Abstract
Sinapine (O-sinapoylcholine) is the predominant phenolic compound in a complex group of sinapate esters in seeds of oilseed rape (Brassica napus). Sinapine has antinutritive activity and prevents the use of seed protein for food and feed. A strategy was developed to lower its content in seeds by expressing an enzyme that hydrolyzes sinapine in developing rape seeds. During early stages of seedling development, a sinapine esterase (BnSCE3) hydrolyzes sinapine, releasing choline and sinapate. A portion of choline enters the phospholipid metabolism, and sinapate is routed via 1-O-sinapoyl-β-glucose into sinapoylmalate. Transgenic oilseed rape lines were generated expressing BnSCE3 under the control of a seed-specific promoter. Two distinct single-copy transgene insertion lines were isolated and propagated to generate homozygous lines, which were subjected to comprehensive phenotyping. Sinapine levels of transgenic seeds were less than 5% of wild-type levels, whereas choline levels were increased. Weight, size, and water content of transgenic seeds were significantly higher than those of wild-type seeds. Seed quality parameters, such as fiber and glucosinolate levels, and agronomically important traits, such as oil and protein contents, differed only slightly, except that amounts of hemicellulose and cellulose were about 30% higher in transgenic compared with wild-type seeds. Electron microscopic examination revealed that a fraction of the transgenic seeds had morphological alterations, characterized by large cavities near the embryonic tissue. Transgenic seedlings were larger than wild-type seedlings, and young seedlings exhibited longer hypocotyls. Examination of metabolic profiles of transgenic seeds indicated that besides suppression of sinapine accumulation, there were other dramatic differences in primary and secondary metabolism. Mapping of these changes onto metabolic pathways revealed global effects of the transgenic BnSCE3 expression on seed metabolism.
Brassicaceous plants, such as oilseed rape (Brassica napus) and Arabidopsis (Arabidopsis thaliana), are characterized by the pronounced formation of soluble sinapate esters (Bouchereau et al., 1992). Seeds of this plant family accumulate sinapine, the choline ester of sinapate (O-sinapoylcholine; Henry and Garot, 1825; Gadamer, 1897), as the major component of complex patterns of sinapate esters, including various sinapoylated Glcs, gentiobioses, and kaempferol glycosides (Baumert et al., 2005; Wolfram et al., 2010). When oilseed rape seeds are processed, a high-quality vegetable oil is produced, and most of the sinapine is found in the residual protein meal. However, this compound is unpalatable and antinutritive (Ismail et al., 1981), making the meal unsuitable for human or animal consumption. Oilseed rape could be a valuable source of protein as well as oil if sinapine and related sinapate esters were reduced or eliminated from the seeds.
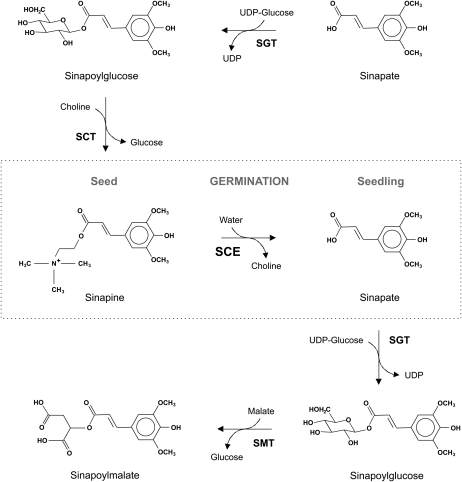
The enzymes involved in the brassicaceous sinapate ester metabolism include UDP-Glc:sinapate glucosyltransferase (EC 2.4.1.120), which forms 1-O-sinapoyl-β-Glc (Nurmann and Strack, 1981; Wang and Ellis, 1998). In seeds, sinapoylglucose is accepted by sinapoylglucose:choline sinapoyltransferase (EC 2.3.1.91), forming sinapine (Strack et al., 1983), while in seedlings, sinapoylglucose is a substrate for sinapoylglucose:l-malate sinapoyltransferase (EC 2.3.1.92), forming sinapoylmalate (Tkotz and Strack, 1981; Milkowski et al., 2004). Metabolic linking of sinapate esters between seeds and seedlings is achieved by the expression of sinapine esterase (SCE; EC 3.1.1.49) during seed germination; SCE hydrolyzes sinapine, liberating sinapate and choline (Nurmann and Strack, 1979; Strack et al., 1980). Sinapate feeds, via sinapoylglucose, into the biosyntheses of sinapoylmalate, and choline feeds into phosphatidylcholine (PC; Strack, 1981). Thus, the sinapoyl moiety is transferred from the seed component sinapine to sinapoylmalate of the seedlings. Figure 1 illustrates the sinapate ester metabolism in brassicaceous plants.
Figure 1.
Scheme of sinapate ester metabolism in brassicaceous plants. SCT, Sinapoylgluose:choline sinapoyltransferase (EC 2.3.1.91); SGT, UDP-Glc:sinapate glucosyltransferase (EC 2.4.1.120); SMT, sinapoylglucose:malate sinapoyltransferase (EC 2.3.1.92).
The gene encoding one of the three SCE isoforms has recently been isolated from oilseed rape seedlings (BnSCE3) and was found to be identical to the previously cloned BnLIP2 (GenBank accession no. AY870270; Ling et al., 2006; Clauss et al., 2008). This gene is a member of the large family of GDSL lipase genes (Akoh et al., 2004). In vitro, BnSCE3 exhibits unusually broad substrate specificity, acting on various phenolic choline esters and, surprisingly, PCs as well (Clauss et al., 2008). The enzyme also accepts p-nitrophenyl laurate (Ruiz et al., 2004), indicating general lipase activity (data not shown). Hence, to verify the specific role of BnSCE3 in sinapine metabolism in planta, Arabidopsis plants were transformed with a BnSCE3 expression cassette under the control of the seed-specific napin promoter. Expression of this gene led to a reduction of sinapine accumulation by more than 90% compared with control plants, confirming that BnSCE3 is indeed involved in sinapine hydrolysis in planta (Clauss et al., 2008). Whether BnSCE3 is also involved in other metabolic areas, such as the metabolism of PCs and other choline esters in seeds, remains an open question.
In this work, we describe changes in seed metabolism of two transgenic oilseed rape lines (OE1 and OE2). Compared with wild-type seeds, the best-performing overexpressor line exhibited reduction of sinapine accumulation by 95% along with dramatic alterations in primary and secondary metabolism. The lowered levels of sinapine and other sinapate esters in the transgenic seeds represent an important step toward the production of oilseed rape seed protein suitable for human and animal consumption.
RESULTS
Generation of Transgenic Lines of Oilseed Rape Expressing BnSCE3 in Seeds
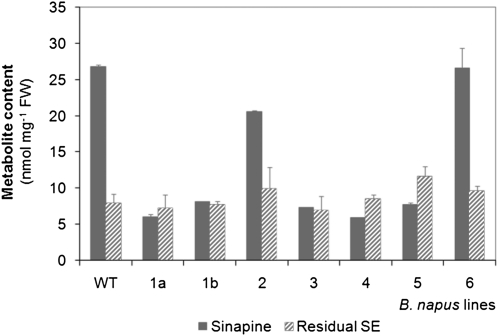
Oilseed rape plants were transformed with a BnSCE3 expression cassette harboring the full-length open reading frame of the BnSCE3 gene under transcriptional control of the seed-specific napin promoter. As a selection marker, the bacterial bar gene provided resistance to the herbicide phosphinothricin (PPT). Transformed plants (T1 generation) regenerated from calli were screened for PPT resistance as described earlier (Hüsken et al., 2005a). T1 plants were selfed to produce the first transgenic seed generation (T2 seeds). As an initial screening for the low-sinapine trait, 10 T2 seeds from each of the seven PPT-resistant T1 plants were pooled and subjected to quantification of sinapine and sinapate esters in methanolic extracts (Fig. 2). In the bulk analysis of segregating T2 seeds, six of the transgenic lines showed a significant decrease of the sinapine content, compared with the wild-type, whereas the total amount of residual sinapate esters was not significantly altered. To identify transgenic lines carrying single-copy transgene insertions, segregation of PPT resistance within the T2 seedlings (from each of the seven T1 plants) was characterized. Four independent transformant lines (1a, 1b, 4, and 5; Fig. 2) exhibited a 3:1 inheritance of PPT resistance, suggesting that these transformants contained a single T-DNA locus. Based on these initial results, the transgenic lines 1a (denominated as BnSCE3 overexpressor 1 [OE1]), 4 (OE2), and 5 (OE3) were selected for further analyses.
Figure 2.
Contents of sinapine and remaining sinapate esters (Residual SE) in segregating T2 seeds of transgenic oilseed rape lines (1a, 1b, 2, 3, 4, 5, 6) and the parental cv Lisora (wild type [WT]). For bulk analysis, 10 normally shaped seeds from each line were pooled and subjected to methanolic extraction. Extracts were analyzed by HPLC. The values are means ± sd of three biological replicates. FW, Fresh weight.
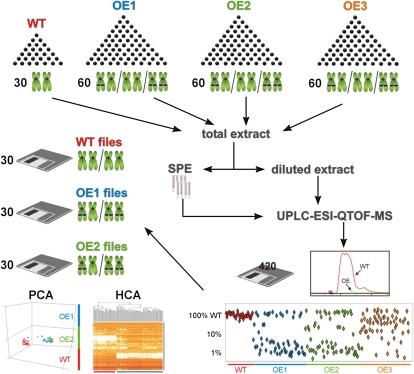
To verify single-copy transgene insertions within T2 seeds, a “single-seed metabolite profiling approach” was established (Fig. 3). T2 seeds from the assumed BnSCE3 overexpressors (OE1, -2, and -3) and the parental oilseed rape line, cv Lisora (wild type), were initially screened by HPLC for sinapine content as a biochemical marker. The individual transgenic seeds showed a sinapine reduction of about 99% in comparison with the wild type. To comply with the statistical requirements of T2 segregation analysis, 30 individual seeds of the wild type and 60 individual seeds of each of the individual OE lines were analyzed. To measure the abundant choline esters in a linear range without losing the ability to determine the residual seed metabolome components present at low levels, a two-step extraction/fractionation method was employed (Böttcher et al., 2011). Each of the resulting methanolic extracts of the individual grains were analyzed via a nontargeted liquid chromatography-mass spectrometry (LC-MS) platform (Böttcher et al., 2009) to determine profiles of secondary metabolites. In a first step, the biomarker sinapine was quantified, and segregation analysis, based on intensity of sinapine signals, was performed. Due to the dominant nature of the transgene in the homozygous and heterozygous allelic state, OE grains containing approximately 1% of wild-type sinapine levels were considered to contain the transgene and express the corresponding protein. Two of the three transgenic lines investigated (OE1 and OE2) showed a clear 3:1 distribution of the high- to low-sinapine chemotype, as would be expected for a single-copy insertion of the dominant napin promoter::BnSCE3 fusion. In the final step, 30 LC-MS profiles of each class (wild type, OE1, and OE2) were subjected to nontargeted analyses. Instead of using only single ion traces, as with the biomarker screening, the entire set of information in the raw data files, including chromatographic and spectral precursor and fragmentation data, was used in the analysis. The results of the exploratory data analyses, principal component analysis (PCA) or hierarchical cluster analysis (HCA), were used to examine clustering and separation of the sample classes. For the total extracts, PCA and HCA were based on 1,235 aligned mass signals and showed a clear separation of wild-type and OE lines (Fig. 3; Supplemental Fig. S1, A and B). The HCA could not differentiate between the two OE lines, indicating a close relationship caused by the transgene-mediated BnSCE3 expression. Profiling of the choline ester-depleted fractions led to 2,788 aligned mass signals, mirroring the results of the total extracts (Supplemental Fig. S1, C and D). Again, the transgenic lines resembled each other, clearly clustering apart from the wild-type line. The HCA also revealed that nearly 50% of the detectable mass signals generated by the seed metabolome differed in the BnSCE3 transgenic line compared with the wild-type line.
Figure 3.
Single seed segregation analysis scheme. Thirty wild-type seeds (WT) and 180 mutant seeds (OE1, OE2, and OE3, representing allelic mutations) were each extracted independently and analyzed via the LC-MS platform. The resulting data files were screened for reduction of sinapine in the transgenic seeds. Thirty each of those data files belonging to seeds of OE1 and OE2, which showed less than 1% of the wild-type level of sinapine, were included in further nontargeted bioinformatic analyses. The resulting PCA and HCA show a clear separation of wild-type and mutant samples. Red lines in the HCA analysis represent mass signals of highest intensities.
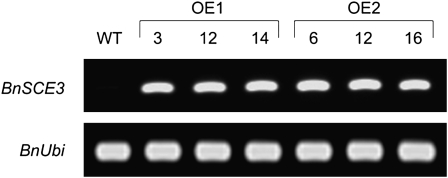
For segregation analysis, T2 plants of OE1 and OE2 were propagated and selfed to produce T3 seeds. Homozygous BnSCE3-overexpressing lines were isolated by characterization of inheritance of PPT resistance among T3 seedlings. To verify BnSCE3 overexpression, semiquantitative reverse transcription (RT)-PCR was carried out with RNA from seeds of wild-type and homozygous OE1 and OE2 lines (Fig. 4). PCR-derived signals revealed strong expression of BnSCE3 during seed development of OE1 and OE2, whereas no expression was detectable in wild-type seeds. The corresponding enzyme activities, analyzed as described previously (Clauss et al., 2008), were found to be 0.53 and 0.62 nkat mg−1 seed protein of OE1 and OE2, respectively.
Figure 4.
RT-PCR analysis of the expression of BnSCE3 in homozygous transgenic T3 seeds of three independent plants of the overexpression lines OE1 (plants 3, 12, and 14) and OE2 (plants 6, 12, and 16). Expression of the ubiquitin gene is given as a constitutive control. WT, Wild type.
Profiling of Secondary Metabolites in Seeds
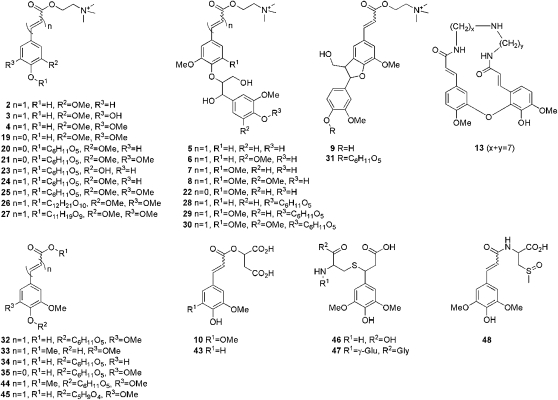
The results obtained by metabolite profiling of the methanolic extracts of OE1 and OE2 via the nontargeted LC-MS platform are summarized in Tables I and II. Table I lists all compounds from the total methanolic extracts that were found to accumulate differentially in the transgenic seeds compared with wild-type seeds. Table II lists the compounds annotated from the corresponding choline ester-depleted extracts. Metabolites that did not show any marked quantitative differences were not included (change > 2-fold; P < 0.05 in both comparisons: wild type/OE1 and wild type/OE2). The 50 mostly putatively identified compounds included hydroxybenzoate, hydroxycinnamate, a few flavonoid (kaempferol) conjugates, and 12 compounds that could only partially be annotated by their elemental composition, including eight choline esters and two sulfated substances. Eight compounds were exclusively detected in the choline ester-depleted extracts: 2-O-feruloylmalate, methyl sinapate 4-O-hexoside, sinapate 4-O-pentoside, glutathione and Cys sinapate conjugates, as well as N-sinapoyl-S-methylcysteine sulfoxide. Figure 5 shows the structures of 33 hydroxybenzoate- and hydroxycinnamate-related compounds annotated from the LC-MS-based metabolite profiling. Complete mass spectrometric data (elemental composition, retention time, quasimolecular ions or observed fragment ions) with references for some of the known compounds are given in Supplemental Table S1.
Table I. Putatively annotated metabolites differentially accumulating in wild-type and BnSCE3 OE seeds as identified by comparative ultraperformance liquid chromatography (UPLC)/ESI(+)-QTOF-MS-based metabolite profiling using methanolic total extracts.
Metabolites with threshold levels of change fold factors 20 (4.322; log2) and 0.1 (–3.322; log2) in both lines are set in boldface and boxed in Figure 10. G, Guaiacyl moiety; S, syringyl moiety; UC, unknown compound.
| No. | Metabolite | Annotation Level | Elemental Composition | Retention Time | Quantifier Ion | Relative Metabolite Intensity (log2) |
|
| OE1/Wild Type | OE2/Wild Type | ||||||
| s | m/z | log2 (ratio) | |||||
| 1 | Benzoylcholine | 1 | C12H18NO2+ | 125 | 149.06 | −3.68 | −1.55 |
| 2 | Feruloylcholine (FC) | 1 | C15H22NO4+ | 133 | 221.08 | −8.91 | −8.91 |
| 3 | 5-Hydroxy-FC | 2 | C15H22NO5+ | 109 | 237.08 | −8.30 | −6.93 |
| 4 | Sinapine (SC) | 1 | C16H24NO5+ | 135 | 310.16 | −10.82 | −9.56 |
| 5 | FC(4-O-8′)G | 1 | C25H34NO8+ | 143/146 | 476.23 | −1.37 | −1.41 |
| 6 | FC(4-O-8′)S | 2 | C26H36NO9+ | 141/146 | 506.24 | −1.10 | −0.67 |
| 7 | SC(4-O-8′)G | 2 | C26H36NO9+ | 171/186 | 506.24 | −2.99 | −2.75 |
| 8 | SC(4-O-8′)S | 2 | C27H38NO10+ | 168/183 | 536.25 | −3.07 | −2.27 |
| 9 | FC(5-8′)G | 1 | C25H32NO7+ | 215 | 458.22 | −3.47 | −3.01 |
| 10 | 2-O-Sinapoylmalate | 1 | C15H16O9 | 193 | 207.07 | −1.78 | −2.12 |
| 11 | Hydroxybenzoate O-hexoside | 2 | C13H16O8 | 92 | 323.07 | −3.75 | −3.49 |
| 12 | Caffeate di-O-hexoside | 2 | C21H28O14 | 97 | 527.14 | −4.04 | −2.65 |
| 13 | Cyclic spermidine conjugate | 2 | C27H33N3O6 | 180/186 | 496.24 | −1.39 | −0.72 |
| 14 | UC#1 (choline ester) | 3 | C18H34NO4+ | 177 | 328.25 | −3.34 | −3.78 |
| 15 | UC#2 (choline ester) | 3 | C25H31N2O6+ | 192 | 455.22 | −5.13 | −4.76 |
| 16 | UC#3 (phenolic choline ester) | 3 | C26H36NO8+ | 175 | 490.24 | −6.80 | −5.74 |
| 17 | UC#4 (sulfated) | 4 | C24H22N2O12S | 145 | 505.12 | −2.59 | −0.96 |
| 18 | UC#5 (sulfated) | 4 | C24H22N2O11S | 181 | 489.13 | −4.77 | −1.45 |
| 19 | Syringoylcholine (SyC) | 1 | C14H22NO5+ | 89 | 225.08 | 6.74 | 7.41 |
| 20 | Vanilloylcholine 4-O-hexoside | 2 | C19H30NO9+ | 64 | 416.19 | 2.35 | 2.83 |
| 21 | SyC 4-O-hexoside | 2 | C20H32NO10+ | 73 | 446.20 | 4.40 | 4.73 |
| 22 | SyC(4-O-8′)G | 2 | C24H34NO9+ | 134/148 | 480.22 | 6.73 | 6.66 |
| 23 | Caffeoylcholine 4-O-hexoside | 2 | C20H30NO9+ | 83/94 | 428.19 | 3.12 | 2.82 |
| 24 | FC 4-O-hexoside | 2 | C21H32NO9+ | 90/105 | 442.21 | 2.93 | 4.06 |
| 25 | SC 4-O-glucoside | 1 | C22H34NO10+ | 96/104 | 472.22 | 3.58 | 4.38 |
| 26 | SC 4-O-dihexoside | 2 | C28H44NO15+ | 90/99 | 634.27 | 8.91 | 9.37 |
| 27 | SC 4-O-(hexoside-pentoside) | 2 | C27H42NO14+ | 101 | 604.26 | 6.55 | 7.19 |
| 28 | FC(4-O-8′)G 4-O′-hexoside | 2 | C31H44NO13+ | 111/124 | 638.28 | 2.64 | 3.04 |
| 29 | SC(4-O-8′)G 4-O′-hexoside | 2 | C32H46NO14+ | 128/140 | 668.29 | 4.02 | 4.62 |
| 30 | SC(4-O-8′)S 4-O′-hexoside | 2 | C33H48NO15+ | 133/143 | 698.30 | 3.89 | 4.78 |
| 31 | FC(5-8′)G 4-O′-hexoside | 2 | C31H42NO12+ | 153 | 620.27 | 1.41 | 2.33 |
| 32 | Sinapate 4-O-glucoside | 1 | C17H22O10 | 118 | 409.11 | 4.22 | 4.66 |
| 33 | Methyl sinapate | 1 | C12H14O5 | 314 | 207.07 | 4.89 | 4.52 |
| 34 | Ferulic acid 4-O-hexoside | 2 | C16H20O9 | 110 | 379.10 | 3.84 | 4.47 |
| 35 | Syringate 4-O-hexoside | 1 | C15H20O10 | 94 | 383.09 | 3.48 | 3.61 |
| 36 | Kaempferol 3-O-sophoroside | 1 | C27H30O16 | 169 | 287.06 | 3.06 | 4.08 |
| 37 | Kaempferol 3-O-sinapoylsophoroside | 1 | C38H40O20 | 185 | 369.12 | 3.16 | 2.97 |
| 38 | UC#6 (phenolic choline ester) | 3 | C30H44NO13+ | 128 | 626.28 | 4.26 | 5.34 |
| 39 | UC#7 (phenolic choline ester) | 3 | C15H26NO4+ | 175 | 225.11 | 2.94 | 3.20 |
| 40 | UC#8 (phenolic choline ester) | 3 | C17H28NO4+ | 229 | 251.13 | 2.58 | 2.52 |
| 41 | UC#9 (phenolic choline ester) | 3 | C26H42NO12+ | 188 | 560.27 | 2.46 | 2.43 |
| 42 | UC#10 (phenolic choline ester) | 3 | C22H34NO11+ | 97 | 488.21 | 4.19 | 5.10 |
Table II. Putatively annotated metabolites differentially accumulating in wild-type and BnSCE3 OE seeds as identified by comparative UPLC/ESI(+)-QTOF-MS-based metabolite profiling using choline ester-depleted and concentrated methanolic total extracts.
The compounds are putatively annotated as stated in Table I, except those identified earlier (see Supplemental Table S1). Metabolites with threshold levels of change fold factors 20 (4.322; log2) and 0.1 (–3.322; log2) in both lines are set in boldface and boxed in Figure 10. UC, Unknown compound.
| No. | Metabolite | Annotation Level | Elemental Composition | Retention Time | Quantifier Ion | Relative Metabolite Intensity (log2) |
|
| OE1/Wild Type | OE2/Wild Type | ||||||
| s | m/z | log2 (ratio) | |||||
| 10 | 2-O-Sinapoylmalate | 1 | C15H16O9 | 193 | 363.07 | −1.13 | −1.53 |
| 11 | Hydroxybenzoate O-hexoside | 2 | C13H16O8 | 92 | 323.07 | −4.86 | −5.80 |
| 12 | Caffeate di-O-hexoside | 2 | C21H28O14 | 97 | 527.14 | −3.33 | −1.95 |
| 17 | UC#4 (sulfated) | 4 | C24H22N2O12S | 145 | 505.12 | −2.59 | −1.05 |
| 18 | UC#5 (sulfated) | 4 | C24H22N2O11S | 181 | 489.13 | −5.51 | −1.52 |
| 43 | 2-O-Feruloylmalate | 2 | C14H14O8 | 191 | 333.06 | 5.07 | 5.21 |
| 32 | Sinapate 4-O-glucoside | 1 | C17H22O10 | 118 | 409.11 | 4.04 | 3.72 |
| 33 | Methyl sinapate | 1 | C12H14O5 | 314 | 207.07 | 7.99 | 8.48 |
| 34 | Ferulate 4-O-hexoside | 2 | C16H20O9 | 110 | 379.10 | 4.02 | 4.16 |
| 35 | Syringate 4-O-hexoside | 1 | C15H20O10 | 94 | 383.09 | 2.55 | 3.39 |
| 36 | Kaempferol 3-O-sophoroside | 1 | C27H30O16 | 169 | 287.06 | 5.35 | 4.97 |
| 37 | Kaempferol 3-O-sinapoylsophoroside | 1 | C38H40O20 | 185 | 369.12 | 5.25 | 3.61 |
| 44 | Methyl sinapate 4-O-hexoside | 2 | C18H24O10 | 200 | 423.13 | 8.16 | 9.19 |
| 45 | Sinapate 4-O-pentoside | 2 | C16H20O9 | 130 | 379.10 | 1.91 | 2.31 |
| 46 | Sinapate-Cys conjugate | 2 | C14H19NO7S | 90 | 346.10 | 3.92 | 4.51 |
| 47 | Sinapate-glutathione conjugate | 2 | C21H29N3O11S | 101 | 308.09 | 2.23 | 3.16 |
| 48 | Sinapoyl-methylcysteine sulfoxide | 2 | C15H19NO7S | 128 | 358.10 | 6.67 | 10.15 |
| 49 | UC#11 | 4 | C19H32O9 | 185 | 427.19 | 2.33 | 1.46 |
| 50 | UC#12 | 4 | C12H11NO3 | 218/242 | 240.06 | 2.71 | 2.35 |
Figure 5.
Scheme of secondary phenolic structures from oilseed rape seeds analyzed by UPLC/ESI-QTOF-MS (Tables I and II; Supplemental Table S1).
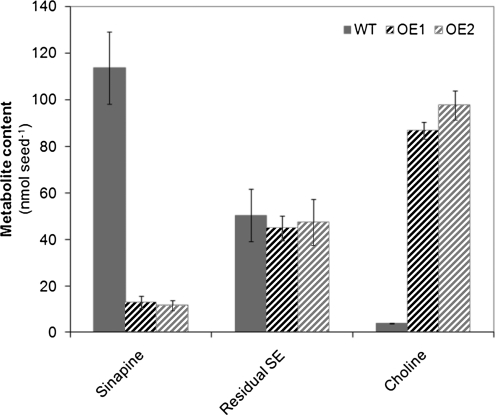
In general, both OE lines behaved similarly with regard to changes in the pattern of secondary metabolites in seeds compared with wild-type plants. As expected from initial analyses, the most striking effect triggered by seed-specific overexpression of BnSCE3 was a severe depletion of sinapine and related esters, including feruloylcholine, 5-hydroxyferuloylcholine, and some unknown phenolic choline esters (Table I; Fig. 6). However, the hydrolysis product sinapate, liberated by enzymatic BnSCE3 activity, did not accumulate. Instead, levels of major sinapate conjugates, sinapate 4-O-glucoside, methyl sinapate and its 4-O-hexoside, N-sinapoyl-S-methylcysteine sulfoxide, and two syringoylcholine conjugates, were higher. A portion of sinapine, obviously not accessible by BnSCE3 activity, accumulated in various glycosylated forms, such as 4-O-glucoside, 4-O-di-hexoside, 4-O-hexoside-pentoside, and even in guaiacyl and syringyl conjugates. The latter was also observed with feruloylcholine besides accumulating ferulate 4-O-glucoside. Interestingly, also the level of two flavonoid conjugates, kaempferol 3-O-sophoroside and kaempferol 3-O-sinapoylsophoroside, increased significantly. The observed accumulation of related phenolic compounds, however, could not compensate quantitatively for the net loss of sinapate moieties in the BnSCE3-overexpressing lines. As a direct effect of induced sinapine hydrolysis, the transgenic OE lines showed a significant accumulation of free choline, resulting in increased seed amounts by a factor of about 25 compared with the wild type (Fig. 6).
Figure 6.
Content of sinapine, remaining sinapate esters (Residual SE), and choline in wild-type (WT) seeds and homozygous T3 seeds of the transgenic oilseed rape (nmol seed−1) overexpression lines OE1 and OE2. The values are means ± sd of three biological replicates.
Profiling of Primary Metabolites in Seeds
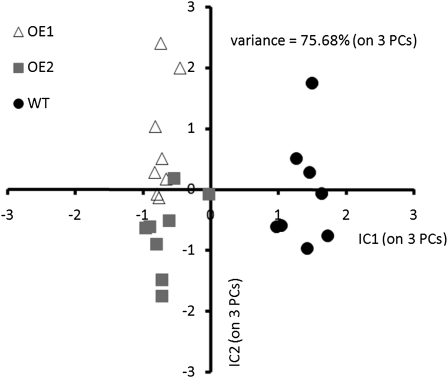
The data from the gas chromatography (GC)-MS-based analysis of the primary metabolite profiles revealed that overexpression of BnSCE3 in the seeds not only affected secondary metabolism but, in addition, led to significant changes in various areas of primary metabolism, including organic acids, polyols, sugars, amino acids, and lipids. Independent component analysis of the profiles of polar primary metabolites in seeds (Fig. 7) resulted in clear discrimination of wild-type and transgenic OE lines.
Figure 7.
Independent component analysis (ICA) of polar primary metabolite profiles from oilseed rape wild-type (WT) seeds (black circles) and BnSCE3-overexpressing seeds (lines OE1 [gray squares] and OE2 [white triangles]). Analysis of independent component analysis scores demonstrates common differences in seeds in response to seed-specific BnSCE3 overexpression.
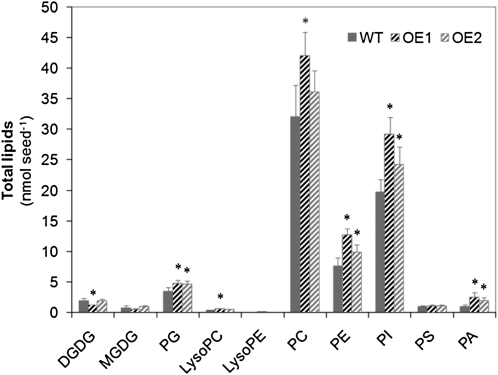
The changes in the seed levels of polar primary metabolites caused by overexpression of BnSCE3 are summarized in Table III. Among the compounds related to the citric acid cycle, the amount of succinate was significantly increased, whereas fumarate showed a strong decrease. Likewise, an increase in Asp was accompanied by a decrease in the levels of Asn and pipecolate, and Phe and Ser were markedly reduced. Among sugar polyols, galactinol, mannitol, and sorbitol showed increased amounts. The oligosaccharide raffinose, however, was found in significantly lower amounts in OE1 and OE2, whereas the glycolysis-derived glycerol 3-phosphate was increased in the BnSCE3-overexpressing lines. With regard to the various classes of lipids, the seeds of OE lines contained more phospholipids per seed than the wild type (Fig. 8). PC was increased in OE1 and tended to be increased in OE2, while phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), and phosphatidic acid (PA) were increased in both OE lines.
Table III. GC-MS analysis of polar primary metabolites from seeds of oilseed rape wild type and two homozygous BnSCE3 OE lines (OE1 and OE2).
Metabolites were analyzed via routine GC-MS-based metabolite profiling and are presented as fold change of genetically modified over wild-type samples (n = 8 pools of 20 seeds). Data were subjected to ANOVA to determine the significance of differences between means of the wild type and samples overexpressing BnSCE3 (OE1 and OE2). Ratios with statistically significant P values (P ≤ 0.05) of both transgenic lines are set in boldface. Metabolites with threshold levels of change fold factors 1.5 and 0.7 in both lines are boxed in Figure 10. NA, KEGG compound identifier not available.
| Metabolite | Elemental Composition | KEGG Identifier | Relative Metabolite Intensity |
|||
| OE1/Wild Type |
OE2/Wild Type |
|||||
| Ratio | P | Ratio | P | |||
| Organic acids | ||||||
| Arabinoate | C5H10O6 | C00545 | 0.90 | 1.8E-01 | 0.82 | 4.1E-03 |
| Citrate | C6H8O7 | C00158 | 0.78 | 3.4E-03 | 0.82 | 3.1E-03 |
| Erythronate | C4H8O5 | NA | 1.24 | 1.9E-04 | 1.65 | 8.6E-04 |
| Fumarate | C4H4O4 | C00122 | 0.15 | 8.4E-08 | 0.11 | 5.1E-08 |
| Galactonate | C6H12O7 | C00880 | 1.14 | 6.4E-02 | 0.72 | 1.0E-04 |
| Gluconate | C6H12O7 | C00257 | 1.31 | 2.1E-01 | 1.44 | 2.9E-01 |
| Glutarate, 2-OH- | C5H8O5 | C03196 | 0.92 | 1.6E-01 | 0.87 | 1.7E-02 |
| Glycolate | C2H4O3 | C00160 | 0.96 | 5.1E-01 | 0.85 | 6.8E-02 |
| Gulonate | C6H12O7 | C00800 | 1.23 | 1.0E-03 | 1.24 | 7.5E-03 |
| Hexadecanoate | C16H32O2 | C00249 | 1.05 | 2.0E-01 | 1.11 | 3.5E-03 |
| Hexanoate | C6H12O2 | C01585 | 1.26 | 7.4E-02 | 1.30 | 6.4E-02 |
| Ribonate | C5H10O6 | C01685 | 0.97 | 6.2E-01 | 0.93 | 2.8E-01 |
| Saccharate | C6H10O8 | C00767 | 1.03 | 7.5E-01 | 1.20 | 3.0E-02 |
| Stearate | C18H36O2 | C01530 | 1.09 | 6.0E-02 | 1.12 | 6.3E-03 |
| Succinate | C4H6O4 | C00042 | 1.65 | 3.5E-05 | 1.40 | 5.6E-06 |
| Threonate | C4H8O5 | C01620 | 0.62 | 4.2E-05 | 0.88 | 1.4E-01 |
| Polyols | ||||||
| Arabitol | C5H12O5 | C01904 | 1.10 | 9.0E-02 | 1.85 | 1.7E-10 |
| Ethanolamine | C2H7NO | C00189 | 1.22 | 1.8E-01 | 1.17 | 2.4E-01 |
| Galactinol | C12H22O11 | C01235 | 2.02 | 3.9E-07 | 2.11 | 1.4E-08 |
| Glycerol | C3H8O3 | C00116 | 1.16 | 6.3E-01 | 0.81 | 4.5E-02 |
| Inositol, myo- | C6H12O6 | C00137 | 0.85 | 1.2E-01 | 0.77 | 1.4E-02 |
| Maltitol | C12H24O11 | NA | 1.56 | 3.1E-01 | 1.71 | 2.6E-01 |
| Mannitol | C6H14O6 | C00392 | 1.96 | 2.4E-11 | 2.28 | 4.4E-09 |
| Sorbitol | C6H14O6 | C00794 | 1.74 | 7.1E-03 | 1.29 | 1.3E-02 |
| Monosaccharides | ||||||
| Glc | C6H12O6 | C00031 | 0.66 | 1.4E-05 | 1.66 | 4.2E-01 |
| Fru | C6H12O6 | C00095 | 1.13 | 8.0E-01 | 1.73 | 5.5E-01 |
| Oligosaccharides | ||||||
| Saccharose | C12H22O11 | C00089 | 1.02 | 8.8E-01 | 0.94 | 6.9E-01 |
| Raffinose | C18H32O16 | C00492 | 0.60 | 2.7E-04 | 0.61 | 8.8E-04 |
| Amino Acids | ||||||
| γ-Amino butyric acid | C4H9NO2 | C00334 | 1.25 | 2.2E-01 | 1.14 | 3.5E-01 |
| Asn | C4H8N2O3 | C00152 | 0.54 | 2.6E-06 | 0.59 | 6.0E-06 |
| Asp | C4H7NO4 | C00049 | 1.68 | 3.4E-05 | 1.27 | 8.0E-03 |
| Glu | C5H9NO4 | C00025 | 1.10 | 7.8E-02 | 1.04 | 4.4E-01 |
| Gln | C5H10N2O3 | C00064 | 0.70 | 2.4E-01 | 0.56 | 5.3E-02 |
| His | C6H9N3O2 | C00135 | 1.54 | 9.6E-03 | 1.03 | 8.2E-01 |
| Ile | C6H13NO2 | C00407 | 1.05 | 7.3E-01 | 0.76 | 1.1E-01 |
| Leu | C6H13NO2 | C00123 | 1.05 | 6.9E-01 | 0.88 | 3.8E-01 |
| Lys | C6H14N2O2 | C00047 | 1.84 | 9.7E-04 | 1.31 | 6.3E-02 |
| Phe | C9H11NO2 | C00079 | 0.59 | 1.0E-07 | 0.60 | 3.0E-07 |
| Pipecolate | C6H11NO2 | C00408 | 0.12 | 1.1E-02 | 0.22 | 2.7E-02 |
| Pro | C5H9NO2 | C00148 | 0.61 | 4.5E-01 | 0.38 | 1.8E-01 |
| Pyroglutamate | C5H7NO3 | C02238 | 1.18 | 4.0E-03 | 1.06 | 2.5E-01 |
| Ser | C3H7NO3 | C00065 | 0.41 | 4.3E-09 | 0.51 | 1.3E-07 |
| Thr | C4H9NO3 | C00188 | 1.13 | 2.5E-01 | 0.88 | 1.8E-01 |
| Trp | C11H12N2O2 | C00078 | 1.01 | 9.4E-01 | 0.61 | 6.5E-05 |
| Tyr | C9H11NO3 | C00082 | 0.70 | 1.2E-01 | 0.65 | 5.4E-02 |
| Val | C5H11NO2 | C00183 | 1.13 | 4.8E-01 | 0.80 | 2.2E-01 |
| Phosphates | ||||||
| Guanosine-5-phosphate | C10H14N5O8P | C00144 | 1.07 | 7.9E-01 | 1.05 | 8.6E-01 |
| Glycerol-3-phosphate | C3H9O6P | C00093 | 1.30 | 3.1E-03 | 1.30 | 3.3E-03 |
| Glycerophosphoglycerol | C6H15O8P | C03274 | 1.12 | 1.6E-01 | 1.30 | 1.7E-02 |
| Phosphate | H3O4P | C00009 | 1.20 | 6.7E-02 | 1.10 | 2.5E-01 |
| N Compounds | ||||||
| Nicotinate | C6H5NO2 | C00253 | 1.30 | 1.6E-03 | 1.07 | 1.9E-01 |
| 2-Pyridone | C5H5NO | C02502 | 1.11 | 2.1E-01 | 1.01 | 8.9E-01 |
Figure 8.
Total amount of lipid classes (nmol seed−1) of wild-type seeds (WT; black bars) in comparison with homozygous BnSCE3 overexpression seeds (OE1 and OE2; gray bars). The data represent means of five biological replicates (each sample consisted of six seeds). Values indicated by asterisks were significantly different from wild-type data (P ≤ 0.05 by t test).
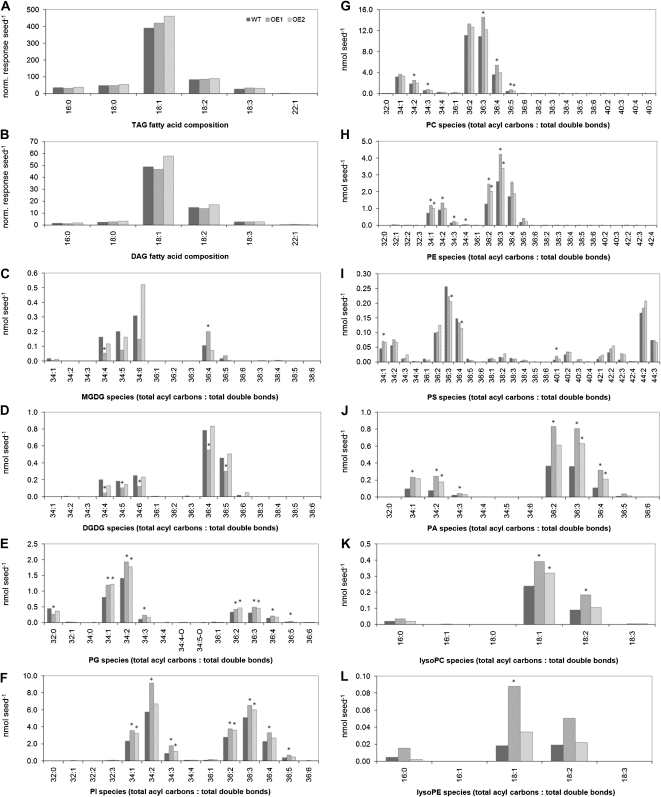
Fatty acid compositional analysis indicated that the total fatty acid (esterified plus nonesterified) composition of the wild-type seeds was 4% 16:0, 2% 18:0, 72% 18:1, 14% 18:2, and 4% 18:3. The fatty acid composition of the transgenic seeds was not significantly different from the wild type (data not shown). The nonpolar lipids, triacylglycerol (TAG) and diacylglycerol (DAG), contained largely 18:1 species, followed by 18:2 chains, reflecting the overall seed fatty acid composition. The analysis of polar lipid molecular species showed that the higher levels of phospholipids per seed observed in the overexpressors were due to higher levels of many phospholipid species rather than to species with any particular acyl combinations (Figs. 8 and 9). Examination of the apparent phospholipid molecular species in seeds revealed that in the major phospholipid classes, PC, 36:2 and 36:3 (total acyl carbon:total acyl carbon-carbon double bonds) molecular species were most common, accounting for almost 70% of the total PC (Fig. 8). Given the fatty acid composition, these PC designations most likely represent 18:1 to 18:1 and 18:1 to 18:2 acyl combinations. The other extraplastidially produced phospholipids, PE, PS, and PI, were also rich in molecular species, containing a total of 36 acyl carbons. This was particularly notable for PI, as the PI species differ from those of Arabidopsis, which contains mainly 34-C combinations in all tissues examined (Devaiah et al., 2006). Seed galactolipids (monogalactosyldiacylglycerol [MGDG] and digalactosyldiacylglycerol [DGDG]) produced in the plastids were similar in acyl composition to those of Arabidopsis, with 34:6 and 34:5 the most abundant MGDG species. 34:6 MGDG represents an 18:3 to 16:3 combination, consistent with oilseed rape being a “16:3 plant” (Benhassaine-Kesri et al., 2002).
Figure 9.
Lipid molecular species of wild-type seeds (WT) in comparison with homozygous BnSCE3-overexpressing seeds (OE1 and OE2). The data represent means of five biological replicates. Values indicated by asterisks were significantly different from wild-type data (P ≤ 0.05 by t test).
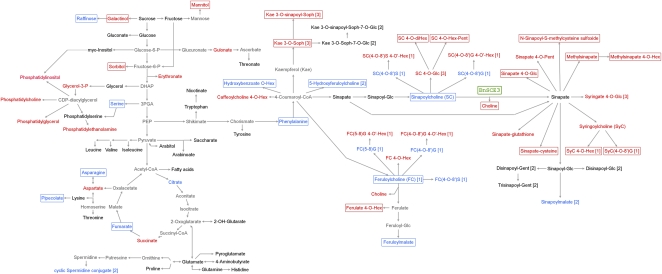
The quantitative changes observed in primary and secondary metabolites indicate large metabolic shifts in seed metabolism. These global alterations are visualized in a metabolic map (Fig. 10).
Figure 10.
Changes in the metabolic network (primary and secondary metabolites) of transgenic BnSCE3 seeds. Sinapate leading to sinapine and sinapate derived from BnSCE3 activity in the transgenic seeds might be present within the same metabolite pool. Red compound names indicate significantly increased amounts, blue names indicate reduced amounts, black names indicate unchanged amounts, and gray names indicate amounts not determined. Boxed names point to compounds with largest quantitative changes in both transgenic lines (OE1 and OE2) compared with the wild-type plants, with threshold levels for primary metabolites (Table III) of change fold factors 1.5 and 0.7 and for secondary metabolites (Tables I and II) of change fold factors 20 (4.322; log2) and 0.1 (−3.322; log2). For increased amounts of phospholipids, see Figure 8. Arrows indicate both one-step and multistep reactions, based on conceivable and well-known biochemical reactions. The reaction steps of the general phenylpropanoid metabolism are defined according to Humphreys and Chapple (2002), Boerjan et al. (2003), and Weng et al. (2010). G, Guaiacyl moiety; Gent, gentiobiose; Hex, unknown hexose moieties (commonly Glc); Pent, pentose; S, syringyl moiety; Soph, sophorose; [1], Böttcher et al. (2009); [2], Baumert et al. (2005); [3], Wolfram et al. (2010).
Changes of Seed Morphology and Physiology
Seeds of BnSCE3-overexpressing lines were increased in size by about 16%. This was accompanied by increases in seed weight by up to 60% and in seed moisture by about 26% (Table IV).
Table IV. Comparison of morphological/physiological properties and data from near-infrared spectroscopy of wild-type and OE lines.
Seed size and length of seedling hypocotyls (6 d old) are expressed in mm, seed weight in mg, and moisture in percentage of seed mass (mean ± sd; n = 30). Contents of oil, protein, hemicellulose, and cellulose are expressed in percentage of seed mass (mean ± sd; four biological replicates with four technical replicates each). Data were subjected to Student’s t test to determine the significance of differences between means of the wild type and overexpression samples (OE1 and OE2).
| Property | Wild Type | OE1 | P | OE2 | P |
| Size | 2.18 ± 0.17 | 2.53 ± 0.15 | 8.8E-04 | 2.55 ± 0.24 | 7.8E-04 |
| Weight | 3.70 ± 0.69 | 5.99 ± 0.85 | 5.3E-23 | 5.90 ± 0.91 | 1.7E-23 |
| Hypocotyl | 1.69 ± 0.29 | 2.14 ± 0.36 | 4.7E-10 | 2.49 ± 0.50 | 1.3E-17 |
| Moisture | 4.60 ± 0.25 | 5.62 ± 0.35 | 3.1E-02 | 5.94 ± 0.17 | 9.1E-03 |
| Oil | 43.3 ± 0.37 | 42.5 ± 0.53 | 3.5E-04 | 43.5 ± 1.56 | 7.9E-01 |
| Protein | 27.0 ± 0.35 | 25.9 ± 0.51 | 1.5E-05 | 25.4 ± 1.37 | 3.0E-03 |
| Hemicellulose | 4.08 ± 0.05 | 5.43 ± 0.13 | 1.7E-18 | 5.19 ± 0.18 | 1.3E-11 |
| Cellulose | 4.75 ± 0.08 | 6.48 ± 0.17 | 1.5E-17 | 6.28 ± 0.12 | 2.5E-16 |
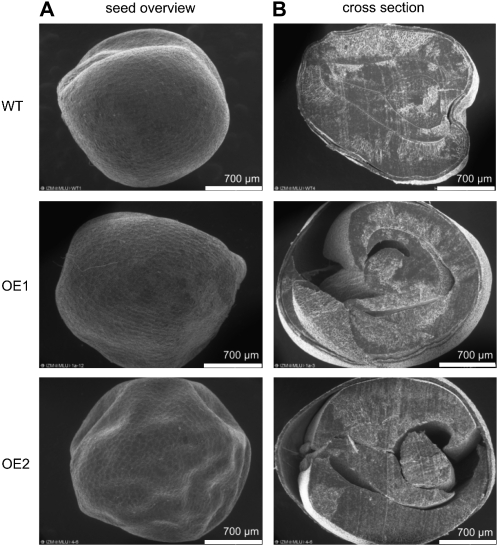
Comparative environmental scanning electron microscopy showed marked differences in the cross-section images between wild-type and OE seeds. In contrast to the usual compact tissues of the wild-type seeds, large cavities between the seed coat and the embryo were detected in some of the transgenic OE1 and OE2 seeds (Fig. 11). Some agronomically important properties of the transgenic seeds were analyzed by near-infrared spectroscopy (Table IV), but no significant differences could be detected between BnSCE3-overexpressing lines OE1 and OE2 and the wild type. Seed oil and protein contents with 43% and 26% of seed mass, respectively, did not change. However, hemicelluloses and cellulose both showed increases of about 30%. This probably leads, together with increased water content, to the observed higher seed weight.
Figure 11.
Environmental scanning electron microscopy images of transgenic (OE1 and OE2) and wild-type (WT) oilseed rape seeds. Representative images show the seed coat (A) and seed cross-section (B). Analyses were done with six seeds of two independent homozygous transgenic plants of the two overexpression lines OE1 and OE2. In comparison, we analyzed four wild-type seeds. Bars = 700 μm.
Seed germination rate and seedling development were slightly enhanced in the OE lines. Six-day-old OE seedlings showed 20% longer hypocotyls compared with wild-type seedlings at the same age.
Metabolic Changes in Developing Seedlings
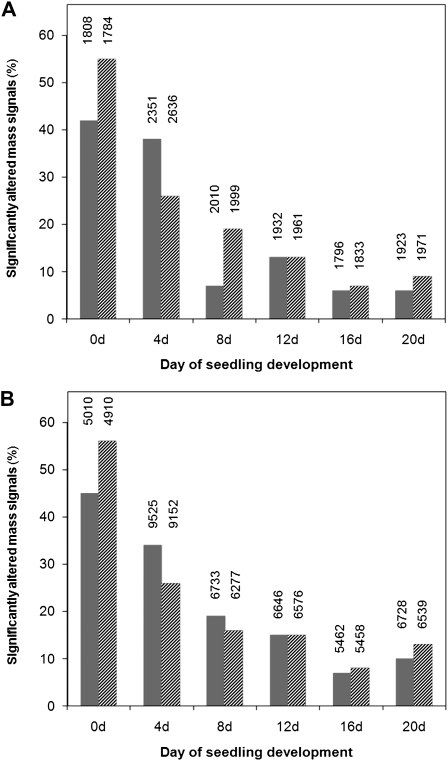
To determine how the dramatic changes in BnSCE3-overexpressing seeds influence the metabolism of developing seedlings, nontargeted metabolic profiles were determined during seed germination and seedling development in the wild-type and OE lines. The percentage of significantly changed mass signals for OE1 and OE2 relative to the wild type was calculated. As a result, the analyses indicated that the degree of metabolic differences between OE1 and the wild type and between OE2 and the wild type decreased progressively during the first 20 d of seedling development, from 50% to about 10%. This effect was obvious in both the total and the choline ester-depleted extracts, although the latter contained about 5-fold more mass signals (Fig. 12). Thus, the chemical composition of fully developed seedlings and adult plants did not vary significantly between OE lines and the wild type.
Figure 12.
A, Calculation of the percentage of significantly altered mass signals in the total extracts of OE1 (dark gray bars) and OE2 (light gray bars) lines in comparison with the parental line in seeds and developing seedlings. Data are derived from a nontargeted profiling approach. The exact numbers and the total amount of mass signals are presented for each sample class. B, Calculation of the percentage of significantly altered mass signals in the choline derivative reduced fractions of OE1 (dark gray bars) and OE2 (light gray bars) lines in comparison with the parental line.
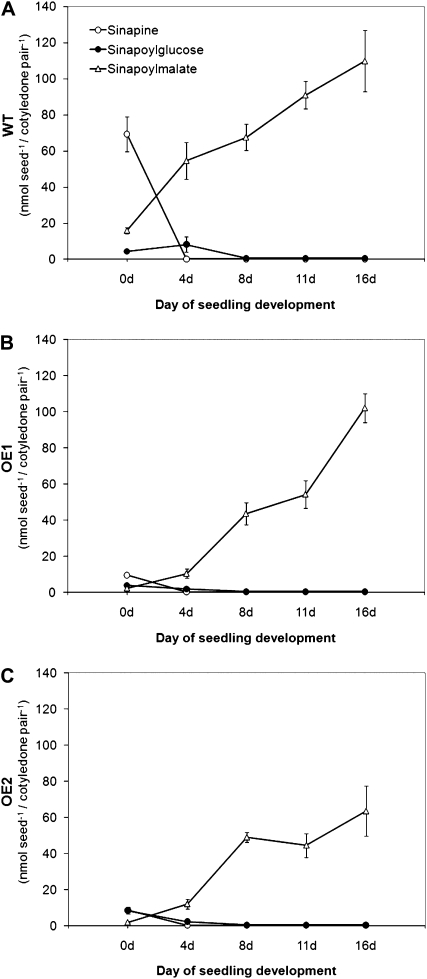
This was reflected by the accumulation kinetics of the predominating sinapate esters during seedling development (Fig. 13; Supplemental Fig. S2). In the wild type, the seed storage compound sinapine is hydrolyzed by enzymatic BnSCE3 activity during early seed development. The liberated sinapate is conjugated via 1-O-sinapoylglucose to l-malate, resulting in sinapoylmalate accumulation (Fig. 13A). OE1 and OE2 showed accumulation patterns of sinapoylglucose and sinapoylmalate that resembled those from the wild-type plants, although the low-sinapine seeds could not provide sinapoyl moieties (Fig. 13, B and C). In contrast to the wild type, however, accumulation of sinapoylmalate was retarded in OE1 and OE2 during early seedling development up to 11 d after sowing.
Figure 13.
Accumulation pattern of sinapine, sinapoylglucose, and sinapoylmalate in cotyledons of developing seedlings of oilseed rape. The cotyledons were harvested pairwise at 4, 8, 11, or 16 d after sowing and analyzed by UPLC-diode array detection. The graphs show the amount of metabolites in seeds and cotyledons of wild-type (WT) plants (A), of the homozygous overexpression line 1 (B; OE1), and of the homozygous overexpression line 2 (C; OE2). The data represent means ± sd of five biological replicates.
DISCUSSION
BnSCE3 Expression Prevents the Accumulation of Sinapine in Seeds
During the past decade, comprehensive genetic and biochemical approaches unraveled the complex metabolic pathway for synthesis of the accumulating soluble phenolic compounds mainly derived from sinapate in Arabidopsis and Brassica (for review, see Milkowski and Strack, 2004). Identification of genes encoding enzymes involved in sinapine biosynthesis laid the foundation for transgenic strategies aimed at reducing this antinutritive phenolic metabolite in seeds of the valuable crop plant oilseed rape. This reduction is one of the major requirements for the efficient use of oilseed rape protein as an animal feed and human food supplement. Previous strategies to suppress sinapine biosynthesis in oilseed rape involved antisense or double-stranded RNA interference-mediated silencing of early metabolic steps, such as ferulate-5-hydroxylase (Nair et al., 2000), cinnamate-4-hydroxylase, 4-coumarate ester-3-hydroxylase, and caffeate O-methyltransferase (Bhinu et al., 2009). However, quantification of seed phenolics revealed that suppression of the upper part of the phenylpropanoid pathway was not effective in decreasing the sinapine content. Among the late steps of sinapine biosynthesis, silencing of UDP-Glc:sinapate glucosyltransferase (UGT84A9) turned out to be a useful strategy (Hüsken et al., 2005b). By this approach, sinapine was decreased by 72% and total sinapate ester content was decreased by 76% in T3 seeds, and there were no significant differences in important agronomic traits, such as oil, protein, or fatty acid content and composition. UGT84A9-suppressing plants have been propagated to the T6 generation with stable inheritance of the low-sinapine trait (Wolfram et al., 2010). Another suppression strategy aimed at silencing the gene encoding sinapoylglucose:choline sinapoyltransferase, the final enzyme in sinapine biosynthesis, led to a significant reduction in the level of sinapine. Unfortunately, the decrease in sinapine content was counterbalanced by an increase in the level of sinapoylglucose, the metabolic precursor of sinapine (Weier et al., 2008; Bhinu et al., 2009).
In this work, we describe a transgenic approach aimed at induced degradation as a novel strategy to reduce the sinapine content in seeds of oilseed rape. Based on the identification of BnSCE3 as a sinapine esterase, the expression of which is strongly induced upon seed germination (Clauss et al., 2008), we expressed the corresponding cDNA during seed development with the aim of synchronizing the biosynthesis and hydrolysis of sinapine. As expected from a previous “proof-of-concept” study in Arabidopsis (Clauss et al., 2008), this strategy led to a strong depletion of sinapine. Expression of BnSCE3 in seeds resulted in a decrease in sinapine content by 95%, thus providing the best-performing strategy reported so far to establish the “low-sinapine trait” in oilseed rape. Together with its dominant inheritance, the strong effect of BnSCE3 expression allowed a biochemical T2 segregation analysis with sinapine content as a chemical marker. This selection strategy for the transgenic lines avoids employing undesired herbicide resistance as a marker when generating low-sinapine lines of oilseed rape. In BnSCE3-expressing seeds, sinapine depletion was accompanied by a significant increase in the amount of free choline, liberated from sinapine upon hydrolysis. The high amount of choline could potentially contribute to an increased nutritive value of the transgenic seeds (Zeisel, 2000). On the other hand, it would be interesting to determine, whether the high amount of choline can be efficiently converted into betaine by the introduction of a gene encoding choline oxidase (Rozwadowski et al., 1991). So far, choline oxidase-mediated betaine production has been limited by low levels of free choline as a precursor, combined with choline homoeostasis in seeds (Nuccio et al., 1998; Huang et al., 2000, 2008). In future experiments, the hypothesis that the high-choline chemotype of seeds produced by BnSCE3 expression can overcome the substrate limitation in betaine production can be tested. Since betaine is proven to be protective against various abiotic stress factors, its accumulation in seeds could potentially increase plant vigor during seed germination and early seedling development under adverse environmental conditions.
BnSCE3 Expression Affects Seed Metabolism
This study describes comprehensive metabolite profiling of primary and secondary products in transgenic low-sinapine seeds of oilseed rape, providing insight into changes of the metabolic network (Fig. 10). With regard to secondary metabolism, the parallel examination of both the total methanolic seed extract and the choline ester-depleted seed extract allowed quantification of the predominating sinapine and related compounds as well as the analysis of minor phenolic compounds, which were frequently masked by the sinapine peak in conventional analyses (Böttcher et al., 2011). Besides the strong depletion of sinapine, the profiling approach identified significant changes in the amount of related phenolic compounds. These involve the appearance of glycosylated derivatives of sinapine like sinapoylcholine 4-O-glucoside, sinapoylcholine 4-O-dihexoside, and sinapoylcholine 4-O-hexoside-pentoside (Table I) or sinapate 4-O-glucoside (Tables I and II). This confirmed the general role of glycosylation as a means to avoid the accumulation of reactive compounds potentially interfering with essential pathways in plant metabolism. Likewise, the accumulation of phenolic glucosides was observed in transgenic poplar (Populus spp.) plants designed to produce modified lignin (Meyermans et al., 2000; Suzuki et al., 2010) as well as in Arabidopsis ugt84A2 mutants defective in the enzyme UDP-Glc:sinapate glucosyltransferase (Meissner et al., 2008). On the other hand, the appearance of glycosylated sinapoylcholine in OE1 and OE2 suggests that the BnSCE3 enzymatic activity was not able to act on the whole sinapine pool in the seeds of the transgenic oilseed rape lines described in this study.
Another striking feature of the altered secondary metabolism in BnSCE3-overexpressing seeds of oilseed rape was the appearance of sinapate conjugated to several other small molecules, including methyl moieties (33 and 44; Tables I and II), Cys (46 and 48; Table II), and kaempferol-3-O-sophoroside (37; Tables I and II). These metabolites revealed that various conjugation reactions contribute to the chemical conversion of sinapate that was transiently liberated from the sinapine pool by BnSCE3-catalyzed hydrolysis. Unlike the suppression of sinapine biosynthesis via double-stranded RNA interference-mediated silencing of UGT84A9 (Hüsken et al., 2005b), BnSCE3-overexpressing seeds did not show a dramatic depletion of the characteristic minor sinapate esters like other sugar esters or kaempferol glycoside esters (Baumert et al., 2005), thus pointing to a different mechanism of sinapine depletion. As with the UGT84A9 suppression approach, however, the molecular basis of sinapine reduction by BnSCE3-mediated cleavage remains unclear. So far, it might be possible that transiently liberated sinapate triggers a negative metabolic feedback effect that leads to down-regulation of sinapate biosynthesis (Milkowski and Strack, 2004).
Beyond secondary metabolism, the comprehensive profiling approach detected various alterations in primary metabolism in BnSCE3-overexpressing seeds (Table III; Fig. 10). This demonstrated the potential of the GC-MS technique to analyze the primary metabolism, as has been recently pointed out in a related article (Trenkamp et al., 2009). The affected compounds in BnSCE3-overexpressing seeds of oilseed rape were derived from several pathways, including glycolysis, the citric acid cycle, and the metabolism of amino acids and phospholipids, thereby indicating the global effects of BnSCE3 expression. Given the diversity of the metabolites with altered levels, it is difficult to attribute them to individual changes within the complex seed metabolic network, but possible mechanisms can be suggested for some of the alterations. For example, the increased amount of choline in the OE lines (Fig. 6) may lead to an increased level of CDP-choline, which could be combined with diacylglycerol to produce the increased level of PC found in the transgenic seeds. A larger pool of PC might increase the amount of fatty acyl chains removed from PC by acyl editing (Bates et al., 2009). This might feed into the pool of fatty acyl-CoA available for the formation of other phospholipids. Moreover, an increased metabolic pool of glycerol 3-phosphate, derived from the glycolytic pathway, could feed via CDP-diacylglycerol into biosynthesis of phospholipids. Electrospray ionization-tandem mass spectrometry (ESI-MS/MS) analysis of the polar lipids (membrane lipids) in the seeds showed that members of the PC class were the most abundant phospholipids. This was previously found to be the case in oilseed rape oil bodies (Tzen et al., 1993; Katavic et al., 2006). In agreement with the data of Tzen et al. (1993), PI was also identified by the analyses presented in this work as a predominant class of lipids in oil bodies.
The finding that about 50% of the detectable signals in seeds of oilseed rape were changed by overexpression of BnSCE3 demonstrated the need to study metabolism as a network instead of analyzing individual pathways. Especially the overexpression of genes encoding enzymes with low substrate specificity like BnSCE3 combined with a possible mistargeting in the host cells might impart a variety of unpredicted changes in plant metabolism. Multiple effects on plant metabolism have also been shown in a recent study with a sinapoylglucose:choline sinapoyltransferase mutant of Arabidopsis (Huang et al., 2009). It revealed profound changes in global large-scale gene expression, resulting in pleiotropic responses to environmental cues, illustrated by increased susceptibility to white mold (Sclerotinia sclerotiorum). This effect was correlated with reduced amounts of defense compounds like salicylic acid and glucosinolates, obviously caused by an unknown metabolic cross talk with sinapine biosynthesis. In contrast, the seed-specific overexpression of BnSCE3 described herein did not affect agronomically important traits like content and composition of oil, protein, and glucosinolates. This makes BnSCE3 overexpression a promising strategy to generate transgenic low-sinapine oilseed rape lines.
Given the high percentage of altered mass signals in BnSCE3-overexpressing seeds of oilseed rape, the results of our profiling studies showed again the need to implement accessory metabolite screening procedures into each transgenic approach designed to increase the value of crop plants. Since metabolite profiling has proven a powerful tool for biochemical phenotyping of plants, it seems to be in order for it to become a requirement to assess the “substantial equivalence” of transgenic cultivars to the wild-type plants (Chen et al., 2003).
“Low-Sinapine Trait” Affects Seedling Development
During early seedling development, the seed sinapate reserves, stored as sinapine and related esters, feed into the biosynthesis of sinapoylmalate via a metabolic pathway including SCE, UDP-Glc:sinapate glucosyltransferase, and sinapoylglucose:l-malate sinapoyltransferase. Sinapoylmalate accumulates in vacuoles of the epidermis (Strack et al., 1985) and contributes to the protection of epidermal guard cells and subepidermal tissues against the deleterious action of UV-B radiation. This led us to investigate how the lack of sinapine in the BnSCE3 overexpression lines OE1 and OE2 might affect the phenylpropanoid pattern in young seedlings. On the other hand, given the global character of metabolic alterations in seeds, it was necessary to evaluate the seed specificity of the transgenic approach to exclude interference with the plant metabolism during vegetative development, which potentially affects plant vigor and performance. Surprisingly, even in the absence of sinapine in the seeds, the transgenic lines OE1 and OE2 produced sinapoylmalate in the range of wild-type seedlings, distinguished only by a slight retardation in the accumulation kinetics (Fig. 13). Considering the observed decrease in the mass signal intensities of some of the newly accumulated sinapate esters from seeds of OE1 and OE2 within the first week of seedling development (Supplemental Fig. S2), it is conceivable that sinapate derived from degradation of these esters feeds into sinapoylmalate biosynthesis. Two conjugates of sinapine [sinapoylcholine 4-O-Glc and sinapoylcholine (4-O-8′)G 4-O′-Hex] and two of sinapate (sinapate 4-O-Glc and methylsinapate) that are metabolized during seed germination and development of young seedlings might be the major sinapoyl suppliers for sinapoylmalate formation. However, these novel sinapate derivatives do not compensate for the lack of major sinapate esters of the wild type in a quantitative way. This raises the question of whether de novo biosynthesis of sinapate via the shikimate/phenylpropanoid pathway started earlier in sinapine-depleted OE1 and OE2 lines than in the wild type. Time-course quantification of altered mass signals in OE lines revealed that the characteristic wild-type metabolism recovered within 16 d after the start of seedling development (Fig. 12). This gives rise to the assumption that seedling vigor was not affected by our transgenic approach, at least under greenhouse conditions. The slightly increased germination rate as well as the faster seedling growth during early developmental stages might be a consequence of the increased water content, probably caused by the high concentration of the hygroscopic choline in OE seeds (Table IV).
MATERIALS AND METHODS
Transgenic Plant Material and Growth Conditions
Seeds of summer oilseed rape (Brassica napus var napus ‘Lisora’) were obtained from the Norddeutsche Pflanzenzucht (http://norddeutschepflanzenzucht.de). For transformation and seed-specific expression of BnSCE3, the BnSCE3 cDNA (Clauss et al., 2008) was fused to the seed-specific promoter of the napin gene from oilseed rape and cloned into plasmid pBINPLUS (van Engelen et al., 1995). The expression cassette, consisting of the nap promoter, coding sequence, and nos transcription terminator, was transferred into Agrobacterium tumefaciens strain C58C1 (Koncz and Schell, 1986). The vector also contained the bar gene mediating resistance against the herbicide PPT and expressed from the cauliflower mosaic virus 35S promoter for selection of transformed plants. The BnSCE3 coding sequence was PCR amplified from cloned cDNA using primers BNSCEnap_F (5′-ACTGCCCGGGGAAAATGGCTTCTTCACTAAAGAAGCTC-3′) and BNSCEnap_R (5′-GCAGGATCCTCAACTACCGAAAGAAGATTCGTTATC-3′), designed to introduce an upstream SmaI restriction site and a downstream BamHI site (underlined) into the PCR product. PCR was performed using Platinum PCR SuperMix High Fidelity (Invitrogen). The coding sequence was inserted as a SmaI-BamHI fragment into vector pLH7000. The stem disc transformation of oilseed rapewas performed by Saaten-Union BIOTEC according to the protocol described by De Block et al. (1989). The wild-type and transgenic seeds were germinated under greenhouse conditions at 15°C to 18°C on soil GS 90 (Patzer) in closed plastic boxes under a 16-h-light regimen. After germination, the plastic cap was removed and plants were grown as described earlier (Clauss et al., 2008).
Segregation Analysis by PPT Resistance
After transformation and regeneration, PPT-resistant plants of the first transgenic generation (T1) were transferred to the greenhouse and selfed to produce seeds of the second transgenic generation (T2 seeds). To identify transgenic lines with single-copy transgene insertions, the inheritance of PPT resistance was characterized within the segregating T2 seedlings. Seedlings were selected for PPT resistance by spraying with a commercially available herbicide solution (BASTA; 0.04 mg mL−1) several times starting at 4 d after sowing. The inheritance of PPT resistance with a ratio of 3:1 (resistant:nonresistant) was indicative of single-copy transgene insertion. Selected T2 plants were propagated and selfed in the greenhouse to produce T3 seeds. To isolate homozygous lines, inheritance of PPT resistance was analyzed within T3 seedlings. T2 plants producing exclusively PPT-resistant T3 descendents were considered homozygous (Hüsken et al., 2005a).
BnSCE3 Transcript Analysis
Extraction of total RNA from oilseed rape wild-type and BnSCE3-overexpressing seeds was performed by selective adsorption onto a silica matrix using a kit for plant RNA purification (Qiagen; http://www.qiagen.com/). RT reactions were performed with 1 μg of total oilseed rape RNA using the Omniscript RT kit (Qiagen) according to the manufacturer’s protocol. Fifty-fold dilutions of cDNA samples were used for each PCR amplification performed with Platinum PCR SuperMix High Fidelity (Invitrogen; http://www.invitrogen.com/) and primer pair LIP2RT_F (5′-CAGTTGGATGTAGCGCCGCGTATC-3′) and LIP2RT_R (5′-CTTCTGGTGTGCGGCTTCGGTTATATG-3′) for amplification of BnSCE3. Control experiments were done using a primer pair recognizing the oilseed rape sequence encoding ubiquitin (UbiRT_F, 5′-AAGCCAAGATCCAGGACAAAG-3′; UbiRT_R, 5′-CGAGCCAAAGCCATCAAAGAC-3′). The RT-PCR protocol consisted of 25 to 35 cycles, depending on the linear range of PCR amplification for the individual genes. Each cycle included denaturation at 94°C for 15 s, annealing (55°C, 30 s), and elongation (72°C, 42 s). A final incubation step at 72°C for 10 min allowed trimming of incomplete polymerization products. PCR mixtures were separated by electrophoresis on ethidium bromide-stained agarose gels. PCR products were visualized by fluorescence under UV light in a transilluminator.
UPLC-Diode Array Detection Analysis of Sinapate Esters
For UPLC-diode array detection analysis, seeds (20 mg) or cotyledons (from 16 mg at day 4 to 466 mg at day 16) were ground to a fine powder with beads (3 mm diameter; Retsch) using a cryo mill (MM 400; Retsch) and extracted twice with 300 μL of 80% aqueous methanol by incubation in an ultrasonic bath for 5 min. Centrifugation was performed after vortexing at 11,000g for 5 min at room temperature, and the supernatants were combined. Metabolites were analyzed by LC using an ACQUITY UPLC system (Waters) equipped with binary solvent manager, sample manager, and photodiode array detector. Eluent A was 0.1% aqueous trifluoroacetic acid and eluent B was 98% aqueous acetonitrile with 0.1% trifluoroacetic acid. Methanolic extracts (3 μL) were injected onto a BEH C18 column (2 mm × 50 mm, 1.7 μm; Waters), and metabolites were eluted with 5% eluent B for 0.5 min, 27% eluent B for 1.3 min, followed by a linear gradient to 95% eluent B for 0.2 min using a flow rate of 500 μL min−1. Compounds were photometrically detected at 330 nm. Identification and external quantification of metabolites were performed using authentic external standards. Data were collected and processed using the Empower Software System, version 2 (Waters). Analysis of total sinapate ester content and saponification were performed as described by Wolfram et al. (2010). In short, methanolic seed extracts were treated with 10 n KOH. The solution was neutralized with aqueous acetic acid, centrifuged, and the supernatant subjected to HPLC analysis. The amount of “residual sinapate esters” (Fig. 5) was calculated by subtracting the amount of sinapine from total sinapate ester content.
Analysis of Choline
Quantification of choline was performed using the Choline/Acetylcholine Quantification Kit (BioCat) according to the manufacturer’s instructions. For each plant sample, 20 seeds were ground to a fine powder in a mortar with liquid nitrogen. One hundred microliters of the provided choline assay buffer was added to 3 mg of the seed powder. The sample was mixed, homogenized for 10 min in an ultrasonic bath, and centrifuged at 11,000g for 2 min. Ten microliters of wild-type extracts or 1 μL of BnSCE3 overexpression extracts was added to the reaction mixture. After 30 min of incubation at room temperature (light protected), the fluorescence of the samples was measured at excitation/emission 535/590 nm in a CytoFluor II Microwell Fluorescence Reader (PerSeptive Biosystems). A standard curve (authentic choline) was used to calculate the amount of choline in the seed extracts.
Analysis of Seed Morphological Properties
To compare the morphological/physiological properties of oilseed rape wild-type and transgenic BnSCE3-overexpressing T3 seeds, single seed size (maximal diameter; mm), single seed weight (mg), moisture content (%), and hypocotyl length of 6-d-old seedlings were determined. For analysis of the moisture content, a seed pool of 3 × 50 seeds per sample was heated to 80°C for 48 h. The seed samples were weighed before and after the heating process to detect the loss of water. Intact and half-section seeds were fixed on a dish and investigated with an environmental scanning electron microscope (Philips ESEM XL 30 FEG; Philips Electron Optics) in wet mode (chamber pressure, 1.2 mbar; voltage acceleration, 12 kV) by detecting secondary electrons. In this study, four wild-type seeds and six transgenic seeds of two independent BnSCE3 overexpression lines (OE1 and OE2) were analyzed with help from Frank Syrowatka (Center of Material Sciences, Martin-Luther-University Halle-Wittenberg).
Near-Infrared Spectroscopy of Seed Traits
Intact seeds were scanned in a NIR Systems model 6500 spectrophotometer (FOSS-NIRSystems) to perform near-infrared spectroscopy analyses. Spectra were recorded in reflectance mode, equipped with a transport module, acquiring their spectra at 2-nm intervals over a wavelength range of 400 to 2,500 nm. In this mode, a ceramic standard is placed in the radiant beam and the diffusely reflected energy is measured at each wavelength before and after reading the sample. Spectra of the samples were recorded four times from each of the four biological replicates. The spectra were obtained as an average of 32 scans. The ceramic and the sample spectra were used to generate the final log (1/R) spectrum. Seed samples were placed for the analysis in a 3-cm-diameter round cell holder (10-mL volume). This cell is composed of quartz glass and anodized aluminum to avoid absorption. The calibration process and validation of the samples were performed as described elsewhere (Font et al., 2005).
Extraction of Single Seeds for LC-MS Profiling
The weights of 30 wild-type seeds and seeds of three transgenic lines (OE1, OE2, and OE3; 60 each) were determined ranging from 1.6 to 8.1 mg. Together with two steel balls (2 mm diameter), the seeds were individually placed in 1.5-mL tubes (Eppendorf) and cooled down in liquid nitrogen. Using a ball mill, the material was ground for 2 min at 30 Hz without defrosting (Retsch). After addition of 50 μL of precooled (−20°C) solvent solution (50% [v/v] aqueous methanol, containing 2 μm o-anisic acid as internal standard), the samples were immediately vortexed for 15 s, sonicated for 10 min at room temperature, and centrifuged for 10 min at 19,000g. All solvents used for extraction and the LC-MS gradients were of LC-MS quality (Fluka). The supernatants were transferred to new tubes, and the remaining pellet was subjected to a second extraction using 50 μL of 80% (v/v) aqueous methanol. The combined extracts were diluted with water by a factor of 3. In order to be able to measure the phenolic choline esters in a linear range, an aliquot of 50 μL was taken and again diluted by the same procedure before being filtered using 0.2-mm polytetrafluoroethylene membrane devices (Whatman). This was defined as the total extract fraction. The rest of the diluted combined extracts were subjected to solid-phase extraction for depletion of choline conjugates using an adapted protocol of Böttcher et al. (2011). In short, 30-mg Strata-X-CW33 columns (Phenomenex) were solvated with 1 mL of methanol and equilibrated with the same amount of water. After sample loading, the columns were washed using 300 μL of 25 mm ammonium acetate and 300 μL of methanol. The flow-through and the wash volume were combined and evaporated to near dryness at reduced pressure. They were reconstituted in 50 μL of 10% aqueous methanol, sonicated for 10 min, and centrifuged using the above-mentioned parameters and defined as the choline ester-depleted fraction. Calculating the various dilution and concentration steps of the different fractions at the end, the choline ester-depleted fraction showed an approximately 15 times higher concentration factor in comparison with the total extract fraction.
Extraction of Developing Seedlings for LC-MS Profiling
In order to compare seeds and seedlings with different water contents, the ratio between fresh weight and dry weight had to be determined for each sample class. Pools of seeds and seedlings for 0, 4, 8, 12, 16, and 20 d after germination were freeze dried and the fresh weight-dry weight ratios were calculated. To 25-mg-aliquot samples, 250 μL of precooled (–20°C) solvent solution (50% [v/v] aqueous methanol, containing 2 μm o-anisic acid as internal standard) was added and the sample was immediately vortexed for 15 s, sonicated for 10 min at room temperature, and centrifuged for 10 min at 19,000g. The supernatants were transferred to new tubes, and the remaining pellet was subjected to a second extraction using 250 μL of 80% (v/v) aqueous methanol. The combined extracts were diluted with water by a factor of 3. An aliquot of 250 μL was taken and again diluted by the same procedure before filtration. The rest of the diluted combined extracts were subjected to solid-phase extraction as stated above. Three technical replicates for each developing time point and oilseed rape seed line were included in the experimental setup.
UPLC/ESI-Quadrupole Time-of-Flight-MS
Chromatographic separations were performed on an Acquity UPLC system (Waters) equipped with a modified C18 column (HSS T3; 1.0 × 100 mm, particle size of 1.8 μm; Waters) applying the following binary gradient at a flow rate of 150 μL min−1: 0 to 1 min, linear from 95% A (water and 0.1% aqueous formic acid) to 85% A; 1 to 7 min, linear from 85% A to 55% A; 7 to 7.5 min, linear from 55% A to 45% A; 7.5 to 8 min, linear from 45% A to 5% A; 8 to 11 min, isocratic 95% B (acetonitrile and 0.1% aqueous formic acid); 11.5 to 14 min, isocratic 95% A. The injection volume was 3.1 μL (full-loop injection). Eluted compounds were detected from mass-to-charge ratio (m/z) 100 to 1,000 by a MicrOTOF-Q hybrid quadrupole time-of-flight (QTOF) mass spectrometer (Bruker Daltonics) equipped with an Apollo II electrospray ion source in positive ion mode using the following instrument settings: nebulizer gas, nitrogen, 1.6 bar; dry gas, nitrogen, 6 L min−1, 190°C; capillary, 25,500 V (+4,000 V); end plate offset, 2,500 V; funnel 1 radio frequency (RF), 200 V; funnel 2 RF, 200 V; in-source collision-induced dissociation energy, 0 V; hexapole RF, 100 V; quadrupole ion energy, 5 eV; collision gas, argon; collision energy, 10 eV; collision RF, 200/400 V (timing 50/50); transfer time, 70 ms; prepulse storage, 5 ms; pulser frequency, 10 kHz; spectra rate, 3 Hz. Internal mass calibration of each analysis was performed by infusion of 20 μL min−1 10 mm lithium formate in isopropanol:water (1:1, v/v) at a gradient time of 12 min using a diverter valve. For the acquisition of collision-induced fragmentation spectra, ions were isolated with a width of ±3 D. Collision energy was chosen in the range of 10 to 30 eV according to the substance in question. Prior to automatic data processing, the quality of sample preparation, chromatographic separation, and mass spectrometric detection was checked by determination of the reproducibility of retention times and responses of internal standards. In addition, in about every 20th place in the sample batch, a standard mixture was included for performance control of the LC-MS platform (for composition of the standard mixture, see Supplemental Table S2).
Data Deconvolution and Bioinformatical Analysis of Single Seed Analysis
For biomarker screening, the relative amount of sinapine was determined using the extracted ion current of the quantifier ion m/z 251.09 (sinapine fragment C13H15O5+). Thirty raw data files for each sample class were converted to mz Data format using the vendor-specific CompassXPort (http://www.bdal.de) and processed using the XCMS package (http://metlin.scripps.edu/; http://www.bioconductor.org) The raw data files were arranged in three separate sample classes (wild type, OE-1, and OE-2). Chromatographic peak detection was performed using the centWave algorithm (Tautenhahn et al., 2008), applying the following parameter settings: sntresh = 3, prefilter = c(3,100), ppm = 25, and peak width = c(5,12). LC-MS data sets were aligned in a first step with retention time correction using the XCMS group function (minfrac = 0.7, bw = 2.5 [estimated from maximum retention time shifts], mzwid = 0.05) and retcor (plottype = “mdevden”, span = 1, missing = 0, extra = 0). The parameters for the second alignment step were minfrac = 0.9, bw = 2, mzwid = 0.05. After filling in missing features using the fillPeaks routine of XCMS, a data matrix was exported into Microsoft Excel. Each m/z-retention time feature was normalized using the seed weight of the respective sample. Median intensity and the presence of each feature in each sample class were determined. A feature was called present within a sample class if it was detected in 90% of the sample files. For pairwise comparisons of sample classes, fold changes and P values (Student’s t test, two sided) were calculated. A feature was assumed as significantly induced or reduced in a sample class if its presence was 90% or greater, P < 0.05, the fold change was greater than a factor of 2 or less than 0.5, respectively, and it showed the same parameter behavior in both transgenic OE lines in relation to the wild-type line. The R-based functions heatmap2 and PCA were used to calculate heat maps and PCA plots (methods: dist, hclust, svdPCA).
Data Deconvolution and Bioinformatical Analysis of Seedlings
Chromatographic separation and MS analysis were performed as described above. Raw data files were converted to mz Data format and processed using the XCMS package. The raw data files were arranged in two separate sample classes (wild type and OE line). The data analysis of the two OE lines was performed independently. Chromatographic peak detection was performed essentially using the same parameters stated above with two exceptions: a higher prefilter (of 3,500) to reduce the amount of low-intensity peaks and a lower minfrac of 0.15, reflecting the six time points per group. Missing features were filled in the exported data matrix with random numerical values between 1 and 10 for calculating P values. For the determination of average area numbers for the three technical replicates, missing features were filled with the numerical value of 1. A feature was called present within a sample class if it was detected in 100% of the sample files. For pairwise comparisons between wild-type and OE lines, average intensity and P values (Student’s t test, two sided) were determined. The area values of the m/z-retention time pairs were normalized using o-anisic acid as internal standard. In addition, numbers for the dry weight per individual (seed or germinated seedling) ratio were used for a second round of normalization. An m/z-retention time feature was assumed as significantly induced or reduced in a sample class if the fold change was greater than or equal to a factor of 2 or less than or equal to 0.5, respectively, and P ≤ 0.05. Based on these calculations, the total amount of altered mass signals per individual with respect to the total amount of aligned mass signals per time point (%) was determined for each OE line.
GC-MS of Polar Primary Compounds and Network Visualization
After homogenization of 20 seeds per sample in liquid nitrogen, the wild-type samples (average weight of 74.87 mg) were extracted with 360 μL of precooled (−20°C) extraction solution (absolute methanol, 10% of a methylnonadecanoate stock solution [2 mg mL−1 methanol]; Sigma-Aldrich) and 10% of a 13C6-sorbitol stock solution (0.2 mg mL−1 methanol; Sigma-Aldrich). The ratio of wild-type or genetically modified seed fresh weight over extraction volume was kept constant at 1:4.8 (w/v). The preparation of the polar metabolite fraction, the subsequent chemical derivatization (i.e. subsequent methoxyamination and trimethylsilylation), and the instrumental GC-TOF-MS metabolite profiling analyses were performed essentially as described previously (Erban et al., 2007) using a 30-m length, 0.25-mm i.d., and 0.25-μm film thickness capillary column from Varian (VF-5ms). Peak heights of mass (m/z) fragments were normalized using the amount of the sample fresh weight and 13C6-sorbitol for the internal standardization of variations in sample amount and volume errors. Automated standardized chromatography data preprocessing and annotation of peak identity were manually supervised using the TagFinder software (Luedemann et al., 2008) and the reference mass spectra and retention time index collection of the Golm Metabolome Database (Hummel et al., 2010). Metabolites were identified using the chemical abstracts system identifiers and compound codes issued by the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2004). Significant differences of relative metabolite pool size changes were determined by ANOVA and Tukey’s honestly significant difference test. Independent component analysis with automated missing value replacement (Scholz et al., 2004) was applied to the metabolite profile compiled in Table IV. Logarithmic transformation of response ratios approximated prerequisite Gaussian normal distribution of metabolite profiling data for statistical analyses. The data are represented in Table III and visualized in the context of biological networks with the VANTED software (Junker et al., 2006), adding the pathways of secondary metabolites (Fig. 7).
ESI-MS/MS Profiling of Seed Oil (Nonpolar Lipids)
An automated ESI-MS/MS approach was used for seed oil analysis. The samples were dissolved in 1 mL of chloroform. An aliquot of 10 μL of extract in chloroform was used. Precise amounts of internal standards (Avanti Polar Lipids) were added in the following quantities: 3.1 nmol of tri17:1 TAG and 4.6 nmol of di15:0 DAG. The sample and internal standard mixture were combined with solvents, such that the ratio of chloroform:methanol:300 mm ammonium acetate in water was 300:665:35, and the final volume was 1.4 mL.
Unfractionated lipid extracts were introduced by continuous infusion into the ESI source on a triple quadrupole MS apparatus (API4000; Applied Biosystems). Samples were introduced using an autosampler (LC Mini PAL; CTC Analytics) fitted with the required injection loop for the acquisition time and presented to the ESI needle at 30 μL min−1. TAG and DAG were detected by a series of neutral loss scans that detected TAG and DAG species as [M + NH4]+ ions. The scans targeted losses of various fatty acids as neutral ammoniated fragments: NL 285.2 (17:1, for the TAG internal standard); NL 273.2 (16:0); NL 301.2 (18:0); NL 299.2 (18:1); NL 297.2 (18:2); NL 295.2 (18:3); NL 355.3 (22:1). The scan speed was 100 units s−1. The collision energy, with nitrogen in the collision cell, was +20 V, declustering potential was +100 V, entrance potential was +14 V, and exit potential was +14 V. Sixty continuum scans were averaged in multiple channel analyzer mode.
For all analyses, the collision gas pressure was set on “low” and the mass analyzers were adjusted to a resolution of 0.7 units full width at half height. The source temperature (heated nebulizer) was 100°C, the interface heater was on, +5.5 kV was applied to the electrospray capillary, the curtain gas was set at 20 (arbitrary units), and the two ion source gases were set at 45 (arbitrary units). For all analyses, the background of each spectrum was subtracted, the data were smoothed, and peak areas were integrated using a custom script and Applied Biosystems Analyst software.
For TAG and DAG analysis, data from the neutral loss scans were corrected for overlap of isotopic variants (A + two peaks). Because there is variation in ionization efficiency among acyl glycerol species with different fatty acyl groups (Han and Gross, 2001), the ratio of the MS response of the TAG and DAG species to the MS response of the TAG or DAG internal standard does not provide a value that is directly proportional to the TAG or DAG content of each species. TAG and DAG amounts are thus expressed as relative mass spectral signal mg−1 dry weight seed−1, where a signal of 1.0 represents a signal equal to the signal of 1 nmol of tri17:1 TAG (the internal standard).
ESI-MS/MS Profiling of Polar Lipid
An automated ESI-MS/MS approach was used, and data acquisition and analysis and acyl group identification were carried out as described previously (Devaiah et al., 2006) with modifications. The samples were dissolved in 1 mL of chloroform. An aliquot of 6 μL of extract in chloroform was used. Precise amounts of internal standards, obtained and quantified as described previously (Welti et al., 2002), were added in the following quantities (with some small variation in amounts in different batches of internal standards): 0.6 nmol of di12:0-PC, 0.6 nmol of di24:1-PC, 0.6 nmol of 13:0-lysoPC, 0.6 nmol of 19:0-lysoPC, 0.3 nmol of di12:0-PE, 0.3 nmol of di23:0-PE, 0.3 nmol of 14:0-lysoPE, 0.3 nmol of 18:0-lysoPE, 0.3 nmol of di14:0-PG, 0.3 nmol of di20:0(phytanoyl)-PG, 0.3 nmol of 14:0-lysoPG, 0.3 nmol of 18:0-lysoPG, 0.3 nmol of di14:0-PA, 0.3 nmol of di20:0(phytanoyl)-PA, 0.2 nmol of di14:0-PS, 0.2 nmol of di20:0(phytanoyl)-PS, 0.23 nmol of 16:0 to 18:0-PI, 0.16 nmol of di18:0-PI, 2.01 nmol of 16:0 to 18:0-MGDG, 0.39 nmol of di18:0-MGDG, 0.49 nmol of 16:0 to 18:0-DGDG, and 0.71 nmol of di18:0-DGDG. The sample and internal standard mixture was combined with solvents, such that the ratio of chloroform:methanol:300 mm ammonium acetate in water was 300:665:35, and the final volume was 1.4 mL.
Unfractionated lipid extracts were introduced by continuous infusion into the ESI source on a triple quadrupole MS/MS device (API 4000; Applied Biosystems). Samples were introduced using an autosampler (LC Mini PAL; CTC Analytics) fitted with the required injection loop for the acquisition time and presented to the ESI needle at 30 μL min−1.
Sequential precursor and neutral loss scans of the extracts produce a series of spectra with each spectrum revealing a set of lipid species containing a common head group fragment. Lipid species were detected with the following scans: PC and lysoPC, [M + H]+ ions in positive ion mode with precursor of 184.1 (Pre 184.1); PE and lysoPE, [M + H]+ ions in positive ion mode with neutral loss of 141.0 (NL 141.0); PG, [M + NH4]+ in positive ion mode with NL 189.0 for PG; lysoPG, [M – H]− in negative mode with Pre 152.9; PI, [M + NH4]+ in positive ion mode with NL 277.0; PS, [M + H]+ in positive ion mode with NL 185.0; PA, [M + NH4]+ in positive ion mode with NL 115.0; MGDG, [M + NH4]+ in positive ion mode with NL179.1; and DGDG, [M + NH4]+ in positive ion mode with NL 341.1. The scan speed was 50 or 100 units s−1. The collision gas pressure was set at 2 (arbitrary units). The collision energies, with nitrogen in the collision cell, were +28 V for PE, +40 V for PC, +25 V for PI, PS, and PA, +20 V for PG, +21 V for MGDG, and +24 V for DGDG. Declustering potentials were +100 V for PE, PC, PA, PG, PI, and PS and +90 V for MGDG and DGDG. Entrance potentials were +15 V for PE, +14 V for PC, PI, PA, PG, and PS, and +10 V for MGDG and DGDG. Exit potentials were +11 V for PE, +14 V for PC, PI, PA, PG, and PS, and +23 V for MGDG and DGDG. The mass analyzers were adjusted to a resolution of 0.7 units full width at half height. For each spectrum, nine to 150 continuum scans were averaged in multiple channel analyzer mode. The source temperature (heated nebulizer) was 100°C, the interface heater was on, +5.5 kV or −4.5 kV was applied to the electrospray capillary, the curtain gas was set at 20 (arbitrary units), and the two ion source gases were set at 45 (arbitrary units).
The background of each spectrum was subtracted, the data were smoothed, and peak areas were integrated using a custom script and Applied Biosystems Analyst software. The lipids in each class were quantified in comparison with the two internal standards of that class. The first and typically every 11th set of mass spectra were acquired on the internal standard mixture only. Peaks corresponding to the target lipids in these spectra were identified, and molar amounts were calculated in comparison with the internal standards on the same lipid class. To correct for chemical or instrumental noise in the samples, the molar amount of each lipid metabolite detected in the “internal standards-only” spectra was subtracted from the molar amount of each metabolite calculated in each set of sample spectra. The data from each internal standards-only set of spectra were used to correct the data from the following 10 samples. Finally, the data were corrected for the fraction of the sample analyzed and normalized to the sample “dry weights” to produce data in units of nmol mg−1.
GC of Total Fatty Acids
A 15:0 internal standard was added to samples. Total fatty acids were converted to fatty acid methyl esters by incubation of extracts in 3 m methanolic hydrochloric acid at 78°C for 30 min followed by addition of water. Samples were extracted with pentane and analyzed by GC using a Supelco SP-2380 GC capillary column (30 m × 0.25 mm i.d., 0.20 μm; no. 24110-U; Sigma-Aldrich). Injection temperature was 120°C, and the oven temperature was 120°C for 2 min, then increased by 4°C min−1 to 230°C, at which it was held for 2 min. Detection was by flame ionization at 260°C. Fatty acids were identified by comparison of retention times with standard compounds and quantified in comparison with the internal standard.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY870270.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PCA and HCA of LC-MS profiling data.
Supplemental Figure S2. Decrease of selected sinapate conjugates in developing oilseed rape seedlings.
Supplemental Table S1. MS data of annotated secondary compounds.
Supplemental Table S2. Standard mixture for LC-MS performance control.
Supplementary Material
Acknowledgments
Oilseed rape seeds were kindly provided by the Norddeutsche Pflanzenzucht.
References
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43: 534–552 [DOI] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M. (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert A, Milkowski C, Schmidt J, Nimtz M, Wray V, Strack D. (2005) Formation of a complex pattern of sinapate esters in Brassica napus seeds, catalyzed by enzymes of a serine carboxypeptidase-like acyltransferase family? Phytochemistry 66: 1334–1345 [DOI] [PubMed] [Google Scholar]
- Benhassaine-Kesri G, Aid F, Demandre C, Kader JC, Mazliak P. (2002) Drought stress affects chloroplast lipid metabolism in rape (Brassica napus) leaves. Physiol Plant 115: 221–227 [DOI] [PubMed] [Google Scholar]
- Bhinu VS, Schäfer UA, Li R, Huang J, Hannoufa A. (2009) Targeted modulation of sinapine biosynthesis pathway for seed quality improvement in Brassica napus. Transgenic Res 18: 31–44 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Roepenack-Lahaye E, Scheel D. (2011) Resources for metabolomics. Bancroft I, Schmidt R, , Genetics and Genomics of the Brassicaceae. Springer-Verlag, Berlin, pp 469–503 [Google Scholar]
- Böttcher C, Westphal L, Schmotz C, Prade E, Scheel D, Glawischnig E. (2009) The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchereau AJ, Hamelin J, Renard M, Larher F. (1992) Structural changes in sinapic acid conjugates during development of rape. Plant Physiol Biochem 30: 467–475 [Google Scholar]
- Chen F, Duran AL, Blount JW, Sumner LW, Dixon RA. (2003) Profiling phenolic metabolites in transgenic alfalfa modified in lignin biosynthesis. Phytochemistry 64: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Clauss K, Baumert A, Nimtz M, Milkowski C, Strack D. (2008) Role of a GDSL lipase-like protein as sinapine esterase in Brassicaceae. Plant J 53: 802–813 [DOI] [PubMed] [Google Scholar]
- De Block M, De Brouwer D, Tenning P. (1989) Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the bar and neo genes in the transgenic plants. Plant Physiol 91: 694–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. (2006) Quantitative profiling of polar glycerolipid species and the role of phospholipase Dα1 in defining the lipid species in Arabidopsis tissues. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Erban A, Schauer N, Fernie AR, Kopka J. (2007) Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography-mass spectrometry metabolite profiles. Methods Mol Biol 358: 19–38 [DOI] [PubMed] [Google Scholar]
- Font R, Wittkop B, Badani AG, del Rio-Celestino M, Friedt W, Lühs W, de Haro-Bailón A. (2005) The measurements of acid detergent fibre in rapeseed by visible and near-infrared spectroscopy. Plant Breed 124: 410–412 [Google Scholar]
- Gadamer J. (1897) Über die Bestandteile des schwarzen und weissen Senfsamens. Arch Pharm 235: 44–114 [Google Scholar]
- Han X, Gross RW. (2001) Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem 295: 88–100 [DOI] [PubMed] [Google Scholar]
- Huang J, Bhinu VS, Li X, Dallal Bashi Z, Zhou R, Hannoufa A. (2009) Pleiotropic changes in Arabidopsis f5h and sct mutants revealed by large-scale gene expression and metabolite analysis. Planta 230: 1057–1069 [DOI] [PubMed] [Google Scholar]
- Huang J, Hirji R, Adam L, Rozwadowski KL, Hammerlindl JK, Keller WA, Selvaraj G. (2000) Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: metabolic limitations. Plant Physiol 122: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Rozwadowski K, Bhinu VS, Schäfer U, Hannoufa A. (2008) Manipulation of sinapine, choline and betaine accumulation in Arabidopsis seed: towards improving the nutritional value of the meal and enhancing the seedling performance under environmental stresses in oilseed crops. Plant Physiol Biochem 46: 647–654 [DOI] [PubMed] [Google Scholar]
- Hummel J, Strehmel N, Selbig J, Walther D, Kopka J. (2010) Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 6: 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Chapple C. (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5: 224–229 [DOI] [PubMed] [Google Scholar]
- Hüsken A, Baumert A, Milkowski C, Becker HC, Strack D, Möllers C. (2005a) Resveratrol glucoside (Piceid) synthesis in seeds of transgenic oilseed rape (Brassica napus L.). Theor Appl Genet 111: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Hüsken A, Baumert A, Strack D, Becker HC, Möllers C, Milkowski C. (2005b) Reduction of sinapate ester content in transgenic oilseed rape (Brassica napus) by dsRNAi-based suppression of BnSGT1 gene expression. Mol Breed 16: 127–138 [Google Scholar]
- Ismail F, Vaisey-Genser M, Fyfe B. (1981) Bitterness and astringency of sinapine and its components. J Food Sci 46: 1241–1244 [Google Scholar]
- Junker BH, Klukas C, Schreiber F. (2006) VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Agrawal GK, Hajduch M, Harris SL, Thelen JJ. (2006) Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6: 4586–4598 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Ling H, Zuo K, Zhao J, Qin J, Qiu C, Sun X, Tang K. (2006) Isolation and characterization of a homologous to lipase gene from Brassica napus. Russ J Plant Physiol 53: 366–372 [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Meissner D, Albert A, Böttcher C, Strack D, Milkowski C. (2008) The role of UDP-glucose:hydroxycinnamate glucosyltransferases in phenylpropanoid metabolism and the response to UV-B radiation in Arabidopsis thaliana. Planta 228: 663–674 [DOI] [PubMed] [Google Scholar]
- Meyermans H, Morreel K, Lapierre C, Pollet B, De Bruyn A, Busson R, Herdewijn P, Devreese B, Van Beeumen J, Marita JM, et al. (2000) Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Milkowski C, Baumert A, Schmidt D, Nehlin L, Strack D. (2004) Molecular regulation of sinapate ester metabolism in Brassica napus: expression of genes, properties of the encoded proteins and correlation of enzyme activities with metabolite accumulation. Plant J 38: 80–92 [DOI] [PubMed] [Google Scholar]
- Milkowski C, Strack D. (2004) Serine carboxypeptidase-like acyltransferases. Phytochemistry 65: 517–524 [DOI] [PubMed] [Google Scholar]
- Nair RB, Joy RW, Kurylo E, Shi X, Schnaider J, Datla RS, Keller WA, Selvaraj G. (2000) Identification of a CYP84 family of cytochrome P450-dependent mono-oxygenase genes in Brassica napus and perturbation of their expression for engineering sinapine reduction in the seeds. Plant Physiol 123: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio ML, Russell BL, Nolte KD, Rathinasabapathi B, Gage DA, Hanson AD. (1998) The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J 16: 487–496 [DOI] [PubMed] [Google Scholar]
- Nurmann G, Strack D. (1979) Sinapine esterase. Part I. Characterization of sinapine esterase from cotyledons of Raphanus sativus. Z Naturforsch C 34c: 715–720 [Google Scholar]
- Nurmann G, Strack D. (1981) Formation of 1-sinapoylglucose by UDP-glucose:sinapic acid glucosyltransferase from cotyledons of Raphanus sativus. Z Pflanzenphysiol 102: 11–17 [Google Scholar]
- Rozwadowski KL, Khachatourians GG, Selvaraj G. (1991) Choline oxidase, a catabolic enzyme in Arthrobacter pascens, facilitates adaptation to osmotic stress in Escherichia coli. J Bacteriol 173: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C, Falcocchio S, Xoxi E, Pastor FIJ, Diaz P, Saso L. (2004) Activation and inhibition of Candida rugosa and Bacillus-related lipases by saturated fatty acids, evaluated by a new colorimetric microassay. Biochim Biophys Acta 1672: 184–191 [DOI] [PubMed] [Google Scholar]
- Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J. (2004) Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics 20: 2447–2454 [DOI] [PubMed] [Google Scholar]
- Strack D. (1981) Sinapine as a supply of choline for the biosynthesis of phophatidylcholine in Raphanus sativus seedlings. Z Naturforsch C 36c: 215–221 [Google Scholar]
- Strack D, Knogge W, Dahlbender B. (1983) Enzymatic synthesis of sinapine from 1-O-sinapoyl-β-d-glucose and choline by a cell-free system from developing seeds of red radish (Raphanus sativus L. var. sativus). Z Naturforsch C 38c: 21–27 [Google Scholar]
- Strack D, Nurmann G, Sachs G. (1980) Sinapine esterase. Part II. Specificity and change of sinapine esterase activity during germination of Raphanus sativus. Z Naturforsch C 35c: 963–966 [Google Scholar]
- Strack D, Pieroth M, Scharf H, Sharma V. (1985) Tissue distribution of phenylpropanoid metabolism in cotyledons of Raphanus sativus L. Planta 164: 507–511 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Sakakibara N, Li L, Umezawa T, Chiang VL. (2010) Profiling of phenylpropanoid monomers in developing xylem tissues of transgenic aspen (Populus tremuloides). J Wood Sci 56: 71–76 [Google Scholar]
- Tautenhahn R, Böttcher C, Neumann S. (2008) Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkotz N, Strack D. (1981) Enzymatic synthesis of sinapoyl-l-malate from 1-sinapoylglucose and l-malate by a protein preparation from Raphanus sativus cotyledons. Z Naturforsch C 35c: 835–837 [Google Scholar]
- Trenkamp S, Eckes P, Busch M, Fernie AR. (2009) Temporally resolved GC-MS-based metabolic profiling of herbicide treated plants reveals that changes in polar primary metabolites alone can distinguish herbicides of differing mode of action. Metabolomics 5: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen JTC, Cao Y, Laurent P, Ratnayake C, Huang A. (1993) Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol 101: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Wang SX, Ellis BE. (1998) Enzymology of UDP-glucose:sinapic acid glucosyltransferase from Brassica napus. Phytochemistry 49: 307–318 [Google Scholar]
- Weier D, Mittasch J, Strack D, Milkowski C. (2008) The genes BnSCT1 and BnSCT2 from Brassica napus encoding the final enzyme of sinapine biosynthesis: molecular characterization and suppression. Planta 227: 375–385 [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. (2002) Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Weng JK, Akiyama T, Bonawitz ND, Li X, Ralph J, Chapple C. (2010) Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram K, Schmidt J, Wray V, Milkowski C, Schliemann W, Strack D. (2010) Profiling of phenylpropanoids in transgenic low-sinapine oilseed rape (Brassica napus). Phytochemistry 71: 1076–1084 [DOI] [PubMed] [Google Scholar]
- Zeisel SH. (2000) Choline: an essential nutrient for humans. Nutrition 16: 669–671 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.