Abstract
Introduction
As a group, women who carry germline mutations in partner and localizer of breast cancer 2 susceptibility protein (PALB2) are at increased risk of breast cancer. Little is known about by how much or whether risk differs by mutation or family history, owing to the paucity of studies of cases unselected for family history.
Methods
We screened 1,403 case probands for PALB2 mutations in a population-based study of Australian women with invasive breast cancer stratified by age at onset. The age-specific risk of breast cancer was estimated from the cancer histories of first- and second-degree relatives of mutation-carrying probands using a modified segregation analysis that included a polygenic modifier and was conditioned on the carrier case proband. Further screening for PALB2 c.3113G > A (W1038X) was conducted for 779 families with multiple cases of breast cancer ascertained through family cancer clinics in Australia and New Zealand and 764 population-based controls.
Results
We found five independent case probands in the population-based sample with the protein-truncating mutation PALB2 c.3113G > A (W1038X); 2 of 695 were diagnosed before age 40 years and 3 of 708 were diagnosed when between ages 40 and 59 years. Both of the two early-onset carrier case probands had very strong family histories of breast cancer. Further testing found that the mutation segregated with breast cancer in these families. No c.3113G > A (W1038X) carriers were found in 764 population-based unaffected controls. The hazard ratio was estimated to be 30.1 (95% confidence interval (CI), 7.5 to 120; P < 0.0001), and the corresponding cumulative risk estimates were 49% (95% CI, 15 to 93) to age 50 and 91% (95% CI, 44 to 100) to age 70. We found another eight families carrying this mutation in 779 families with multiple cases of breast cancer ascertained through family cancer clinics.
Conclusions
The PALB2 c.3113G > A mutation appears to be associated with substantial risks of breast cancer that are of clinical relevance.
Introduction
An increasing number of so-called moderate-risk [1] breast cancer susceptibility genes have been identified. Protein-truncating mutations in ataxia telangiectasia mutated (ATM), breast cancer type 1 susceptibility protein (BRCA1)-interacting protein 1 (BRIP1) and partner and localizer of the breast cancer 2 early onset protein (BRCA2) (PALB2), as well as CHK2 checkpoint homolog (CHEK2), have been observed to be nearly 10 times more likely in patients with strong breast cancer family histories than in unaffected controls [2-4]. The implications of these observations are not straightforward, so it is difficult to use the cancer histories of these families to make informative estimates of risk (penetrance) when the reason for studying them in the first place is their family cancer history [2-5]. Inference is more informative when based on testing families unselected for family history, but there is a paucity of such data for these genes.
Predicated on the assumption that the increased risk associated with a mutation multiplies a woman's underlying familial genetic risk, for these genes it has been estimated that rare protein-truncating mutations are associated with, on average, two- to threefold increases in risk [1-4]. Therefore, this increased risk must be interpreted by taking into account a carrier's family history. Consequently, for a given gene, the absolute risk is not the same for all carriers of such mutations [5].
It is also possible that the increased risk is not the same for all mutations. Some mutations in moderate-risk genes are high risk. For example, population-based studies have shown that the 7217T > G variant in ATM is associated with a substantially increased risk comparable to that for mutations in BRCA1 and BRCA2 [6], as has a subset of rare substitutions in ATM [7]. Another example is the Finnish founder PALB2 mutation; on the basis of studying cases unselected for family history, it has been estimated to be associated with a sixfold increased risk and a cumulative breast cancer risk of 40% by age 70 years [8].
We tested Australian women affected and unaffected by breast cancer and selected and unselected for family history for PALB2 mutations to estimate the prevalence and penetrance of these mutations in the Australian population.
Materials and methods
Participants
The Australian Breast Cancer Family Registry (ABCFR) is a population-based, case control family study of breast cancer with an emphasis on early-onset breast cancer that is being carried out in Melbourne and Sydney, Australia [9-11], and is part of the Breast Cancer Family Registry [12]. All adult women living in the metropolitan areas of Melbourne and Sydney who were diagnosed with a histologically confirmed first primary cancer of the breast were invited to participate in the ABCFR. From January 1, 1992, through September 30, 1999, in Melbourne and from January 1, 1993, through December 31, 1998, in Sydney, women younger than 40 years of age at diagnosis were selected; after January 1, 1996, random samples of women ages 40-49 years and 50-59 years at diagnosis were selected. All recruitment was done regardless of family history.
Cases were identified by use of the Victoria and New South Wales cancer registries, to which notification of cancer diagnoses is a legislative requirement. Overall, we approached 2,303 eligible individuals (ages at diagnosis: <40 years, 1,208; 40-49 years, 551; 50-59 years, 544), which resulted in 1,610 cases, and blood samples were collected from 94% of these cases. Cancer in relatives was verified by cancer registry reports, medical records or death certificates.
Previous mutation screening performed on the germline DNA of these women included screening for BRCA1, BRCA2, TP53 and ATM using a variety of mutation detection techniques [13-20]. These identified BRCA1 (n = 47), BRCA2 (n = 48), ATM (n = 1) and tumor protein 53 (TP53) (n = 5) mutation carriers who were not excluded from PALB2 mutation screening performed in this study. Six hundred ninety-five women under the age of 40 years diagnosed with breast cancer were available from the ABCFR for high-resolution melting (HRM) curve analysis of PALB2, and 708 women ages 40 years and over and diagnosed with breast cancer were screened for PALB2 c.3113G > A using the Custom TaqMan SNP Genotyping Assay (Applied Biosystems, Carlsbad, CA, USA). The study was approved by the ethics committees of The University of Melbourne and The Cancer Council Victoria, and all participants provided written informed consent for participation in the study.
The Kathleen Cuningham Foundation Consortium for Research in Familial Breast Cancer (kConFab) collected the same epidemiological, family history and lifestyle data, as well as biospecimens, as the ABCFR from more than 1,500 Australasian families with multiple cases of breast encompassing 10,000 individuals gathered from family cancer clinics in Australia and New Zealand [21]. For 778 families we obtained a DNA sample from the youngest affected member who had provided a blood sample and who had been tested and found not to carry a BRCA1 or BRCA2 mutation. All participants provided written informed consent for participation in the study.
High-resolution melting curve analysis and sequencing analysis of PALB2
The PALB2 genomic sequence was obtained from the National Center for Biotechnology Information (reference sequence number NG_007406.1). Primers (Geneworks, Hindmarsh, South Australia, Australia) (Additional file 1) were designed by using Primer3 software (Whitehead Institute and Howard Hughes Medical Institute, Cambridge, MA, USA). For optimal performance of the HRM curve analysis, primers were designed to amplify DNA products between 100 and 310 bp. A total of 35 fragments were designed to cover the coding and flanking intronic regions of PALB2. The primer sequences and annealing temperatures are listed in Additional file 2. Initially, 96 DNA were Sanger-sequenced as described in Tischkowitz et al. [22]. These data and the corresponding DNA were then used to establish the optimal conditions for HRM curve analysis. DNA extracted from peripheral blood samples provided by 1,473 case probands (695 from women diagnosed with breast cancer under the age of 40 years participating in the ABCFR and 778 from kConFab) were screened for germline PALB2 mutations using HRM curve analysis [23,24]. DNA was then systematically screened using this established method. HRM reactions were carried out in 15-μL volumes and included 1.5 μL of 10 × polymerase chain reaction (PCR) buffer (Applied Biosystems, Victoria, Australia), a 3 mM final concentration of MgCl2 (Applied Biosystems), a 100 μM final concentration of deoxyribonucleotide triphosphate (dNTP) (Bioline, Alexandria, New South Wales, Australia), a 200 nM final concentration of each primer (Geneworks) (Additonal File 2), a 2.3 μM final concentration of Syto9 (Invitrogen, Victoria, Australia), 0.25 U of AmpliTaq Gold (Applied Biosystems) and 3 μL of Q solution (Qiagen, Victoria, Australia). Each reaction underwent a hold of 10 minutes at 95°C and 40 cycles of amplification of 30 s at 95°C and 1 minute at annealing temperature followed by melting to dissociate double-stranded DNA. The temperature range for melting was set at ± 10°C of the melting temperature of each amplicon with a rise in temperature of 0.05°C/s. HRM analysis was performed using Rotor-Gene 6000 Series Software 1.7 (Qiagen). Fragments displaying aberrant melt curves were sequenced to determine potential underlying genetic variations. For sequencing reactions, we utilized larger amplicons than those generated during HRM curve analysis (Additional file 1). Sequencing was carried out in 10-μL reactions, which included 1 μL of 10 × PCR buffer (Applied Biosystems), a 3 mM final concentration of MgCl2 (Applied Biosystems), a 100 μM final concentration of dNTP (Bioline), a 200 nM concentration of each primer (Geneworks), 0.25 U of AmpliTaq Gold (Applied Biosystems) and 3 μL of Q solution (Qiagen). PCR products were purified and analyzed on a 3130xl Genetic Analyser (Applied Biosystems) and the results were viewed using Chromas 1.45 (Technelusium, Tewantin, Queensland, Australia).
TaqMan assays
Using the methods of Orlando et al. [25] and Ratnasinghe et al. [26], DNA extracted from the peripheral blood of 708 ABCFR probands diagnosed with breast cancer over the age of 40 years, 403 probands selected from the kConFab resource and 764 unaffected population controls from the ABCFR were analyzed for PALB2, c.3113G > A using the TaqMan assay. Each 10-μL reaction contained 5 μL of 2 × TaqMan Genotyping Master Mix (Applied Biosystems) and 0.125 μL of 40 × Custom Taqman SNP Genotyping Assay mix (Applied Biosystems), and the reactions were performed using a LightCycler 480 SW1.5 (Roche, Penzberg, Germany).
Statistical methods for penetrance analyses
The age-specific hazard ratio (HR) for breast cancer (that is, the ratio of the age-specific breast cancer incidence rates for carriers of the mutation to that for noncarriers) was estimated using modified segregation analysis [27]. Models were fitted by the method of maximum likelihood using the statistical package MENDEL version 3.2 [28] (Department of Human Genetics, UCLA School of Medicine, Los Angeles, California, USA). To adjust for ascertainment, the likelihood for each pedigree was conditioned on the proband's phenotype (breast cancer status and age of onset) and genotype. Each pedigree involved data for the carrier proband and her first- and second-degree relatives.
A mixed model was employed which incorporates an unmeasured polygenic factor to model the effect on breast cancer risk of a large number of unmeasured genes in addition to the measured major gene [29]. The polygenic part of this model was implemented via a hypergeometric polygenic model with four loci [30] and postulates a normally distributed random variable G for each person so that these variables are correlated within families (see section 8.9 of Lange et al. [31]). A woman's age at breast cancer diagnosis was modeled as a random variable whose HR was, for noncarriers, exp(G) times the Australian breast cancer incidence rate for 1992-2002 [32] or, for carriers, the product of this HR multiplied by the age-specific HR. As in Antoniou et al. [33], the variance of G was chosen to be 1.67, and the mean was chosen so that the average HR for noncarriers equaled the population incidence. When testing for an age dependence of the HR, the model with a constant HR was compared to one where the HR was a continuous, piecewise linear function of age which was constant before age 40 years, linear between ages 40 and 60 years and constant after age 60 years.
All models assumed Hardy-Weinberg equilibrium at the PALB2 locus, a dominant action of PALB2 c.3113 G > A on breast cancer risk, conditional independence of all phenotypes given genotypes and an allele frequency of 0.001 for the variant in the Australian population. Two-sided P values for the modified segregation analyses were based on the likelihood ratio test.
Age-specific cumulative risk estimates were calculated from these HRs as one minus the exponential value of the cumulative HR, and the corresponding confidence intervals were calculated using a parametric bootstrap with 5,000 replications. More specifically, 5,000 draws were taken from the normal distribution that the parameter estimates would be expected to follow under asymptotic likelihood theory. For each age, corresponding values of the cumulative risk were calculated, and the 95% CI was taken to be the 2.5th and 97.5th percentiles of this sample. To test the accuracy of this estimate, we compared an appropriate weighted average of the corresponding survival functions (which are one minus the cumulative risk) with the Kaplan-Meier survival curve [33].
Results
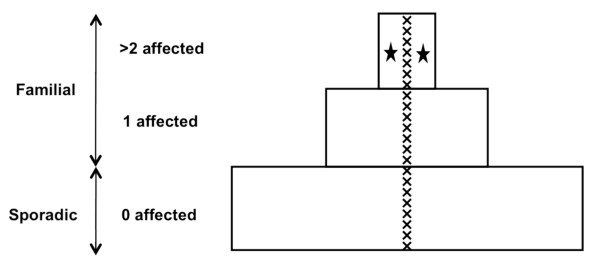
We tested for PALB2 mutations using HRM curve analysis to scan DNA extracted from peripheral blood samples taken from a sample unselected for family history of women diagnosed with breast cancer before the age of 40 years (probands) from a population-based case control family study (ABCFR). Two of the 695 probands diagnosed before the age of 40 years were found to carry the PALB2 c.3113G > A (W1038X) mutation, and both had strong family histories of breast cancer. Figure 1 shows that only 8% of all tested probands had two or more first- or second-degree relatives with breast or ovarian cancer [13], yet both these carriers were in this extreme group.
Figure 1.
Women diagnosed with breast cancer under age 40 years: The Australian Breast Cancer Family Registry. The majority of breast cancer patients diagnosed before the age of 40 years have no affected family members (zero affected; the "sporadic" breast cancer group), some probands have one affected family member (one affected), and ~8% have a stronger family history (the "familial" breast cancer group). A strong family history is defined as the case proband having two or more first- or second-degree relatives affected by breast or ovarian cancer. x represents the 22 partner and localizer of the breast cancer 2 early onset protein (BRCA2) mutation carriers identified by previous testing in the ABCFR [13,19]. The filled black stars represent the two probands found to carry PALB2 c.3113G > A who both have a very strong family history of breast cancer. Adapted from Hopper [42].
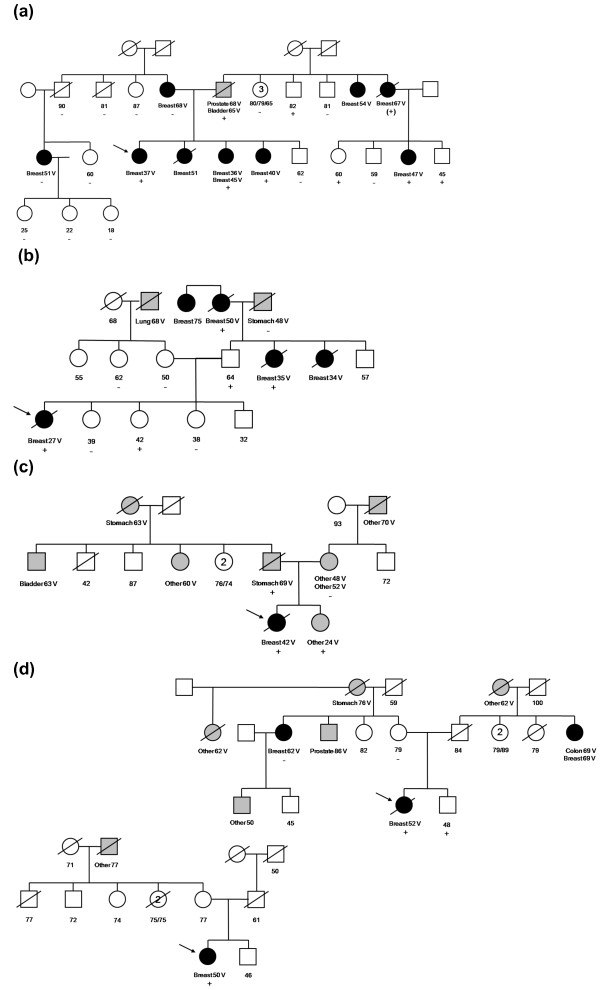
Proband 1 was diagnosed at the age of 37 years with a grade 3 infiltrating ductal carcinoma. Figure 2a shows that proband 1 had three sisters who had all been diagnosed with breast cancer (one had two primary diagnoses) at the ages of 36, 40, 45 and 51 years. Her mother had been diagnosed with breast cancer at the age of 68 years, and a maternal cousin had a breast cancer diagnosis at the age of 51 years. The proband's father had been diagnosed with bladder cancer at the age of 65 years and prostate cancer at the age of 68 years. Two paternal aunts had breast cancer diagnoses at ages 67 and 54 years, and a paternal cousin (the daughter of an affected paternal aunt) had been diagnosed with breast cancer at the age of 47 years. Predictive testing was performed for this PALB2 mutation using blood samples collected from 22 family members. Four affected female relatives were found to carry this mutation (one was an obligate carrier) that involved the paternal lineage. The proband's mother, maternal uncle and a maternal cousin with breast cancer did not carry the mutation.
Figure 2.
Pedigrees of the PALB2 c.3113G > A mutation-carrying families. Pedigrees of the PALB2 c.3113G > A mutation carrying families with (a) probands (indicated by arrows) and (b) families with probands diagnosed under the age of 40 years identified in the population-based study. (c) Family with probands diagnosed under the age of 50 years and (d) families diagnosed under the age of 60 years. Breast cancer is indicated by black filled symbols, and other cancers are indicated by gray filled symbols. All primary cancer diagnoses are indicated for each individual. Numbers within symbols represent multiple individuals. Breast, breast cancer; V, cancer verified; +, PALB2 c.3113G > A-positive; -, PALB2 c.3113G > A-negative; (+), obligate carrier;/, deceased.
Proband 2 was diagnosed with a grade 3 infiltrating ductal carcinoma at the age of 27 years. Figure 2b shows that she had two paternal aunts diagnosed with breast cancer at ages 34 and 35 years and a paternal grandmother diagnosed at the age of 50 years (all deceased). Predictive testing was performed for this PALB2 mutation using blood samples collected from seven family members as well as paraffin-embedded tissue from three of the affected relatives (one paternal aunt and two paternal grandparents). The proband's father and two affected female relatives related to the father carried the mutation. One unaffected sister of the proband, age 42 years at last contact, carried PALB2 c.3113G > A. The proband's mother, a maternal aunt and two sisters, all unaffected, did not carry the mutation.
We then used a TaqMan assay to screen women diagnosed with breast cancer between the ages of 40 and 59 years and control probands participating in the ABCFR for the PALB2 c.3113G > A mutation. One of the 360 probands diagnosed at age 40-49 years and 2 of 348 diagnosed at ages 50-59 years were found to carry the mutation. Figures 2c-2d show that all of these carriers had a family cancer history, and one included verified reports of breast cancer in relatives. None of 764 tested population-based controls carried this mutation.
Using the five population-based mutation carrier families, we estimated the breast cancer HR using the cancer histories of first- and second-degree relatives and found it to be 30.1 (95% CI, 7.5-120; P < 0.0001), independent of age (P = 0.8; though because of low power, a modest age dependence cannot be ruled out). These data corresponded to estimated age-specific cumulative risk ratios (penetrance) of 49% (95% CI, 15-93) to age 50 years and 91% (95% CI, 44-100) to age 70 years (see Figure 3). Our confidence in these estimates was enhanced by their high degree of agreement with the Kaplan-Meier survival curves for female first- and second-degree relatives of the probands, which accounted for the fact that these relatives are a mix of carriers and noncarriers (see Figure 4). The HR corresponding to the PALB2 W1038X variant could not be distinguished from the average of the age-specific HRs for BRCA2 mutations reported by Antoniou et al. [34] of 13.6 (P = 0.3). It was higher than a HR of 2.3, which is roughly equivalent to the odds ratio of 2.3 for PALB2 variants reported by Rahman et al. [4] (P = 0.0003).
Figure 3.
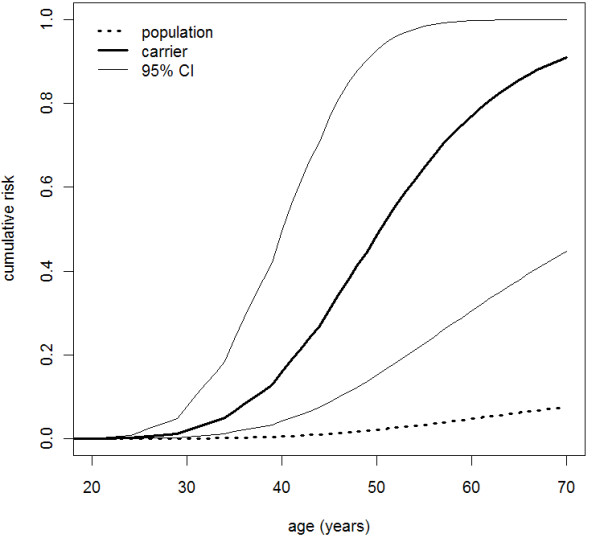
Age-specific cumulative risks for PALB2 c.3113G > A mutation carriers. Age-specific cumulative risks of breast cancer for women carrying the PALB2 c.3113G > A mutation (unbroken lines) and for women in the general Australian population (dotted line).
Figure 4.
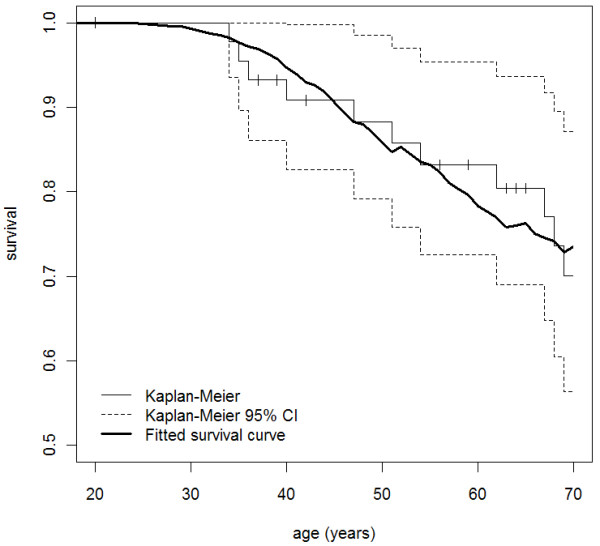
The Kaplan-Meier survival curves for PALB2 c.3113G > A mutation carriers. The Kaplan-Meier survival curve with breast cancer as the outcome for female first- and second-degree relatives of the probands with the fitted survival curve overlaid. The fitted survival curve was calculated as the average of the survival curves for carriers and noncarriers, each of these being equal to the exponential minus the appropriate cumulative incidences. For the averaging of survival curves, we used age-specific weights equal to the expected proportion of relatives at risk of breast cancer at a given age who carry (for the carrier survival curve) or do not carry (for the noncarrier survival curve) the partner and localizer of breast cancer 2 susceptibility protein (PALB2) mutation. Carrier probabilities for the relatives were calculated from known genotypes and family relationships, but not from affected status or other phenotypes, using a modified version of Mendel 3.2 software [28].
HRM screening for PALB2 mutations for 779 families with multiple cases of breast cancer recruited through Australian and New Zealand clinics by the kConFab consortium identified a further eight (1.0%) families that carried the PALB2 c.3113G > A mutation. We did not attempt to estimate penetrance using these family histories, since they were nonsystematically selected by their family histories of cancer.
Discussion
Studies conducted in the United Kingdom, Finland, Italy, Spain and Canada have shown that, while rare, mutations in PALB2 are far more common in breast cancer patients with a strong family history than in unaffected population-based controls. In the UK study, 10 (1.1%) of 923 strongly familial cases, compared with 0 of 1,084 unaffected controls, were mutation carriers (P = 0.004) [4]. The Finnish recurrent mutation PALB2 (c.1592delT) was carried by 3 (2.7%) of 113 familial breast cancer cases compared with 6 (0.2%) of 2,501 controls, an 11-fold difference (95% CI, 3-44) [35]. A French-Canadian founder mutation, c.2323C > T, resulting in Q775X, was found in 2 (0.6%) of 356 unselected early-onset cases compared with 1 (2%) of 50 familial cases and none (0%) of 6442 newborns [36]. Papi et al. [37] screened 132 Italian families with breast cancer without BRCA1 or BRCA2 mutations and identified one protein-truncating mutation (c.2257C > T, R753X) that was not observed in 300 control DNA. Garcia et al. [38] reported one protein-truncating mutation (c.1056-1057delGA, K3531X) in a screen of 95 families with multiple cases of breast cancer without BRCA1 or BRCA2 mutations. These observations, while showing that PALB2 mutations are associated with an increased risk of breast cancer, cannot be used to obtain direct or precise estimates of the magnitude of increased risk [4,5].
Estimation of the average increased risk associated with PALB2 mutations has been performed previously using an indirect method by presuming a polygenic modifier model. This method estimated that, on average, PALB2 protein-truncating mutations conferred a 2.3-fold increased risk of breast cancer (95% CI, 1.4-3.9; P = 0.0025) [4]. For one specific Finnish founder mutation, a direct estimate was made from a sample of population-based cases unselected for family history and screened for the PALB2 1592delT mutation [8]. Eighteen (0.9%) of the 1,918 cases carried the mutation. It was estimated from their family histories that carriers in this setting are at about six times the general population risk (95% CI, 2-17; P = 0.01), equivalent to a breast cancer risk of 40% by age 70 years (95% CI, 17-77), which is comparable to the 45% estimate found for BRCA2 mutation carriers in a large study of unselected carrier families [34].
In Australia, a PALB2 mutation has been found, c.3113G > A, carried by 5 (0.4%) of 1,403 unselected population-based patients diagnosed before age 60 years, 8 (1%) of 779 cases from families with multiple cases of breast cancer, and none of 764 unaffected population-based controls. This mutation was found twice in a UK study [4] that screened 923 cases from families with multiple cases of breast cancer. The Australian carrier families reported here are predominantly of Australian, English and Scottish heritage. These are the only reports of this mutation we know of, so it is possible that this mutation originated in the United Kingdom.
We estimated from analyses of case carrier families that carriers of the PALB2 c.3113G > A mutation who are relatives of unselected case carriers have a high risk of developing breast cancer: about 50% to age 50 years and 90% to age 70 years. The lower bound of the 95% CI of the latter estimate is about the same as the average risk for BRCA2 mutation carriers estimated using the same design and statistical methodology [39]. Therefore, this mutation is associated with as high a risk as other mutations being tested for by cancer family genetics services across the world.
For families with multiple cases of breast cancer currently attending cancer genetics services in Australia, at most 20% of those screened for BRCA1 and BRCA2 are found to carry mutations [21], which is similar to the rates in the United Kingdom [40] and the United States [41]. Although PALB2 c.3113G > A is rare, testing for it is inexpensive and has clinical utility. By virtue of being based on case families, our risk estimate applies to women with at least some family history, so it is appropriate for counseling carriers identified in families with multiple cases of breast cancer who use cancer family genetics services.
Conclusions
Given that carriers of this PALB2 mutation appear to be at least at the conventional level of high risk, thought to apply to BRCA1 and BRCA2 mutation carriers, testing would seem justified in a clinical genetics setting. For the women and their family members who carry these mutations, it is potentially important that they be identified so that they can be offered appropriate prevention, screening and clinical management. There may be other PALB2 mutations that are also as highly penetrant, and this paradigm might apply to other genes, such at ATM, BRIP1 and CHEK2, for which mutations appear to be associated, on average, with moderately increased risks, but for which some mutations might be associated with high risk. Targeted clinical testing of such high-risk mutations in these genes could be justified.
Abbreviations
ABCFS: Australian Breast Cancer Family Study; HR: hazard ratio; HRM: high-resolution melting; kConFab: The Kathleen Cuningham Foundation Consortium for Research in Familial Breast Cancer.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MCS led this study, including the conception, design, acquisition of data, analysis, interpretation of data and preparation of the manuscript. ZLT conducted molecular analyses and contributed to the preparation of the manuscript. JGD led aspects of the analyses and contributed to the preparation of the manuscript. FAO conducted molecular analyses and contributed to the preparation of the manuscript. DJP provided critical input to the molecular analysis and manuscript preparation. MT conducted molecular analyses and contributed to the preparation of the manuscript. NS conducted molecular analyses and contributed to the preparation of the manuscript. GBB contributed to aspects of the analyses and contributed to the preparation of the manuscript. CA contributed data from the Australian Breast Cancer Family Study (ABCFS) resource and contributed to the preparation of the manuscript. IW provided critical input to the molecular analysis and manuscript preparation. LB led aspects of the analyses and contributed to the preparation of the manuscript. GGG, cofounder of the ABCFS resource, contributed data from the ABCFS and contributed to the preparation of the manuscript. DEG provided critical input to the molecular analysis, statistical analyses and manuscript preparation. WDF conducted molecular analyses and contributed to the preparation of the manuscript. JLH, cofounder of the ABCFS resource, contributed data from the ABCFS, contributed to analyses and the preparation of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Sequencing primer sequences. Primers designed for DNA sequencing, including DNA fragment size (bp) and annealing temperatures.
Polymerase chain reaction primer sequences and annealing temperatures. The primer sequences and annealing temperatures designed to amplify 35 fragments (including the coding and flanking intronic regions) of partner and localizer of breast cancer 2 susceptibility protein (PALB2).
Contributor Information
Melissa C Southey, Email: msouthey@unimelb.edu.au.
Zhi L Teo, Email: z.teo@pgrad.unimelb.edu.au.
James G Dowty, Email: jdowty@unimelb.edu.au.
Fabrice A Odefrey, Email: fodefrey@unimelb.edu.au.
Daniel J Park, Email: djp@unimelb.edu.au.
Marc Tischkowitz, Email: marc.tischkowitz@mcgill.ca.
Nelly Sabbaghian, Email: nelly.sabbaghian@mail.mcgill.ca.
Carmel Apicella, Email: capic@unimelb.edu.au.
Graham B Byrnes, Email: byrnesg@iarc.fr.
Ingrid Winship, Email: iwinship@unimelb.edu.au.
Laura Baglietto, Email: Laura.Baglietto@cancervic.org.au.
Graham G Giles, Email: Graham.Giles@cancervic.org.au.
David E Goldgar, Email: david.goldgar@hsc.utah.edu.
William D Foulkes, Email: william.foulkes@mcgill.ca.
John L Hopper, Email: j.hopper@unimelb.edu.au.
kConFab, Email: msouthey@unimelb.edu.au.
Acknowledgements
The Australian Breast Cancer Family Registry was supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the National Cancer Institute, National Institutes of Health, under grant RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry and principal investigators. The University of Melbourne (U01 CA69638) contributed data to this study. The content of this manuscript does not necessarily reflect the views or the policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. government or the Breast Cancer Family Registry. We extend our thanks to the many women and their families who generously participated in the Australian Breast Cancer Family Study and consented to allow us access to their pathology material.
We thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics and the Clinical Follow Up Study (funded by National Health and Medical Research Council (NHMRC) grants 145684, 288704 and 454508) for their contributions to this resource, as well as the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC and the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia.
We thank Margaret McCredie for her key role in the establishment and leadership of the ABCFS in Sydney, Australia. MCS is a National Health and Medical Research Council Senior Research Fellow and Victorian Breast Cancer Research Consortium (VBCRC) Group Leader. JLH is an Australia Fellow of the National Health and Medical Research Council and a VBCRC Group Leader. MT is supported by the Susan G. Komen Foundation for the Cure, the Jewish General Hospital Weekend to End Breast Cancer and Fonds de la Recherche en Santé du Québec (FRSQ). WDF is a Chercheur National of the FRSQ.
References
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, McGuffog L, Evans DG, Eccles D. Breast Cancer Susceptibility Collaboration (UK) Easton DF, Stratton MR, Rahman N. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D. Breast Cancer Susceptibility Collaboration (UK) Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D. Breast Cancer Susceptibility Collaboration (UK) Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes GB, Southey MC, Hopper JL. Are the so-called low penetrance breast cancer genes, ATM, BRIP1, PALB2 and CHEK2, high risk for women with strong family histories? Breast Cancer Res. 2008;10:208. doi: 10.1186/bcr2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JL, Teraoka S, Southey MC, Jenkins MA, Andrulis IL, Knight JA, John EM, Lapinski R, Wolitzer AL, Whittemore AS, West D, Seminara D, Olson ER, Spurdle AB, Chenevix-Trench G, Giles GG, Hopper JL, Concannon P. Population-based estimates of breast cancer risks associated with ATM gene variants c.7271T > G and c.1066-6T > G (IVS10-6T > G) from the Breast Cancer Family Registry. Hum Mutat. 2006;27:1122–1128. doi: 10.1002/humu.20415. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Oefner PJ, Babikyan D, Hartmann A, Healey S, Le Calvez-Kelm F, Lesueur F, Byrnes GB, Chuang SC, Forey N, Feuchtinger C, Gioia L, Hall J, Hashibe M, Herte B, McKay-Chopin S, Thomas A, Vallée MP, Voegele C, Webb PM, Whiteman DC. Australian Cancer Study; Breast Cancer Family Registries (BCFR); Kathleen Cuningham Foundation Consortium for Research into Familial Aspects of Breast Cancer (kConFab) Sangrajrang S, Hopper JL, Southey MC, Andrulis IL, John EM, Chenevix-Trench G. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet. 2009;85:427–446. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkko H, Dowty JG, Nikkilä J, Syrjäkoski K, Mannermaa A, Pylkäs K, Southey MC, Holli K, Kallioniemi A, Jukkola-Vuorinen A, Kataja V, Kosma VM, Xia B, Livingston DM, Winqvist R, Hopper JL. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clin Cancer Res. 2008;14:4667–4671. doi: 10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- McCredie MR, Dite GS, Giles GG, Hopper JL. Breast cancer in Australian women under the age of 40. Cancer Causes Control. 1998;9:189–198. doi: 10.1023/A:1008886328352. [DOI] [PubMed] [Google Scholar]
- Hopper JL, Giles GG, McCredie MR, Boyle P. Background rational and protocol for a case-control-family study of breast cancer. Breast. 1994;3:79–86. doi: 10.1016/0960-9776(94)90003-5. [DOI] [Google Scholar]
- Hopper JL, Chenevix-Trench G, Jolley DJ, Dite GS, Jenkins MA, Venter DJ, McCredie MR, Giles GG. Design and analysis issues in a population-based, case-control-family study of the genetic epidemiology of breast cancer and the Cooperative Family Registry for Breast Cancer Studies (CFRBCS) J Natl Cancer Inst Monogr. 1999;26:95–100. doi: 10.1093/oxfordjournals.jncimonographs.a024232. [DOI] [PubMed] [Google Scholar]
- John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, Ziogas A, Andrulis IL, Anton-Culver H, Boyd N, Buys SS, Daly MB, O'Malley FP, Santella RM, Southey MC, Venne VL, Venter DJ, West DW, Whittemore AS, Seminara D. Breast Cancer Family Registry. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dite GS, Jenkins MA, Southey MC, Hocking JS, Giles GG, McCredie MR, Venter DJ, Hopper JL. Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst. 2003;95:448–457. doi: 10.1093/jnci/95.6.448. [DOI] [PubMed] [Google Scholar]
- Smith LD, Tesoriero AA, Ramus SJ, Dite G, Royce SG, Giles GG, McCredie MR, Hopper JL, Southey MC. BRCA1 promoter deletions in young women with breast cancer and a strong family history: a population-based study. Eur J Cancer. 2007;43:823–827. doi: 10.1016/j.ejca.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey MC, Tesoriero AA, Andersen CR, Jennings KM, Brown SM, Dite GS, Jenkins MA, Osborne RH, Maskiell JA, Porter L, Giles GG, McCredie MR, Hopper JL, Venter DJ. BRCA1 mutations and other sequence variants in a population based sample of Australian women with breast cancer. Br J Cancer. 1999;79:34–39. doi: 10.1038/sj.bjc.6690008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis IL, Anton-Culver H, Beck J, Bove B, Boyd J, Buys S, Godwin AK, Hopper JL, Li F, Neuhausen SL, Ozcelik H, Peel D, Santella RM, Southey MC, van Orsouw NJ, Venter DJ, Vijg J, Whittemore AS. Cooperative Family Registry for Breast Cancer studies. Comparison of DNA- and RNA-based methods for detection of truncating BRCA1 mutations. Hum Mutat. 2002;20:65–73. doi: 10.1002/humu.10097. [DOI] [PubMed] [Google Scholar]
- Chenevix-Trench G, Spurdle AB, Gatei M, Kelly H, Marsh A, Chen X, Donn K, Cummings M, Nyholt D, Jenkins MA, Scott C, Pupo GM, Dörk T, Bendix R, Kirk J, Tucker K, McCredie MR, Hopper JL, Sambrook J, Mann GJ, Khanna KK. Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002;94:205–215. doi: 10.1093/jnci/94.3.205. [DOI] [PubMed] [Google Scholar]
- Apicella C, Andrews L, Hodgson SV, Fisher SA, Lewis CM, Solomon E, Tucker K, Friedlander M, Bankier A, Southey MC, Venter DJ, Hopper JL. Log odds of carrying an Ancestral Mutation in BRCA1 or BRCA2 for a defined personal and family history in an Ashkenazi Jewish woman (LAMBDA) Breast Cancer Res. 2003;5:R206–R216. doi: 10.1186/bcr644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhausen SL, Ozcelik H, Southey MC, John EM, Godwin AK, Chung W, Iriondo-Perez J, Miron A, Santella RM, Whittemore A, Andrulis IL, Buys SS, Daly MB, Hopper JL, Seminara D, Senie RT, Terry MB. Breast Cancer Family Registry. BRCA1 and BRCA2 mutation carriers in the Breast Cancer Family Registry: an open resource for collaborative research. Breast Cancer Res Treat. 2009;116:379–386. doi: 10.1007/s10549-008-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchawar J, Korch C, Byers T, Pitts TM, Li E, McCredie MR, Giles GG, Hopper JL, Southey MC. Population-based estimate of the contribution of TP53 mutations to subgroups of early-onset breast cancer: Australian Breast Cancer Family Study. Cancer Res. 2010;70:4795–4800. doi: 10.1158/0008-5472.CAN-09-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann GJ, Thorne H, Balleine RL, Butow PN, Clarke CL, Edkins E, Evans GM, Fereday S, Haan E, Gattas M, Giles GG, Goldblatt J, Hopper JL, Kirk J, Leary JA, Lindeman G, Niedermayr E, Phillips KA, Picken S, Pupo GM, Saunders C, Scott CL, Spurdle AB, Suthers G, Tucker K, Chenevix-Trench G. Kathleen Cuningham Consortium for Research in Familial Breast Cancer. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 2006;8:R12. doi: 10.1186/bcr1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkowitz M, Sabbaghian N, Ray AM, Lange EM, Foulkes WD, Cooney KA. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in hereditary prostate cancer. Prostate. 2008;68:675–678. doi: 10.1002/pros.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Dumont T, Calvez-Kelm FL, Forey N, McKay-Chopin S, Garritano S, Gioia-Patricola L, De Silva D, Weigel R, Sangrajrang S, Lesueur F, Tavtigian SV. Breast Cancer Family Registries (BCFR); Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) Description and validation of high-throughput simultaneous genotyping and mutation scanning by high-resolution melting curve analysis. Hum Mutat. 2009;30:884–890. doi: 10.1002/humu.20949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez Ide J, Esteban Cardeñosa E, Palanca Suela S, Barragán González E, Bolufer Gilabert P. on behalf of the Group of Cancer Genetic Counselling Program of Valencia Community. Advantages of the high resolution melting in the detection of BRCA1 or BRCA2 mutation carriers. Clin Biochem. 2009;42:1572–1576. doi: 10.1016/j.clinbiochem.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Orlando C, Pinzani P, Pazzagli M. Developments in quantitative PCR. Clin Chem Lab Med. 1998;36:255–269. doi: 10.1515/CCLM.1998.045. [DOI] [PubMed] [Google Scholar]
- Ratnasinghe LD, Abnet C, Qiao YL, Modali R, Stolzenberg-Solomon R, Dong ZW, Dawsey SM, Mark SD, Taylor PR. Polymorphisms of XRCC1 and risk of esophageal and gastric cardia cancer. Cancer Lett. 2004;216:157–164. doi: 10.1016/j.canlet.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Jenkins MA, Baglietto L, Dowty JG, Van Vliet CM, Smith L, Mead LJ, Macrae FA, St John DJ, Jass JR, Giles GG, Hopper JL, Southey MC. Cancer risks for mismatch repair gene mutation carriers: a population-based early onset case-family study. Clin Gastroenterol Hepatol. 2006;4:489–498. doi: 10.1016/j.cgh.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M. Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1998;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PD, McMullan G, Day NE, Ponder BA, Easton D. Evidence for further breast cancer susceptibility genes in additional to BRCA1 and BRCA2 in a population based study. Genet Epidemiol. 2001;21:1–18. doi: 10.1002/gepi.1014. [DOI] [PubMed] [Google Scholar]
- Cannings C, Thompson EA, Skolnick MH. Probability functions on complex pedigrees. Adv Appl Prob. 1978;10:26–61. doi: 10.2307/1426718. [DOI] [Google Scholar]
- Lange K. Mathematical and Statistical Methods for Genetic Analysis. 2. New York: Springer; 2002. [Google Scholar]
- Australian Institute of Health and Welfare and National Breast Cancer Centre. Breast cancer in Australia: an overview. 2006. http://www.aihw.gov.au/publications/can/bca06/bca06.pdf
- Antoniou AC, Pharoah PD, McMullan G, Day NE, Stratton MR, Peto J, Ponder BJ, Easton DF. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkko H, Xia B, Nikkilä J, Schleutker J, Syrjäkoski K, Mannermaa A, Kallioniemi A, Pylkäs K, Karppinen SM, Rapakko K, Miron A, Sheng Q, Li G, Mattila H, Bell DW, Haber DA, Grip M, Reiman M, Jukkola-Vuorinen A, Mustonen A, Kere J, Aaltonen LA, Kosma VM, Kataja V, Soini Y, Drapkin RI, Livingston DM, Winqvist R. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Ghadirian P, Akbari MR, Hamel N, Giroux S, Sabbaghian N, Darnel A, Royer R, Poll A, Fafard E, Robidoux A, Martin G, Bismar TA, Tischkowitz M, Rousseau F, Narod SA. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi L, Putignano AL, Congregati C, Piaceri I, Zanna I, Sera F, Morrone D, Genuardi M, Palli D. A PALB2 germline mutation associated with hereditary breast cancer in Italy. Fam Cancer. 2010;9:181–185. doi: 10.1007/s10689-009-9295-z. [DOI] [PubMed] [Google Scholar]
- García MJ, Fernández V, Osorio A, Barroso A, Llort G, Lázaro C, Blanco I, Caldés T, de la Hoya M, Ramóny Cajal T, Alonso C, Tejada MI, San Román C, Robles-Díaz L, Urioste M, Benítez J. Analysis of FANCB and FANCN/PALB2 Fanconi Anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Res Treat. 2009;113:545–551. doi: 10.1007/s10549-008-9945-0. [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assn. 1958;53:457–481. doi: 10.2307/2281868. [DOI] [Google Scholar]
- Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, Risch HA, Eyfjord JE, Hopper JL, Southey MC, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tryggvadottir L, Syrjakoski K, Kallioniemi OP, Eerola H, Nevanlinna H, Pharoah PD. et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, Wenstrup RJ, Ward BE, Scholl TA, Noll WW. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115:2222–2233. doi: 10.1002/cncr.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper JL. Genetic epidemiology of female breast cancer. Semin Cancer Biol. 2001;11:367–374. doi: 10.1006/scbi.2001.0392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing primer sequences. Primers designed for DNA sequencing, including DNA fragment size (bp) and annealing temperatures.
Polymerase chain reaction primer sequences and annealing temperatures. The primer sequences and annealing temperatures designed to amplify 35 fragments (including the coding and flanking intronic regions) of partner and localizer of breast cancer 2 susceptibility protein (PALB2).