lisa Walker, Diane McLeod, Dorothy Halliday, Sarah Durrell, Barbara Stayner); The Institute of Cancer Research and Royal Marsden NHS Foundation Trust (Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Lucia D'Mello, Elizabeth Page, Audrey Ardern-Jones, Kelly Kohut, Jennifer Wiggins. Elena Castro, Anita Mitra, Lisa Robertson); North Trent Clinical Genetics Service, Sheffield (Jackie Cook, Oliver Quarrell, Cathryn Bardsley); South Essex Cancer Research Network, Southend (Anne Robinson); South West Thames Regional Genetics Service, London (Shirley Hodgson, Sheila Goff, Glen Brice, Lizzie Winchester); and Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton (Diana Eccles, Anneke Lucassen, Gillian Crawford, Emma Tyler, Donna McBride).
The GEMO Study (Cancer Genetics Network Groupe Génétique et Cancer, Fédération Nationale des Centres de Lutte Contre le Cancer, France) is supported by the Ligue National Contre le Cancer, the Association for International Cancer Research Grant (AICR-07-0454), and the Association Le cancer du sein, parlons-en! Award. The authors wish to thank all of the GEMO collaborating groups for their contribution to this study. GEMO collaborating centers include: Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Centre Hospitalier Universitaire de Lyon/Centre Léon Bérard, and UMR5201 CNRS, Université de Lyon, Lyon (Olga Sinilnikova, Laure Barjhoux, Sophie Giraud, Mélanie Léone, Sylvie Mazoyer); INSERM U509, Service de Génétique Oncologique, Institut Curie, Paris (Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Bruno Buecher, Claude Houdayer, Virginie Moncoutier, Muriel Belotti, Antoine de Pauw); Institut Gustave Roussy, Villejuif (Brigitte Bressac-de-Paillerets, Audrey Remenieras, Véronique Byrde, Olivier Caron, Gilbert Lenoir); Centre Jean Perrin, Clermont-Ferrand (Yves-Jean Bignon, Nancy Uhrhammer); Centre Léon Bérard, Lyon (Christine Lasset, Valérie Bonadona); Centre François Baclesse, Caen (Agnès Hardouin, Pascaline Berthet); Institut Paoli Calmettes, Marseille (Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, François Eisinger); Groupe Hospitalier Pitié-Salpétrière, Paris (Florence Coulet, Chrystelle Colas, Florent Soubrier); CHU de Arnaud-de-Villeneuve, Montpellier (Isabelle Coupier, Pascal Pujol); Centre Oscar Lambret, Lille (Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin, Claude Adenis); Centre René Huguenin, St Cloud (Etienne Rouleau, Rosette Lidereau, Liliane Demange, Catherine Nogues); Centre Paul Strauss, Strasbourg (Danièle Muller, Jean-Pierre Fricker); Institut Bergonié, Bordeaux (Michel Longy, Nicolas Sevenet); Institut Claudius Regaud, Toulouse (Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel); CHU de Grenoble (Dominique Leroux, Hélène Dreyfus, Christine Rebischung); CHU de Dijon (Cécile Cassini, Laurence Faivre); CHU de St-Etienne (Fabienne Prieur); Hôtel Dieu Centre Hospitalier, Chambéry (Sandra Fert Ferrer); Centre Antoine Lacassagne, Nice (Marc Frénay); CHU de Limoges (Laurence Vénat-Bouvet); CHU de Nantes (Capucine Delnatte); and. Creighton University, Omaha, USA (Henry T Lynch).
The Iceland Landspitali - University Hospital Study (ILUH) is supported by the Research Fund of Landspitali-University Hospital and the Walk together for breast cancer research.
GC-HBOC is supported by a grant of the German Cancer Aid (grant107054) to RKS. The authors thank Juliane Köhler for her excellent technical assistance and the centres of the GC-HBOC for providing samples and clinical data.
The HEBCS study was supported by Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society and the Sigrid Juselius Foundation. The authors thank Dr Kristiina Aittomäki, Dr Carl Blomqvist and Dr Kirsimari Aaltonen as well as RN Hanna Jäntti for their help with patient data and samples.
The Pisa Breast Cancer Study (PBCS) acknowledges Fondazione Cassa di Risparmio di Pisa, Istituto Toscano Tumori.
The Fox Chase Cancer Center (FCCC) acknowledges Ms JoEllen Weaver, Mr John Malick and Dr Betsy Bove for expert technical assistance. AKG was funded by SPORE P50 CA83638, U01 CA69631, 5U01 CA113916, and the Eileen Stein Jacoby Fund.
The Medical University of Vienna (MUV) collaborators include CF Singer, D Gschwantler-Kaulich, G Pfeiler, and A-C Spiess. This research project has been supported by the Austrian Society for Endocrinological Oncology and by the Comprehensive Cancer Center, Cluster Genetics and Epigenetics.
Georgetown acknowledges Claudine Isaacs and is supported by a National Cancer Institute Cancer Centre Support Grant to the Lombardi Comprehensive Cancer Centre (NCI P30 CA51008-12), Georgetown University, Washington, DC, USA.
LCW is a John Gavin Postdoctoral Fellow (Genesis Oncology Trust), ABS is an NHMRC Senior Research Fellow, and GCT is an NHMRC Senior Principal Research Fellow. ACA is a Cancer Research UK Senior Cancer Research Fellow, and LM, the CIMBA genotyping and data management are funded by Cancer Research - UK.
References
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Iversen ES, Friebel T, Finkelstein D, Weber BL, Eisen A, Peterson LE, Schildkraut JM, Isaacs C, Peshkin BN, Corio C, Leondaridis L, Tomlinson G, Dutson D, Kerber R, Amos CI, Strong LC, Berry DA, Euhus DM, Parmigiani G. Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J Clin Oncol. 2006;24:863–871. doi: 10.1200/JCO.2005.03.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet. 1995;56:265–271. [PMC free article] [PubMed] [Google Scholar]
- Ford D, Easton DF, Peto J. Estimates of the gene frequency of BRCA1 and its contribution to breast and ovarian cancer incidence. Am J Hum Genet. 1995;57:1457–1462. [PMC free article] [PubMed] [Google Scholar]
- Milne RL, Osorio A, Cajal TR, Vega A, Llort G, de la Hoya M, Diez O, Alonso MC, Lazaro C, Blanco I, Sanchez-de-Abajo A, Caldes T, Blanco A, Grana B, Duran M, Velasco E, Chirivella I, Cardenosa EE, Tejada MI, Beristain E, Miramar MD, Calvo MT, Martinez E, Guillen C, Salazar R, San Roman C, Antoniou AC, Urioste M, Benitez J. The average cumulative risks of breast and ovarian cancer for carriers of mutations in BRCA1 and BRCA2 attending genetic counseling units in Spain. Clin Cancer Res. 2008;14:2861–2869. doi: 10.1158/1078-0432.CCR-07-4436. [DOI] [PubMed] [Google Scholar]
- Simchoni S, Friedman E, Kaufman B, Gershoni-Baruch R, Orr-Urtreger A, Kedar-Barnes I, Shiri-Sverdlov R, Dagan E, Tsabari S, Shohat M, Catane R, King MC, Lahad A, Levy-Lahad E. Familial clustering of site-specific cancer risks associated with BRCA1 and BRCA2 mutations in the Ashkenazi Jewish population. Proc Natl Acad Sci USA. 2006;103:3770–3774. doi: 10.1073/pnas.0511301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Sinilnikova OM, Simard J, Leone M, Dumont M, Neuhausen SL, Struewing JP, Stoppa-Lyonnet D, Barjhoux L, Hughes DJ, Coupier I, Belotti M, Lasset C, Bonadona V, Bignon YJ, Rebbeck TR, Wagner T, Lynch HT, Domchek SM, Nathanson KL, Garber JE, Weitzel J, Narod SA, Tomlinson G, Olopade OI, Godwin A, Isaacs C, Jakubowska A, Lubinski J, Gronwald J. et al. RAD51 135G→C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Spurdle AB, Sinilnikova OM, Healey S, Pooley KA, Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N, Hofmann W, Sutter C, Niederacher D, Deissler H, Caldes T, Kampjarvi K, Nevanlinna H, Simard J, Beesley J, Chen X, Neuhausen SL, Rebbeck TR, Wagner T, Lynch HT, Isaacs C, Weitzel J, Ganz PA, Daly MB, Tomlinson G, Olopade OI. et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ, Sinilnikova O, Vierkant RA, Pankratz VS, Fredericksen ZS, Stoppa-Lyonnet D, Coupier I, Hughes D, Hardouin A, Berthet P, Peock S, Cook M, Baynes C, Hodgson S, Morrison PJ, Porteous ME, Jakubowska A, Lubinski J, Gronwald J, Spurdle AB, Schmutzler R, Versmold B, Engel C, Meindl A, Sutter C, Horst J, Schaefer D, Offit K, Kirchhoff T, Andrulis IL. et al. AURKA F31I polymorphism and breast cancer risk in BRCA1 and BRCA2 mutation carriers: a consortium of investigators of modifiers of BRCA1/2 study. Cancer Epidemiol Biomarkers Prev. 2007;16:1416–1421. doi: 10.1158/1055-9965.EPI-06-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnatty SE, Couch FJ, Fredericksen Z, Tarrell R, Spurdle AB, Beesley J, Chen X, Gschwantler-Kaulich D, Singer CF, Fuerhauser C, Fink-Retter A, Domchek SM, Nathanson KL, Pankratz VS, Lindor NM, Godwin AK, Caligo MA, Hopper J, Southey MC, Giles GG, Justenhoven C, Brauch H, Hamann U, Ko YD, Heikkinen T, Aaltonen K, Aittomaki K, Blomqvist C, Nevanlinna H, Hall P. et al. No evidence that GATA3 rs570613 SNP modifies breast cancer risk. Breast Cancer Res Treat. 2009;117:371–379. doi: 10.1007/s10549-008-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, Antoniou AC, Llopis TC, Nevanlinna H, Aittomaki K, Simard J, Spurdle AB, Couch FJ, Pereira LH, Greene MH, Andrulis IL, Pasche B, Kaklamani V, Hamann U, Szabo C, Peock S, Cook M, Harrington PA, Donaldson A, Male AM, Gardiner CA, Gregory H, Side LE, Robinson AC, Emmerson L, Ellis I, Peyrat JP, Fournier J, Vennin P, Adenis C. et al. No association of TGFB1 L10P genotypes and breast cancer risk in BRCA1 and BRCA2 mutation carriers: a multi-center cohort study. Breast Cancer Res Treat. 2009;115:185–192. doi: 10.1007/s10549-008-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Sinilnikova OM, McGuffog L, Healey S, Nevanlinna H, Heikkinen T, Simard J, Spurdle AB, Beesley J, Chen X, Neuhausen SL, Ding YC, Couch FJ, Wang X, Fredericksen Z, Peterlongo P, Peissel B, Bonanni B, Viel A, Bernard L, Radice P, Szabo CI, Foretova L, Zikan M, Claes K, Greene MH, Mai PL, Rennert G, Lejbkowicz F, Andrulis IL. et al. Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska A, Rozkrut D, Antoniou A, Hamann U, Lubinski J. The Leu33Pro polymorphism in the ITGB3 gene does not modify BRCA1/2-associated breast or ovarian cancer risks: results from a multicenter study among 15,542 BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2010;121:639–649. doi: 10.1007/s10549-009-0595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio A, Milne RL, Pita G, Peterlongo P, Heikkinen T, Simard J, Chenevix-Trench G, Spurdle AB, Beesley J, Chen X, Healey S, Neuhausen SL, Ding YC, Couch FJ, Wang X, Lindor N, Manoukian S, Barile M, Viel A, Tizzoni L, Szabo CI, Foretova L, Zikan M, Claes K, Greene MH, Mai P, Rennert G, Lejbkowicz F, Barnett-Griness O, Andrulis IL. et al. Evaluation of a candidate breast cancer associated SNP in ERCC4 as a risk modifier in BRCA1 and BRCA2 mutation carriers. Results from the Consortium of Investigators of Modifiers of BRCA1/BRCA2 (CIMBA) Br J Cancer. 2009;101:2048–2054. doi: 10.1038/sj.bjc.6605416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinilnikova OM, Antoniou AC, Simard J, Healey S, Leone M, Sinnett D, Spurdle AB, Beesley J, Chen X, Greene MH, Loud JT, Lejbkowicz F, Rennert G, Dishon S, Andrulis IL, Domchek SM, Nathanson KL, Manoukian S, Radice P, Konstantopoulou I, Blanco I, Laborde AL, Duran M, Osorio A, Benitez J, Hamann U, Hogervorst FB, van Os TA, Gille HJ, Peock S. et al. The TP53 Arg72Pro and MDM2 309G > T polymorphisms are not associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Br J Cancer. 2009;101:1456–1460. doi: 10.1038/sj.bjc.6605279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LC, Waddell N, Ten Haaf A, Grimmond S, Spurdle AB. Use of expression data and the CGEMS genome-wide breast cancer association study to identify genes that may modify risk in BRCA1/2 mutation carriers. Breast Cancer Res Treat. 2008;112:229–236. doi: 10.1007/s10549-007-9848-5. [DOI] [PubMed] [Google Scholar]
- Cancer Genetic Markers of Susceptibility (CGEMS) http://cgems.cancer.gov/
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzato EM, Tyrer J, Fasching PA, Beckmann MW, Ekici AB, Schulz-Wendtland R, Bojesen SE, Nordestgaard BG, Flyger H, Milne RL, Arias JI, Menendez P, Benitez J, Chang-Claude J, Hein R, Wang-Gohrke S, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Margolin S, Mannermaa A, Kosma VM, Kataja V, Beesley J, Chen X, Chenevix-Trench G, Couch FJ, Olson JE, Fredericksen ZS. et al. Association between a germline OCA2 polymorphism at chromosome 15q13.1 and estrogen receptor-negative breast cancer survival. J Natl Cancer Inst. 2010;102:650–662. doi: 10.1093/jnci/djq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemers FJ, Gunderson KL. Illumina, Inc. Pharmacogenomics. 2005;6:777–782. doi: 10.2217/14622416.6.7.777. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Goldgar DE, Andrieu N, Chang-Claude J, Brohet R, Rookus MA, Easton DF. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol. 2005;29:1–11. doi: 10.1002/gepi.20074. [DOI] [PubMed] [Google Scholar]
- Kramer JL, Velazquez IA, Chen BE, Rosenberg PS, Struewing JP, Greene MH. Prophylactic oophorectomy reduces breast cancer penetrance during prospective, long-term follow-up of BRCA1 mutation carriers. J Clin Oncol. 2005;23:8629–8635. doi: 10.1200/JCO.2005.02.9199. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Lynch HT, Neuhausen SL, Narod SA, Van't Veer L, Garber JE, Evans G, Isaacs C, Daly MB, Matloff E, Olopade OI, Weber BL. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N Engl J Med. 2002;346:1616–1622. doi: 10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- The R Project for Statistical Computing. http://www.r-project.org/index.html
- Preobrazhenska O, Yakymovych M, Kanamoto T, Yakymovych I, Stoika R, Heldin CH, Souchelnytskyi S. BRCA2 and Smad3 synergize in regulation of gene transcription. Oncogene. 2002;21:5660–5664. doi: 10.1038/sj.onc.1205732. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Akhurst RJ. Transforming growth factor-beta in breast cancer: too much, too late. Breast Cancer Res. 2009;11:202. doi: 10.1186/bcr2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WT, Lewis MT, Hess K, Wong H, Tsimelzon A, Karadag N, Cairo M, Wei C, Meric-Bernstam F, Brown P, Arun B, Hortobagyi GN, Sahin A, Chang JC. Decreased TGFβ signaling and increased COX2 expression in high risk women with increased mammographic breast density. Breast Cancer Res Treat. 2010;119:305–314. doi: 10.1007/s10549-009-0350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, Healey S, Morrison J, Kartsonaki C, Lesnick T, Ghoussaini M, Barrowdale D, Peock S, Cook M, Oliver C, Frost D, Eccles D, Evans DG, Eeles R, Izatt L, Chu C, Douglas F, Paterson J, Stoppa-Lyonnet D, Houdayer C, Mazoyer S, Giraud S, Lasset C, Remenieras A, Caron O. et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Genes predicted to modify risk. Genes predicted to modify risk in BRCA1 and/or BRCA2 mutation carriers by Walker and colleagues [18].
Supplementary Table S2. List of 37 candidate BRCA1/2 risk modifier SNPs. Each SNP listed was tagged to a gene and shown to be associated with breast cancer risk from the CGEMS Study version 1.