Abstract
BACKGROUND
Meningomyelocele (MM) results from lack of closure of the neural tube during embryologic development. Periconceptional folic acid supplementation is a modifier of MM risk in humans, leading to an interest in the folate transport genes as potential candidates for association to MM.
METHODS
This study used the SNPlex Genotyping (ABI, Foster City, CA) platform to genotype 20 single polymorphic variants across the folate receptor genes (FOLR1, FOLR2, FOLR3) and the folate carrier gene (SLC19A1) to assess their association to MM. The study population included 329 trio and 281 duo families. Only cases with MM were included. Genetic association was assessed using the transmission disequilibrium test in PLINK.
RESULTS
A variant in the FOLR2 gene (rs13908), three linked variants in the FOLR3 gene (rs7925545, rs7926875, rs7926987), and two variants in the SLC19A1 gene (rs1888530 and rs3788200) were statistically significant for association to MM in our population.
CONCLUSION
This study involved the analyses of selected single nucleotide polymorphisms across the folate receptor genes and the folate carrier gene in a large population sample. It provided evidence that the rare alleles of specific single nucleotide polymorphisms within these genes appear to be statistically significant for association to MM in the patient population that was tested.
Keywords: neural tube, meningomyelocele, folic acid, folate transport genes, single nucleotide polymorphisms
INTRODUCTION
Neural tube defects (NTDs) are common malformations of the brain and spinal cord, and they include all abnormalities resulting from lack of closure of the developing neural tube during embryologic development. The causes of human neural tube defects are largely unknown, but are almost certainly multifactorial, consisting of both genetic and environmental components (Finnell et al., 2000; Cabrera et al., 2004; Detrait et al., 2005; Beaudin and Stover 2009). No single major gene has been implicated in the etiology of these disorders across all populations studied to date. It seems likely that NTDs involve multiple variants across a single gene or variant effects across a number of genes. Furthermore, the variants and genes may differ between ethnic groups. Previously, definitive results have been hampered by the lack of reproducibility of the association studies implicating certain genes (Hirschhorn et al., 2002; Beaudin and Stover 2009; Greene et al., 2009; Copp and Greene 2010). Linkage studies have yielded some interesting results (Rampersaud et al., 2005). In addition, animal models have proved to be a useful tool to gain some insight into the molecular processes of neural tube formation and closure. In clinical severity, NTDs range from mild forms (e.g., spina bifida occulta) that often do not require surgical intervention to lethal forms such as anencephaly (Botto et al., 1999).
Meningomyelocele (MM) is the most severe form of spina bifida that is compatible with survival. In an MM, both the meninges and the spinal cord protrude through a gap in the vertebral column, and the lesion is not covered by the skin. Although these defects can occur at any point along the developing neural tube, lumbosacral lesions are the most common (Hunter et al., 1996). As a result of current surgical and medical interventions, most children born with MM in the United States survive, although in the first year of life the mortality risk of these infants is greater than in the general population. Despite these interventions, children born with an MM almost invariably have profound, life-long disabilities (Detrait et al., 2005).
It is known that maternal folate status is a modifier of NTD risk (Botto et al., 2005). These findings make the folate metabolic pathway genes potential candidate genes for association to meningomyelocele. The folate transport genes are involved in the transport and maintenance of intracellular levels of folate. FOLR1 and FOLR2 are glycosyl phosphatidylinositol (GPI)-anchored proteins that facilitate unidirectional transport of folates (Verma et al., 1992; Henderson et al., 1995), whereas FOLR3 codes for a secreted form of the receptor protein (Shen et al., 1995). SLC19A1 is a cell surface transmembrane protein that participates in bidirectional movement of folate across the membrane (Matherly et al., 2007; Hou and Matherly, 2009). Because of the role that these proteins play in the maintenance of critical levels of intracellular folate, it is logical to hypothesize that functional variants within these genes are associated with MM. In fact, it has been found that polymorphisms in the SLC19A1 gene, such as the A80G variant, appear to confer susceptibility to MM risk in some populations (Shang et al., 2008; Pei et al., 2009). Furthermore, the knockout mouse models of the FOLR1 and SLC19A1 genes result in embryonic lethality. Failure of neural tube closure is one of the abnormal morphologic findings in the null embryos (Peidrahita et al., 1999; Gelineau-van Waes et al., 2008). This finding provides more compelling evidence that these genes play a role in neural tube closure and thereby potentially in MM risk.
The FOLR1 receptors are expressed on the microvillus plasma membrane of the placenta where, in combination with the proton-coupled high affinity folate transporters (PCFT), appear to be responsible for the internalization and cytoplasmic release of folate (Solanky et al., 2010). In contrast, the reduced folate carrier (SLC19A1), a bidirectional transporter of primarily reduced folates, is expressed on both the microvillus plasma membrane and the basolateral plasma membrane of the placenta (Solanky et al., 2010).
The present study focused on the folate transporter genes as candidates for association to MM. The rationale for choosing these genes was based on epidemiologic studies, genetic association studies by other groups, biologic function of candidate genes, and animal models (MRC Vitamin Study Research Group, 1991; Czeizel and Dudas, 1992; Piedrahita et al., 1999; Williams et al., 2002; De Marco et al., 2003; Zhu et al., 2007; Shang et al., 2008; Pei et al., 2009).
MATERIALS AND METHODS
Study Population
The majority of the MM cohort tested in the study consists of Caucasians of European descent and Hispanics of Mexican descent in the United States (Table 1). The MM probands and their parents were enrolled after obtaining informed consent. The patient cohort was recruited primarily from five different sites: Houston, Texas; the Texas-Mexico border area; Lexington, Kentucky; Los Angeles, California; and Toronto, Ontario, Canada. Recruitment took place during clinical visits, hospitalizations, or at parent meetings. The probands were born between 1955 and 2008 (Au et al., 2008), and the ages at enrollment ranged from 6 months to adulthood. The criteria for inclusion were based on whether an individual had an MM or was related to an affected individual. The exclusion criteria were the presence of spina bifida that was not associated with an MM or a syndromic form of spina bifida. No individual was excluded on the basis of race or sex. In the study, 329 affected child-parent trios and 281 affected child-parent duos were tested. The level of defect was determined by the review of medical records and also, in the case of some of the affected individuals, by the review of radiographs. Maternal health history, pregnancy history, maternal exposures, and sociodemographic information were obtained from the parents of the affected children. Information about vitamin supplementation was not obtained, but a study (food frequency questionnaire) is now underway that includes this information. Because family association studies, specifically transmission disequilibrium test (TDT) analyses, were used as the primary statistical tool, the family trios (consisting of the father, mother, and affected child) were the most important component of the population for the study. The project was approved by the Institutional Review Board of the University of Texas Health Science Center at Houston.
Table 1.
Study Population
| Characteristic | Trios | Duos | Total |
|---|---|---|---|
| Race | |||
| Caucasian | 142 | 84 | 226 |
| Mexican American | 168 | 166 | 334 |
| African American | 4 | 14 | 18 |
| Asian American | 2 | 2 | 4 |
| Othera | 7 | 5 | 12 |
| Unknownb | 6 | 10 | 16 |
| Total | 329 | 281 | 610 |
| Sex | |||
| Male | 154 | 126 | 280 |
| Female | 167 | 144 | 311 |
| Unknown | 8 | 11 | 19 |
| MM Lesion Level | |||
| ≥L1 | 81 | 81 | 162 |
| ≤L2 | 203 | 169 | 372 |
| Mixed | 5 | 4 | 9 |
| Unknown | 40 | 27 | 67 |
Native American and other country of origin.
Unknown ethnicity.
A total of 92 anonymous Hispanic subjects with no personal or family history of NTDs were enrolled in the Houston area to obtain appropriate Hispanic control sample frequencies for the tested single nucleotide polymorphisms (SNPs). To obtain SNP frequencies for Caucasian Americans, a Caucasian control DNA panel (HD100CAU) was purchased from the Coriell Institute (Camden, NJ). DNA samples from 30 Caucasian families used in the HapMap project (http://www.hapmap.org) were included in the genotyping as a quality control measure for genotype calls.
DNA Genotyping
A blood sample was obtained from the proband and both parents when available. Genomic DNA was extracted from the pelleted white blood cells using the Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN). The Oragene DNA Preparation Kit (DNA Genotek, Ontario, Canada) was used to prepare DNA from saliva samples when a blood sample could not be obtained.
DNA samples for 610 affected members and 939 parents of the MM population, 90 HD100Cau, and 92 Hispanic controls were genotyped for the selected SNPs. DNA samples from 82 individuals from 15 CEU families were also included. The genotypes of many of these individuals are available in the HapMap project, providing an internal quality control for the genotyping assay.
The testing interrogated SNPs in the folate transport genes. As mentioned previously, these genes are integral players in the delivery and transport of folate and are therefore considered to be good candidate genes for affecting MM formation. In the design of the SNPset, both tag and non-tag SNPs were included to increase coverage (Table 2). The folate receptor genes FOLR1, FOLR2, and FOLR3 map to human chromosome 11q13.4, and the genomic DNA sizes are 6.74 kb, 5.15 kb, and 4.16 kb, respectively. The reduced folate carrier SLC19A1 gene is located on 21q22.3 and is 27.72 kb in size. When possible, SNPs with a minimum heterozygosity of 5%, as reported in the HapMap CEU population, were chosen. The databases that were used for SNPset design were: http://www.ncbi.nlm.nih.gov, http://genome.ucsc.edu, http://www.hapmap.org, http://www.genecards.org, and http://snp.wustl.edu.
Table 2.
SNPs Tested across the Four Folate Transport Genes (FOLR1, FOLR2, FOLR3, SLC19A1)
| Gene Symbol |
Gene Name |
Gene Size (kb) |
Chromosome | SNP | Chromosomal Location |
Significance |
|---|---|---|---|---|---|---|
| FOLR1 | Folate Receptor 1 | 6.74 | 11 | rs2071010 | 71,578,612 | Intron/utr Isoform 3 |
| rs3833748 | 71,579,110 | Intron | ||||
| rs35179028 | 71,584,342 | Val132Val | ||||
| FOLR2 | Folate Receptor 2 | 5.15 | 11 | rs651933 | 71,604,308 | 5′ near gene |
| rs35982790 | 71,606,664 | Intron | ||||
| rs13908 | 71,607,379 | Lys35Glu | ||||
| rs514933 | 71,607,855 | Intron | ||||
| FOLR3 | Folate Receptor 3 | 4.16 | 11 | rs7925055 | 71,522,744 | 5′ near gene |
| rs7925545 | 71,523,189 | 5′ near gene | ||||
| rs7926875 | 71,527,090 | Intron | ||||
| rs7926987 | 71,527,151 | Intron | ||||
| rs508088 | 71,528,304 | Ala173Ala | ||||
| rs34970007 | 71,528,379 | Lys198Lys | ||||
| rs11235449 | 71,528,847 | 3′ near gene | ||||
| SLC19A1 | Solute carrier family 19 member 1 | 27.72 | 21 | rs1888530 | 45,760,851 | Intron |
| rs12482346 | 45,762,055 | Intron | ||||
| rs2838958 | 45,772,995 | Intron | ||||
| rs9282854 | 45,775,894 | Leu262Leu | ||||
| rs914232 | 45,777,178 | Intron | ||||
| rs3788200 | 45,780,999 | Intron |
SNP, Single nucleotide polymorphism.
The genotyping platform used was the SNPlex Genotyping System (Applied Biosystems, Foster City, CA). Based on the SNP sequences submitted, Applied Biosystems designed allele SNP-specific probes for each SNP. The SNPs were submitted as standard SNP Identifiers (SNP IDs) from the common available databases. Based on compatibility of probes representing each SNP, ABI assembled the most compatible SNPs into a SNPSet. The SNPlex platform uses the ligation of these allele-specific probes for each SNP that hybridized to the patient genomic DNA SNP loci, followed by multiplex PCR amplification. The design strategy enables allelic discrimination of SNPs at specific positions in the human genome (Martinez et al., 2009; Shaw et al., 2009). A 5-µl (200 ng) aliquot of genomic DNA was used per reaction, and the standard SNPlex genotyping protocol was used. The subsequent electrophoresis of the SNP probes was performed on the ABI 3730xl DNA analyzer, and genotype calls were made using GeneMapper v4.0.
Data Analyses
Our SNP selection approach yielded 48 potential SNPs, but only 20 SNPs across the four folate transport genes (FOLR1, FOLR2, FOLR3, SLC19A1) met all the criteria to be submitted for statistical analyses (Table 2). Sixteen of the 28 excluded SNPs had clustering errors, four had Mendelian inconsistencies, six were of Hardy Weinberg Equilibrium (HWE), and two did not reach the acceptable (≥90%) genotype concordance call rate with the internal CEU (U.S. residents with northern and western European ancestry collected by the Centre d’Etude du Polymorphisme Humain) controls when the data were available on HapMap (http://hapmap.ncbi.nlm.nih.gov/)
To minimize the effect of missing genotypes on the subsequent association study, only the SNPs that reached an acceptable genotype call rate of ≥90% were considered for statistical analyses. Prior to association analyses, all family units were tested for the presence of Mendelian errors. To control for genotype call error, an SNP was re-examined if 10 families or more (i.e., >10/610 or >1.64%) displayed Mendelian errors. If an SNP did not pass on the first quality control test, it was re-examined to make a second genotype call for an additional quality control test. If it again failed to meet the quality control criteria, it was removed from the association study. HWE was determined based on 237 controls by ethnicity. Therefore, different ethnic groups were not pooled, and it could be determined that the successful SNPs did not deviate from HWE (p < 0.05). The final data analyses involved family-based studies of genetic association, notably the TDT using the TDT component of the PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) whole genome association analysis toolset (Purcell et al., 2007). Our MM patient cohort consists of a number of duo (family unit consisting of either the mother and affected child or the father and affected child). In these cases, the unit is usually missing the father (Martinez et al., 2009). To increase the power in the study, it is important to use as many of the cases as possible. The PLINK algorithm uses the duo families and allows for the missing parents. Furthermore, this analytical method generates counts for transmitted and nontransmitted alleles.
RESULTS
The allele frequencies of the 20 SNPs that passed all quality control criteria are listed in Table 3. A total of 10 SNPs demonstrated significant association with MM risk in this study (Table 4).
Table 3.
Single Nucleotide Polymorphism Allele Frequencies for the Folate Receptor and Carrier Genes (Based on 237 Controls) by Ethnicity
| Gene Symbol |
Marker | Allele 1 | Allele 2 | Caucasian (n = 145) |
Hispanic (n = 92) |
|---|---|---|---|---|---|
| FOLR1 | rs2071010 | A | G | 0.05/0.95 | 0.10/0.90 |
| rs3833748 | delG | G | 0.00/1.00 | 0.05/0.95 | |
| rs35179028 | A | G | mono | 0.01/0.99 | |
| FOLR2 | rs651933 | A | G | 0.47/0.53 | 0.36/0.64 |
| rs35982790 | C | A | 1.00/0.00 | 0.96/0.04 | |
| rs13908 | A | G | 0.90/0.10 | 0.59/0.41 | |
| rs514933 | A | G | 0.57/0.43 | 0.74/0.26 | |
| FOLR3 | rs7925055 | A | G | 0.96/0.04 | 0.94/0.06 |
| rs7925545 | A | G | 0.98/0.02 | 0.98/0.02 | |
| rs7926875 | A | C | 0.05/0.95 | 0.05/0.95 | |
| rs7926987 | C | G | 0.95/0.05 | 0.93/0.07 | |
| rs508088 | C | T | 1.00/0.00 | 0.95/0.05 | |
| rs34970007 | A | G | 0.02/0.98 | 0.04/0.96 | |
| rs11235449 | A | G | 0.43/0.57 | 0.22/0.78 | |
| SLC19A1 | rs1888530 | C | T | 0.15/0.85 | 0.11/0.89 |
| rs12482346 | C | T | 0.61/0.39 | 0.55/0.45 | |
| rs2838958 | A | G | 0.58/0.42 | 0.47/0.53 | |
| rs9282854 | G | A | 0.70/0.30 | 0.72/0.28 | |
| rs914232 | C | T | 0.75/0.25 | 0.60/0.40 | |
| rs3788200 | A | G | 0.38/0.62 | 0.34/0.66 |
Table 4.
Family-Based Association on Total Population for FOLR1, FOLR2, FOLR3, and SLC19A1 Transmission/Disequilibrium Test Analysis (p < 0.05) Performed Using PLINK
| GENE | CHR | SNP | A11 | A22 | T3 | U4 | OR | χ2 value | p value |
|---|---|---|---|---|---|---|---|---|---|
| FOLR1 | 11 | rs35179028 | A | G | 7 | 19 | 0.3684 | 5.538 | 0.0186 |
| FOLR2 | 11 | rs35982790 | C | A | 3 | 21 | 0.1429 | 13.5 | 0.0002386 |
| FOLR2 | 11 | rs13908 | G | A | 39 | 62 | 0.629 | 5.238 | 0.0221 |
| FOLR3 | 11 | rs7925545 | G | A | 7 | 28 | 0.25 | 12.6 | 0.0003857 |
| FOLR3 | 11 | rs7926875 | A | C | 4 | 14 | 0.2857 | 5.556 | 0.01842 |
| FOLR3 | 11 | rs7926987 | G | C | 11 | 25 | 0.44 | 5.444 | 0.01963 |
| FOLR3 | 11 | rs508088 | T | C | 2 | 15 | 0.1333 | 9.941 | 0.001616 |
| FOLR3 | 11 | rs34970007 | A | G | 5 | 19 | 0.2632 | 8.167 | 0.004267 |
| SLC19A | 21 | rs1888530 | C | T | 25 | 62 | 0.4032 | 15.74 | 0.00007284 |
| SLC19A | 21 | rs3788200 | A | G | 37 | 60 | 0.6167 | 5.454 | 0.01953 |
CHR, chromosome; SNP, single nucleotide polymorphism; A11, common allele; A12, minor allele; T3, number of cases with minor allele transmitted; U4, number of cases with minor allele not transmitted; OR, odds ratio.
One SNP in the FOLR1 gene showed significant association with MM. The FOLR1 SNP rs35179028 is a synonymous coding SNP (p.V132V) with a low minor allele frequency (Table 3). The positive result, therefore, has to be interpreted with caution (Table 4).
Two SNPs (rs35982790) and (rs13908) in FOLR2 showed significant association with MM. The intronic SNP rs35982790 has a low minor allele frequency (Table 3), and again the association has to be interpreted with caution (Table 4).
The three significant SNPs—notably rs7925545, rs7926987, and rs7926875 in the FOLR3 gene—are tightly linked in the CEU population (http://www.HapMap.org) (The International HapMap Consortium, 2007) and have TDT p values of 3.85E−04, 1.96E−02, 1.84E−02 respectively (Table 4). The rs7925545 SNP lies in the 5′ region close to the gene, whereas the rs7926987 and rs7926875 variants lie within intron 2. The FOLR3 synonymous coding SNPs rs508088 and rs34970007 are significant, but both SNPs have low minor allele frequencies (Table 3).
Finally, two SNPs (rs1888530 and rs3788200) in SLC19A1 showed significant association with MM. The rs3788200 is located in intron 2 (TDT p value, 1.95E−02). The rs1888530 variant that was significantly associated with MM in the study is located in intron 5 of the SLC19A1 gene (TDT p value, 7.28E−05; Table 4).
DISCUSSION
In our study, a number of SNP variants in the folate receptor genes and the folate carrier gene showed preferential transmission from parent to offspring and are associated with MM in the population tested.
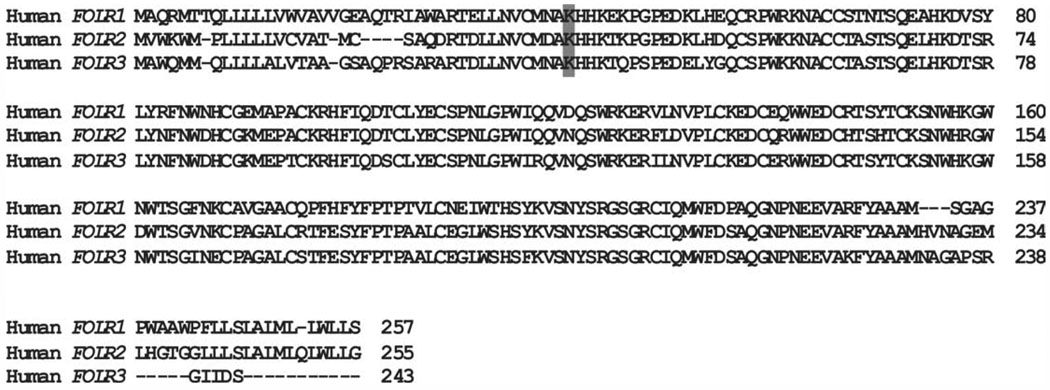
The folate receptor 2 gene (FOLR2) is located on chromosome 11 and is 5.2 kb in size; it consists of five exons and four introns. The variant found in the gene (rs13908) is a nonsynonymous SNP (A → G) located in exon 2 and is the most interesting SNP studied, because of potential functional significance. The variant results in a missense mutation with the lysine (AAG) at this position substituted for a glutamic acid (GAG). The Lys at position 35 is conserved in a multiple species alignment (http://genome.ucsc.edu/) (Kuhn et al., 2007). The amino acid is also conserved among all three folate receptors (FOLR1, FOLR2, FOLR3), again indicating potential functional significance (Fig. 1). The conserved Lys is at amino acid position 35 in the FOLR2 protein and is found as part of a conserved motif consisting of [Lys]-[His]-[His]-[Lys]. The function of this motif is currently unknown, but FOLR2 is a GPI-anchored protein that binds, internalizes, and unloads 5-methyltetrahydrofolate and other folate derivatives to the interior of the cell. The receptor then has to recycle back to the cell surface for an additional round of ligand binding. The variant is not located in the GPI site (Yan and Ratnam, 1995); however, if the motif is involved in any other critical processes and the amino acid substitution (Lys→Glu) disrupts the process, it could potentially lead to decreased levels of intracellular folate during critical periods of embryonic development. It would be useful to further validate the association by replicating the finding in an independent MM population.
Figure 1.
Amino acid sequence alignment between the three human folate receptors. Note: The location of p. Lys35Glu (rs13908) of FOLR2 is shaded to show conservation.
The FOLR1 and FOLR2 receptors are membrane-associated, GPI-anchored receptors (Lacey et al., 1989; Yan and Ratnam, 1995). The FOLR3 receptor, in contrast, is a constitutively secreted form of the folate receptor (Shen et al., 1995). Little has been published about the FOLR3 protein, and there is limited information regarding the role of the FOLR3 receptor in folate transport. The gene is located on chromosome 11 and is approximately 4.2 kb in size. It consists of five exons and four introns. Three SNPs within the FOLR3 gene (rs7925545, rs7926987, and rs7926875), showed statistically significant TDT values. In addition, each of the SNPs had an acceptable genotype concordance call rate with the internal CEU controls (91%, 93%, and 91%, respectively). All four SNPs are in tight linkage disequilibrium in the CEU population (www.HapMap.org); therefore, they would be expected to segregate together and to behave in a similar fashion (i.e., show similar association patterns). The D’ and r2 values were calculated using the CEU data (www.Hap-Map.org) and Haploview v 4.1 (http://www.broadinstitute.org/haploview/haploview).
None of the SNPs in FOLR3 have been evaluated previously for MM risk, and currently no putative function can be assigned. The SNPs rs7926875 and rs7926987 are located in intron 2 of the FOLR3 gene. The rs7925545 SNP is located in the intergenic region approximately 1230 bp upstream from exon 1 of the gene and is conserved among primates (http://genome.ucsc.edu/). Two alternative promoter regions have been suggested for the FOLR3 gene, with both located <500 bp upstream from exon 1 of the gene (http://www.ncbi.nlm.nih.gov/) (The National Center for Biotechnology Information). The rs7925545 SNP may be located within the upstream promoter-enhancer region. The FOLR3 SNPs rs7926987 and rs7926875 are conserved among primates (http://genome.ucsc.edu/).
The reduced folate carrier gene SLC19A1 is located on chromosome 21, and it is is approximately 28 kb in size. The gene consists of six exons and five introns. Six SNPs in this gene were analyzed for association. However, only the rs3788200 located in intron 2 and the rs1888530 SNP located in intron 5 were found to be significantly associated with MM. Although neither of these SNPs have been previously evaluated for MM risk, a nonsynonymous variant in exon 2 (rs1051266) has shown association with NTD risk in three other studies of two population groups (Shang et al., 2008; Franke et al., 2009; Pei et al., 2009). It is important to note that the mouse knockout for the SLC19A1 gene is embryonic lethal. The phenotype can be partially rescued when the pregnant dams receive folic acid supplementation (Gelineau-van Waes et al., 2008).
A major strength of our study is the large population size (Table 1). In addition, the use of family-based association studies, specifically the TDT, as an analytical tool is a strength in addition to the availability of a sizable number of trio families (Table 1) facilitates the use of this approach. The TDT statistical method uses the nontransmitted parental alleles as controls, thereby effectively addressing issues such as population structure, which can be problematic in case versus control analyses. Approximately 60% of the study population is Hispanic of Mexican descent. Hispanics are of particular interest because, in the United States, Hispanics of Mexican descent have a higher risk than any other ethnic group of having a child affected with an NTD (Canfield et al., 1996; Canfield et al., 2009).
Our study involved a SNP screen across the three folate receptor genes (FOLR1, FOLR2, FOLR3) and the reduced folate carrier gene (SLC19A1) in a large population sample consisting of approximately 60% Hispanics of Mexican descent. A number of SNPs across these genes were found to be associated with a protective effect for MM in the population tested. Furthermore, this study is the first to associate the FOLR3 gene with MM. It would be useful to validate our findings in a second, independently ascertained MM population. If the same variants are again found to be associated with MM in a second population, then functional studies should be designed for these SNPs in an attempt to determine their biologic roles.
ACKNOWLEDGMENTS
We thank the patients and their families for their participation in this study.
This work was supported by grants from the National Institutes of Health (PO1 HD35946) and Shriners Hospital for Children (project 8580) to H.N. and K.S.A.
REFERENCES
- Au KS, Tran PX, Tsai CC, et al. Characteristics of a spina bifida population including North American Caucasian and Hispanic individuals. Birth Defects Res A Clin Mol Teratol. 2008;82:692–700. doi: 10.1002/bdra.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin AE, Stover PJ. Insights into metabolic mechanisms underlying folate-responsive neural tube defects: a minireview. Birth Defects Res A Clin Mol Teratol. 2009;85:274–284. doi: 10.1002/bdra.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, Moore CA, Khoury M, Erickson JD. Neural tube defects. N Engl J Med. 1999;341:1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- Botto LD, Lisi A, Robert-Gnansia E, et al. International retrospective cohort study of neural tube defects in relation to folic acid recommendations: Are the recommendations working? Br Med J. 2005;330:571. doi: 10.1136/bmj.38336.664352.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera RM, Hill DS, Etheredge AJ, Finnell RH. Investigations into the etiology of neural tube defects. Birth Defects Res C Embryo Today. 2004;72:330–344. doi: 10.1002/bdrc.20025. [DOI] [PubMed] [Google Scholar]
- Canfield MA, Annegers JF, Brender JD, et al. Hispanic origin and neural tube defects in Houston/Harris County, Texas. Risk Factors. Am J Epidemiol. 1996;143:12–24. doi: 10.1093/oxfordjournals.aje.a008653. [DOI] [PubMed] [Google Scholar]
- Canfield MA, Ramadhani TA, Shaw GM, et al. Anencephaly and spina bifida among Hispanics: maternal, sociodemographic, and acculturation factors in the National Birth Defects Prevention Study. Birth Defects Res A Clin Mol Teratol. 2009;85:637–646. doi: 10.1002/bdra.20582. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND. Genetics and development of neural tube defects. J Pathol. 2010;220:217–230. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeizel AE, Dudas I. Preventation of the first occurrence of neural tube defects by periconceptional vitamin supplementation. N Eng J Med. 1992;327:1832–1835. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- De Marco P, Calevo MG, Moroni A, et al. Reduced folate carrier polymorphism (80A→G) and neural tube defects. Eur J Hum Genet. 2003;11:245–252. doi: 10.1038/sj.ejhg.5200946. [DOI] [PubMed] [Google Scholar]
- Detrait ER, George TM, Etchevers HC, et al. Human neural tube defects: developmental biology, epidemiology, and genetics. Neurotoxicol Teratol. 2005;27:515–524. doi: 10.1016/j.ntt.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnell RH, Gelineau-Van Waes J, Bennett GD, et al. Genetic basis of susceptibility to environmentally induced neural tube defects. Ann N Y Acad Sci. 2000;919:261–277. doi: 10.1111/j.1749-6632.2000.tb06886.x. [DOI] [PubMed] [Google Scholar]
- Franke B, Vermeulen SH, Steegers-Theunissen RP, et al. An association study of 45 folate related genes in spina bifida: involvement of cubilin (CUBN) and tRNA aspartic acid methyltransferase 1 (TRDMT1) Birth Defects Res A Clin Mol Teratol. 2009;85:216–226. doi: 10.1002/bdra.20556. [DOI] [PubMed] [Google Scholar]
- Gelineau-van Waes J, Heller S, Bauer LK, et al. Embryonic development in the reduced folate carrier knockout mouse is modulated by maternal folate supplementation. Birth Defects Res A Clin Mol Teratol. 2008;82:494–507. doi: 10.1002/bdra.20453. [DOI] [PubMed] [Google Scholar]
- Greene ND, Stanier P, Copp AJ. Genetics of neural tube defects. Hum Mol Genet. 2009;18:R113–R129. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson GI, Perez T, Schenker S, et al. Maternal to fetal transfer of 5-methyltetrahydrofolate by the perfused human placental cotyledon: evidence for a concentrative role by placental folate receptors in fetal folate delivery. J Lab Clin Med. 1995;126:184–203. [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Hou Z, Matherly LH. Oligomeric structure of the human reduced folate carrier: identification of homo-oligomers and dominant–negative effects on carrier expression and function. J Biol Chem. 2009;284:3285–3293. doi: 10.1074/jbc.M807206200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AG, Cleveland RH, Blickman JG, Holmes LB. A study of level of lesion, associated malformations and sib occurrence risks in spina bifida. Teratology. 1996;54:213–218. doi: 10.1002/(SICI)1096-9926(199611)54:5<213::AID-TERA1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hinrichs AS, Harte RA, Giardine B, Fujita P, Diekhans M, Dreszer T, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome browser database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey SW, Sanders JM, Rothberg KG, et al. Complementary DNA for the folate binding protein correctly predicts anchoring to the membrane by glycosyl-phosphatidylinositol. J Clin Invest. 1989;84:715–720. doi: 10.1172/JCI114220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez CA, Northrup H, Lin JI, et al. Genetic association study of putative functional single nucleotide polymorphisms of genes in folate metabolism and spina bifida. Am J Obstet Gynecol. 2009;201:394e1–394e11. doi: 10.1016/j.ajog.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matherly LH, Hou Z, Deng Y. Human reduced folate carrier: translation of basic biology to cancer etiology and therapy. Cancer Metastasis Rev. 2007;26:111–128. doi: 10.1007/s10555-007-9046-2. [DOI] [PubMed] [Google Scholar]
- MRC Vitamin Study Research Group. Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. Lancet. 1991;338:131–137. [PubMed] [Google Scholar]
- Pei L, Liu J, Zhang Y, et al. Association of reduced folate carrier gene polymorphism and maternal folic acid use with neural tube defects. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:874–878. doi: 10.1002/ajmg.b.30911. [DOI] [PubMed] [Google Scholar]
- Piedrahita JA, Oetama B, Bennett GD, et al. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat Genet. 1999;23:228–232. doi: 10.1038/13861. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersaud E, Bassuk AG, Enterline DS, et al. Whole genomewide linkage screen for neural tube defects reveals regions of interest on chromosomes 7 and 10. J Med Genet. 2005;42:940–946. doi: 10.1136/jmg.2005.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Zhao H, Niu B, et al. Correlation of polymorphisms of MTHFRS and RFC-1 genes with neural tube defects in China. Birth Defects Res A Clin Mol Teratol. 2008;82:3–7. doi: 10.1002/bdra.20416. [DOI] [PubMed] [Google Scholar]
- Shaw GM, Lu W, Zhu H, et al. 118 SNPs of folate-related genes and risks of spina bifida and conotruncal heart defects. BMC Med Genet. 2009;10:49. doi: 10.1186/1471-2350-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F, Wu M, Ross JF, et al. Folate receptor type gamma is primarily a secretory protein due to lack of an efficient signal for glycosyl-phosphatidylinositol modification: protein characterization and cell type specificity. Biochemistry. 1995;34:5660–5665. doi: 10.1021/bi00016a042. [DOI] [PubMed] [Google Scholar]
- Solanky N, Requena Jimenez A, et al. Expression of folate transporters in human placenta and implications for homocysteine metabolism. Placenta. 2010;31:134–143. doi: 10.1016/j.placenta.2009.11.017. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma RS, Gullapalli S, Antony AC. Evidence that the hydrophobicity of isolated, in situ, and de novo-synthesized native human placental folate receptors is a function of glycosyl-phosphatidylinositol anchoring to membranes. J Biol Chem. 1992;267:4119–4127. [PubMed] [Google Scholar]
- Williams LJ, Mai CT, Edmonds LD, et al. Prevalence of spina bifida and anencephaly during the transition to mandatory folic acid fortification in the United States. Teratology. 2002;66:33–39. doi: 10.1002/tera.10060. [DOI] [PubMed] [Google Scholar]
- Yan W, Ratnam M. Preferred sites of glycosylphosphatidylinositol modification in folate receptors and constraints in the primary structure of the hydrophobic portion of the signal. Biochemistry. 1995;34:14594–14600. doi: 10.1021/bi00044a039. [DOI] [PubMed] [Google Scholar]
- Zhu H, Cabrera RM, Wlodarczyk BJ, et al. Differentially expressed genes in embryonic cardiac tissues of mice lacking Folr1 gene activity. BMC Dev Biol. 2007;7:128. doi: 10.1186/1471-213X-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]