Abstract
The secondary growth of a woody stem requires the formation of a vascular cambium at an appropriate position and proper patterning of the vascular tissues derived from the cambium. Class III homeodomain-leucine zipper (HD ZIP) transcription factors have been implicated in polarity determination and patterning in lateral organs and primary vascular tissues and in the initiation and function of shoot apical meristems. We report here the functional characterization of a Populus class III HD ZIP gene, popREVOLUTA (PRE), that demonstrates another role for class III HD ZIPs in regulating the development of cambia and secondary vascular tissues. PRE is orthologous to Arabidopsis (Arabidopsis thaliana) REVOLUTA and is expressed in both the shoot apical meristem and in the cambial zone and secondary vascular tissues. Transgenic Populus expressing a microRNA-resistant form of PRE presents unstable phenotypic abnormalities affecting both primary and secondary growth. Surprisingly, phenotypic changes include abnormal formation of cambia within cortical parenchyma that can produce secondary vascular tissues in reverse polarity. Genes misexpressed in PRE mutants include transcription factors and auxin-related genes previously implicated in class III HD ZIP functions during primary growth. Together, these results suggest that PRE plays a fundamental role in the initiation of the cambium and in regulating the patterning of secondary vascular tissues.
Secondary growth in woody plant stems requires several fundamental processes, including the initiation of a vascular cambium and the proper patterning of secondary vascular tissues derived from the cambium. Secondary phloem (inner bark) and secondary xylem (wood) are derived from the cambium in predictable positions and provide mechanical support as well as transport of water, nutrients, and photosynthate (Esau, 1960; Larson, 1994). The appearance of secondary vascular growth was a major innovation in the evolution of tracheophytes and enabled the production of massive body types and other innovative growth forms such as woody lianas (Spicer and Groover, 2010). While it is likely that all secondary vascular growth in all extant plant species has a common origin, the molecular and genetic mechanisms regulating secondary growth are only beginning to be defined (Spicer and Groover, 2010).
Secondary growth of trees within the model genus Populus is typical of many perennial woody plants, with primary (elongating) growth at the tip of the stem, followed by a rapid transition to secondary (radial) growth (Groover et al., 2010). Populus stems undergoing primary growth are characterized by primary vascular bundles arranged in a predictable phyllotaxis around the periphery of the stem (Larson and Isebrand, 1974; Larson, 1975, 1976). While the stem is radial in appearance, the vascular bundles can be described in terms of polarity. Within leaf veins, xylem is in an adaxial (top) and phloem is in an abaxial (bottom) position. Following these veins through the petiole and into leaf traces of stems, this positional relationship of xylem to phloem is maintained, and in stems, primary xylem is positioned toward the pith (adaxial) and primary phloem toward the epidermis (abaxial; Larson, 1975). In Populus, the transition to secondary growth is rapid, and a continuous ring of cambium appears nearly simultaneously around the circumference of the stem (Larson and Isebrand, 1974), with the joining of fascicular and interfascicular cambia. Secondary xylem and phloem are produced in the positions relative to the cambium as primary vascular tissues, thus maintaining the same polarity relationships.
Patterning of woody stems does show significant variation in nature, however, as illustrated by so-called cambial variants (also termed anomalous secondary growth; Carlquist, 2001). For example, stems with successive cambia can initiate additional cambia within cortical parenchyma, which can then produce conjunctive tissue as well as secondary xylem and phloem, or even new vascular cambia (Carlquist, 2007). This process is reiterative, leading to a stem composed of increments of secondary xylem and phloem produced by successive cambia within a background of conjunctive tissues. Currently, the mechanisms leading to the formation of successive cambia in cortical tissues is unknown, as are the mechanisms regulating the patterning of secondary vascular tissues.
Class III HD ZIPs constitute a small gene family encoding transcription factors that contain a Leu zipper domain, a putative sterol-binding domain, the DNA-interacting PAS-related MEKHLA domain, and a homeodomain (Baima et al., 1995; Sessa et al., 1997; Mukherjee and Bürglin, 2006). The Arabidopsis (Arabidopsis thaliana) genome contains five class III HD ZIPs: REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV/IFL1), PHABULOSA (PHB), PHAVOLUTA (PHV), ATHB8, and CORONA (ATHB15). The class III HD ZIP family is evolutionarily ancient (Floyd et al., 2006; Prigge and Clark, 2006; Bowman et al., 2007; Floyd and Bowman, 2007) and is negatively regulated at the transcript level by highly conserved microRNAs (miRNAs; Floyd et al., 2006) and at the protein level by LITTLE ZIPPER proteins (Wenkel et al., 2007; Kim et al., 2008). In angiosperms, the involvement of class III HD ZIP genes has been demonstrated for meristem function, polarity of lateral organs (e.g. leaves), and vascular development (Talbert et al., 1995; McConnell and Barton, 1998; McConnell et al., 2001; Prigge et al., 2005; Prigge and Clark, 2006; Floyd and Bowman, 2007; Bowman and Floyd, 2008). The polarity determining the function of class III HD ZIP genes is antagonized by KANADI and YABBY genes (McConnell and Barton, 1998; Eshed et al., 2001; Kerstetter et al., 2001; Emery et al., 2003; Bowman and Floyd, 2008). Most of these functions appear to be acquired, as class III HD ZIPs predate the evolution of vascular tissues and polarized lateral organs (Floyd et al., 2006), and the ancestral role of class III HD ZIP genes may be centered on basic apical meristem initiation and function. The ancient origin of class III HD ZIPs and their subsequent recruitment in diverse developmental processes thus make them of high interest in understanding land plant evolution.
The Arabidopsis class III HD ZIP REV is unique with respect to other family members in that rev loss-of-function mutants show obvious developmental phenotypes. rev loss-of-function mutants have drastic reduction of the band of interfascicular fibers characteristic of Arabidopsis ecotypes Columbia and Landsberg erecta inflorescence stems (Zhong and Ye, 1999; Prigge et al., 2005). rev alleles also display variable loss of lateral meristems from leaf axils leading to alterations in branching architecture and changes in leaf morphology and color (Talbert et al., 1995; Otsuga et al., 2001). REV gain-of-function mutants that lack a functional miRNA recognition site have altered polarity of lateral organs and adaxialized primary vasculature with xylem surrounding phloem (Emery et al., 2003). A number of observations link REV and auxin during development. Polar auxin transport is impaired in rev mutants and is associated with the down-regulation of specific PIN auxin efflux carriers (Zhong and Ye, 2001), loss of proper PIN1 localization may be causative in phenotypic changes in rev phb phv triple mutants (Izhaki and Bowman, 2007), and class III HD ZIPs may function to promote axial cell elongation and auxin canalization (Ilegems et al., 2010). Class III HD ZIPs may also play a role in regulating secondary growth, and previous workers showed that a Populus ortholog of REV is expressed in woody stems of Populus (Schrader et al., 2004). Interestingly, the expression of the Populus mIR166 targeting class III HD ZIP transcripts is inversely proportional to the Populus REV ortholog (Ko et al., 2006).
We describe here the expression and function of a Populus ortholog of REV, popREVOLUTA (PRE), during secondary growth. We show that PRE is normally expressed during secondary growth and plays fundamental roles in cambium initiation and patterning of secondary vascular tissues. PRE misexpression can induce cambium initiation in abnormal positions and lead to patterning defects in derived secondary vascular tissues, including complete polarity reversals. While class III HD ZIPs have been implicated in various aspects of plant development, this report describes a new function for them during secondary growth.
RESULTS
PRE Is a Populus Ortholog of REV
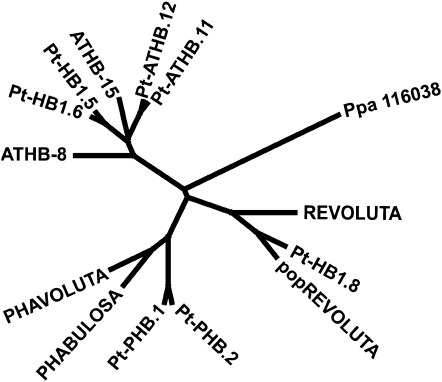
Phylogenetic analysis of Populus class III HD ZIPs indicates a duplication of all family members within the Populus lineage. The full amino acid sequences from all Populus and Arabidopsis class III HD ZIPs were retrieved from the Phytozome database (http://www.phytozome.net) and used to create a phylogenetic tree (see “Materials and Methods”). The relationship of Populus class III HD ZIPs to the well-characterized Arabidopsis orthologs in shown in Figure 1. The Populus genome contains two REV paralogs (Department of Energy Joint Genome Institute Populus genome annotation V1.1 accessions estExt_Genewise1_v1.C_660759 [also known as PtHB1.7; Phytozome accession POPTR_0004s22090] and gw1.IX.4748.1 [also known as PtHB1.8; Phytozome accession POPTR_0009s01990]). The two Populus REV paralogs share 89% identity and 93% similarity to each other. The protein encoded by estExt_Genewise1_v1.C_660759 shares 78% identity and 89% similarity to Arabidopsis REV. A cDNA for estExt_Genewise1_v1.C_660759 was isolated from the sequenced Populus trichocarpa clone Nisqually (Tuskan et al., 2006) using reverse transcription (RT)-PCR (see “Materials and Methods”). Sequencing of the cloned gene, hereafter referred to as PRE, confirmed the fidelity of predicted gene model estExt_Genewise1_v1.C_660759.
Figure 1.
Phylogenetic analysis of PRE and related class III HD ZIP proteins in Arabidopsis and Populus. Entire protein sequences for the indicated genes from Arabidopsis or P. trichocarpa (indicated with Pt prefix) were used in a phylogenetic analysis (see “Materials and Methods”). Genome duplication in Populus is reflected in multiple Populus paralogs for each Arabidopsis gene. Populus has two REV orthologs, PtHB1.8 and PRE. PtATHB.12 (Phytozome accession no. POPTR_0001s18930) corresponds to popCORONA (J. Du and A.T. Groover, unpublished data).
PRE Is Expressed during Both Primary and Secondary Growth
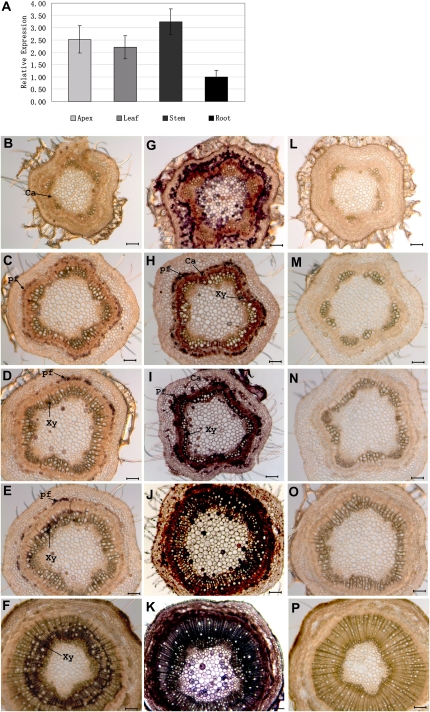
PRE is expressed in apices, leaves, stems, and roots of Populus seedlings (Fig. 2A), with highest expression in stems, as determined by quantitative (Q)-RT-PCR (see “Materials and Methods”) using primers specific to PRE. Q-RT-PCR of PRE paralog PtHB1.8 revealed transcript levels that were not significantly different from PRE in both leaves and shoots (data not shown). Whole mount in situ hybridization was used to further detail PRE expression within a developmental series of transverse stem sections (see “Materials and Methods”). As shown in Figure 2, PRE expression revealed by an antisense PRE probe is weak and diffuse during primary growth (Fig. 2B). The transition to secondary growth is accompanied by stronger PRE expression that is broadly associated with the cambial zone, developing secondary xylem, and phloem fibers (Fig. 2C). As secondary growth proceeds, PRE expression is maintained in the cambial zone, secondary xylem, and phloem fibers, although expression is not uniform around the circumference of the stem (Fig. 2, D and E). Later in secondary growth, nonuniform expression within the secondary xylem is even more apparent (Fig. 2F). Expression extends into older secondary xylem with mature (and thus nonliving) vessel elements, suggesting that PRE expression in these older tissues is from xylem parenchyma and living fibers. Positive control hybridizations with an antisense probe hybridizing to transcripts from a ubiquitously expressed gene encoding a 50S ribosomal subunit provide comparison for differences in staining attributable to variation in cytoplasmic content among cell types (Fig. 2, G–K). A sense probe serving as a negative control shows low background staining, although staining of the epidermis was occasionally seen (Fig. 2, L–P). Because the 109-bp in situ probe has 89% identity with paralog PtHB1.8, it is possible that in situ signals reflect PtHB1.8 expression in addition to PRE.
Figure 2.
Expression of PRE in Populus organs as assayed with Q-RT-PCR (A) and within stem tissues as assayed by in situ hybridization (B–P). A, Q-RT-PCR quantification of PRE transcripts in Populus apices, leaves, stems, and roots, using primers specific to PRE that do not hybridize to paralog PtHB1.8. B to F, Whole mount in situ hybridizations of transverse stem sections with an antisense probe hybridizing to PRE transcripts. Regions of PRE expression are indicated by dark purple staining. Sections are from the following internodes, counting from the first elongating internode: first (B), third (C), fifth (D), seventh (E), and bottom (F). G to K, Whole mount in situ hybridizations of transverse stem sections with an antisense probe hybridizing to pop50S transcripts. Regions of pop50S expression are indicated by dark purple staining. This ubiquitously expressed gene is a positive control that shows differential staining reflective of cytoplasmic density. L to P, Whole mount in situ hybridizations of transverse stem sections with a sense probe that cannot hybridize to pop50S transcripts. This negative control probe shows the level of background staining for this procedure. ca, Cambial zone; pf, phloem fibers; Xy, secondary xylem. Bars = 100 μm.
PRE Transgenic Phenotypes Are miRNA Dependent
Transgenic Populus with altered PRE transcript levels was created to examine the role of PRE during secondary growth. T-DNA constructs were assembled (see “Materials and Methods”) that either targeted PRE transcripts using RNA interference (PRE-RNAi) or an artificial miRNA (PRE-amiRNA) or that overexpressed the PRE coding sequence (35S::PRE) or the PRE coding sequence containing point mutations that abolish the miRNA recognition site (without changing the amino acid coding sequence; 35S::PRE-miRNAd). Constructs were transformed into an easily transformed hybrid aspen genotype using Agrobacterium tumefaciens transformation, and transformants were selected as kanamycin-resistant shoots (see “Materials and Methods”) such that each recovered transgenic line represents an independent transformation event. Individual lines were then tip propagated to produce clones for experimentation. Transcript levels were determined for PRE in transformants from each construct using Q-RT-PCR (see “Materials and Methods”). PRE-RNAi and PRE-amiRNA plants had highly variable PRE transcript levels, but surprisingly, they showed modest up-regulation of PRE transcript levels relative to wild-type controls (Supplemental Fig. S1, A and B). 35S::PRE plants also had highly variable PRE transcript levels that were approximately 2-fold or less higher than the wild type (Supplemental Fig. S1C). Anatomical and morphological analysis of PRE-RNAi, PRE-amiRNA, and 35S::PRE plants failed to reveal any anatomical or histological changes relative to wild-type plants (data not shown).
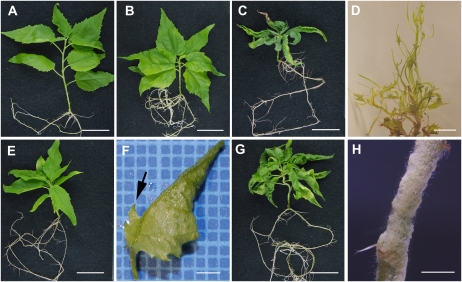
In contrast, 35S::PRE-miRNAd transformants had significant up-regulation of the miRNA-resistant transcript (Supplemental Fig. S1D) and showed obvious phenotypes. As shown in Figure 3, 35S::PRE-miRNAd transformants displayed whole plant phenotypes that range in severity. As compared with wild-type controls (Fig. 3A), lines with less severe phenotypes have shorter internodes (Fig. 3B). Lines with moderately severe phenotypes display shorter internodes and leaves with margins that curl upward (Fig. 3C). The most severe lines recovered had dramatic changes in leaf development, forming leaves that are highly lobed, have reduced blades, and frequently show radialized growth (Fig. 3D). One line with a less severe phenotype occasionally produced leaves with ectopic blades growing perpendicular to the midvein on the abaxial side of the leaf (Fig. 3, E and F). Interestingly, lines with more severe phenotypes frequently produced patches of callus on the surface of stems (Fig. 3, G and H).
Figure 3.
Anatomical phenotypes of transgenic Populus expressing a miRNA-resistant transcript of PRE (35S::PRE-miRNAd). A, Wild-type control plant showing typical anatomy. B, Transgenic line 35S::PRE-miRNAd 3-5-3 presenting a weak phenotype characterized by shorter internode lengths. C, Transgenic line 35S::PRE-miRNAd 3-4-2 presenting a moderately strong phenotype characterized by shorter internode lengths and leaf curling. D, Transgenic line 35S::PRE-miRNAd ε presenting a severe phenotype characterized by short internode length and highly lobed and radialized leaves. E, Transgenic line 35S::PRE-miRNAd 3-4-3 presenting a moderate phenotype characterized by shorter internode lengths and subtle changes to leaf shape. F, Higher magnification of a leaf from 35S::PRE-miRNAd 3-4-3 showing ectopic blade tissue growing from the midvein on the abaxial side of the leaf. G, Transgenic line 35S::PRE-miRNAd 3-8-2 presenting a moderate to severe phenotype characterized by shorter internode lengths, leaf defects, and growth of callus on the surface of the stem. H, Higher magnification of the stem of transgenic line 35S::PRE-miRNAd 3-8-2 showing extensive callus formation on the surface of the stem. Bars = 2 cm (A–C, E, and G), 1 cm (D), 2 mm (F), and 4 mm (H).
Overexpression of a miRNA-Resistant PRE Leads to Variable, Unstable Phenotypes
In addition to variation in phenotypes among independent 35S::PRE-miRNAd transformants, dramatic variation was noted among propagules of individual lines and even within individual plants. Plants with less severe or moderate phenotypes were moved to soil to allow the observation of more extensive vegetative growth. As shown in Figure 4A, an individual 35S::PRE-miRNAd-transformed plant produced a mixture of phenotypically normal leaves and leaves with various degrees of upward curling of the leaf blade around the margins (Fig. 4, B–D). Mutant and normal leaves formed in close proximity to each other (Fig. 4A). A 35S::PRE-miRNAd plant with a stronger phenotype (Fig. 4E) produced leaves that were misshapen, had pockets of overgrowth by interveinal tissues, and were strongly curled upward from the margins (Fig. 4, F and G). The hybrid aspen used for transformation frequently forms root suckers. Surprisingly, root suckers (adventitious shoots emerging from roots) with nearly wild-type leaf morphology were formed by a plant whose primary stem had a strong leaf phenotype (Fig. 4H). Similarly, clonal propagules of 35S::PRE-miRNAd lines showed variation in phenotypes that was either more or less severe after subculture (data not shown).
Figure 4.
Unstable phenotypes presented by Populus expressing a miRNA-resistant transcript of PRE (35S::PRE-miRNAd). A, Greenhouse-grown 35S::PRE-miRNAd line 3-6. Note the presence of phenotypically wild-type leaves and leaves showing varying degrees of upward curling. B to D, Higher magnification of leaves showing various degrees of curling from the plant in A. E, Greenhouse-grown 35S::PRE-miRNAd line 3-8-1. Leaves show a moderately severe phenotype. F and G, Higher magnification of leaves from E showing uneven growth and curling. H, 35S::PRE-miRNAd line 3-8-1 with a phenotypically wild-type root sucker emerging from a plant with a moderately severe phenotype. Bars = 5 cm.
Overexpression of a miRNA-Resistant PRE Leads to Abnormal Cambium Formation and Alterations in Patterning and Polarity of Secondary Vascular Tissues
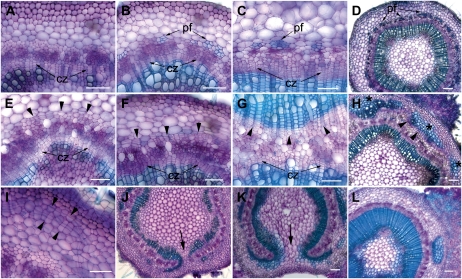
The effect of PRE expression on secondary growth was examined by histological comparison of transverse sections of stems from 35S::PRE-miRNAd and matched wild-type control plants. As shown in Figure 5, stems of wild-type plants make a predictable transition in development from primary through secondary growth. In the fourth internode below the apex, the transition to secondary growth was evidenced by the cell files within the cambial zone (Fig. 5A). At the seventh internode, wild-type stems had a continuous cambium producing secondary xylem toward the center of the stem and secondary phloem toward the outside of the stem (Fig. 5B). Lignified phloem fibers differentiated in groups at the periphery of the phloem (Fig. 5B). At the bottom of the stem, more extensive secondary xylem was present, as were fully differentiated phloem fibers. Overall, the patterning of the stem undergoing secondary growth is characterized by a central pith, encased by successive layers of secondary xylem, cambial zone, secondary phloem with peripheral phloem fibers, cortex, and an outer epidermis (Fig. 5D).
Figure 5.
Histological stem phenotypes presented by Populus expressing a miRNA-resistant transcript of PRE (35S::PRE-miRNAd). A to D, Transverse sections of the stem of a wild-type plant stained with toluidine blue (see “Materials and Methods”). Sections are from the fourth elongating internode (A), seventh internode (B), or bottom internode (C) of a representative in vitro-grown wild-type control. E to G, Transverse sections from the stem of a matched 35S::PRE-miRNAd transgenic line. In the fourth internode (E), a cambial zone (cz) forms normally, but abnormal periclinal divisions are seen in the cortical parenchyma (arrowheads). In the seventh internode (F), more extensive periclinal divisions are giving rise to cell files in the cortical parenchyma. A section from the bottom internode (G) shows extensive production of secondary xylem from the normal cambial zone toward the pith and production of secondary xylem toward the epidermis from the abnormally formed cortical cambium (arrowheads). H, Section from the base of a 35S::PRE-miRNAd stem, showing disjointed regions of abnormal cambial activity (arrowheads) giving rise to secondary xylem toward the epidermis. Note that cortical parenchyma cells adjacent to differentiating secondary xylem show abnormal lignification (asterisks). I, Section from an elongating stem of a 35S::PRE-miRNAd plant showing that the position of abnormal cambium (arrowheads) is not consistent within the cortex. J and K, Sections from middle internodes of 35S::PRE-miRNAd plants showing regions where cambium and derived secondary vascular tissues alter growth and patterning (arrows). L, Section from a 35S::PRE-miRNAd stem showing extensive callus growth from the cortex and/or epidermis. pf, Phloem fibers. Bars = 100 μm.
Transverse sections from 35S::PRE-miRNAd plants revealed defects in the position of cambium initiation and tissue patterning. Sections from a 35S::PRE-miRNAd line with a moderate phenotype had notable morphological changes in the fourth internode, where abnormal periclinal cell divisions appeared within the stem cortex (Fig. 5E, arrowheads). A normal cambial zone was also apparent, as was developing secondary xylem (Fig. 5E). Lower in the stem in internode seven, abnormal periclinal divisions in the cortex produced cell files consistent with cambial activity, and phloem fibers were conspicuously absent (Fig. 5F). At the bottom of the stem, the normal cambial zone was still active and producing files of cells visible in the secondary xylem (Fig. 5G). Strikingly, periclinal divisions in the stem cortex produced secondary xylem toward the outside of the stem (Fig. 5F), a reversal of normal polarity for secondary xylem. Evidence that this was in fact secondary xylem produced by cambial activity was evidenced by the arrangement of lignified cells in files. Significant variation in the severity of stem phenotypes was noted among lines, among clonal propagules, and within individual plants. While some stem sections were characterized by a continuous ring of abnormal cortical cambium and secondary xylem, other stem sections had disjointed sections of abnormal cortical cambium (Fig. 5H). Cortical parenchyma adjacent to the outside of the abnormal secondary xylem frequently showed signs of lignification (Fig. 5H, asterisks). The position of initial formation of abnormal cortical cambia varied from anywhere adjacent to secondary phloem to the epidermis (Fig. 5I). Some sections revealed positions where the cambium and derived secondary xylem curved from normal polarity to reverse polarity (Fig. 5, J and K). The callus noted on some stems (Fig. 3H) was from cell divisions in the outer cortex and epidermis (Fig. 5L). It was not clear if the callus emanated from cambial activity, as the cells apparently continued to divide in different planes outside the boundary of the epidermis and were not arranged in cell files.
Whether defects in primary vascular development played a causative role in secondary growth defects in 35S::PRE-miRNAd stems, or whether secondary growth defects could arise de novo, could not be unequivocally determined. As illustrated by serial sectioning of wild-type and 35S::PRE-miRNAd stems, examples were found within individual stems where there were only subtle defects in primary vascular growth in the first two internodes but very strong phenotypes in older internodes that had transitioned to secondary growth. While it is not possible to image anatomical changes over time in stems, examination of the remnants of primary vascular tissues in older nodes can provide some insight into the progression of developmental events. As illustrated in Supplemental Figure S2K, examples could be found of internodes containing secondary growth defects that had no obvious evidence of defects in the remnants of primary vascular bundles.
Stem Patterning and Polarity Defects in miRNA-Resistant PRE Mutants May Be Exacerbated at the Position of Lateral Organs
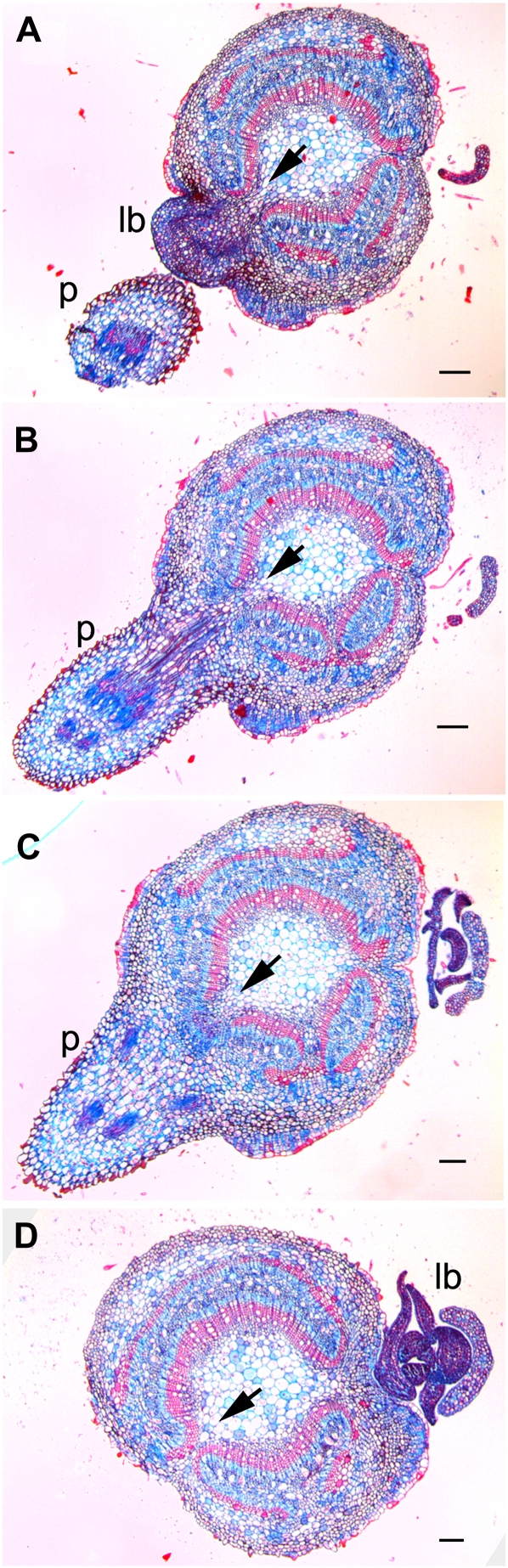
The vasculature of Populus leaves is connected to the stem vasculature by three vascular bundles, or leaf traces. The departure of the leaf trace from the stem vasculature results in a region where the continuity of the vascular cylinder is interrupted by parenchymous tissue, termed a leaf trace gap. Also in Populus, each leaf is associated with a lateral bud in the axil of the leaf. Serial sections revealed correlations between regions where cambium polarity is altered and these lateral organs. As shown in Figure 6, a change of cambial polarity and derived secondary xylem was noted directly adjacent to the position of a lateral bud. Moving farther down the stem, a section through the midpoint of the petiole showed interruption of secondary xylem by a leaf trace gap (Fig. 6, B and C, arrows). This interruption and folding back of the cambium was seen directly below the base of the petiole. A similar sequence of events was seen associated with the lateral bud of the next oldest leaf, with the alteration of cambium polarity occurring above the insertion of the lateral bud in the stem (Fig. 6).
Figure 6.
Serial sections of a stem from Populus expressing a miRNA-resistant transcript of PRE (35S::PRE-miRNAd). Transverse sections of the stem of a 35S::PRE-miRNAd plant stained with alcian blue and safrinin (see “Materials and Methods”) are shown, moving from an apical position (A) toward a basal position (D). A, Section glances through the top of a lateral bud (lb) and through a petiole (p). Note the region of leaf trace gap (arrow) associated with discontinuous cambial activity and the region of abnormal cambial reorientation and change of polarity of secondary growth. B, Section below A, below the lateral bud and glancing the midpoint of insertion of the petiole base into the stem. The leaf trace gap is marked by the arrow. C, Section through the base of a petiole, below B. The leaf trace gap is marked by the arrow. D, Section below the base of the petiole, below C. The arrow marks the region of abnormal cambial reorientation and change of polarity of secondary growth. A lateral bud is visible on the opposite side of the stem that displays a similar ontogeny. Bars = 200 μm.
Overexpression of miRNA-Resistant PRE Leads to Misexpression of Regulatory Genes and Genes Associated with Hormonal and Cell Differentiation Processes
To begin to identify the biological networks in which PRE functions, gene expression in stems of 35S::PRE-miRNAd and matched wild-type control plants was assayed using Populus Affymetrix microarrays (see “Materials and Methods”). Tissues assayed were defoliated stems that included regions of both primary and secondary growth. After normalization and filtering of genes with low expression or low variance among genotypes, a total of 602 genes were identified with statistically significant (t test P < 0.05) and 1.5-fold or greater differences in transcript abundance between 35S::PRE-miRNAd and wild-type stems (see “Materials and Methods”). Analysis of the Gene Ontology classification of genes misregulated in 35S::PRE-miRNAd stems (see “Materials and Methods”) showed strong bias in representation within GOSLIM biological process categories including developmental processes (P = 1.56e-13), cell organization and biogenesis (P = 3.66e-07), transcription (P = 3.05e-09), and other biological processes (P = 3.71e-08). Misexpressed genes overlapped with homologous genes participating in proposed models for class III HD ZIP functions from studies of primary growth in Arabidopsis, as discussed below.
Notably, 92 transcription factors are misexpressed in 35S::PRE-miRNAd (Supplemental Table S1). Of the six transcription factors misregulated in stems of miRNA-resistant overexpression mutants for the related Populus class III HD ZIP, POPCORONA (PCN; J. Du and A.T. Groover, unpublished data). five (all but zinc finger fgenesh4_pg.C_LG_XIV000644) are misregulated in PRE, suggesting an overlap of the transcriptional networks in which PRE and PCN function. Interestingly, several of the previously identified transcription factors that interact physically or genetically with REV are also misregulated in 35S::PRE-miRNAd. SHORT-ROOT (SHR) and SCARECROW (SCR) have been well characterized for their role in regulating stem cells and differentiation in Arabidopsis roots, and SHR has been shown to negatively regulate class III HD ZIPs PHV and PHB (Levesque et al., 2006; Carlsbecker et al., 2010). Orthologs of SHR and SCR have been shown to be expressed during secondary growth in Populus (Schrader et al., 2004), and both genes are down-regulated in 35S::PRE-miRNAd. The Arabidopsis AP2-like transcription factor-encoding genes DORNRÖSCHEN (DRN) and DORNRÖSCHEN-LIKE (DRNL) have been shown to physically interact through heterodimerization with REV and other Arabidopsis class III HD ZIPs and affect the expression and localization of the auxin efflux carrier PIN1 (Chandler et al., 2007). A Populus ortholog of DRNL is up-regulated in 35S::PRE-miRNAd stems. ASYMMETRIC LEAVES2 (AS2) has been shown to be regulated by the class III HD ZIP antagonist, KANADI1 (Wu et al., 2008), and a Populus ortholog of AS2 is down-regulated in 35S::PRE-miRNAd. In addition, several other transcription factors that have not been previously implicated in class III HD ZIP function are misregulated in 35S::PRE-miRNAd, including factors regulating development, cell differentiation, hormone-related processes, and chromatin remodeling (Supplemental Table S1).
Genes related to hormone functions are also misregulated in 35S::PRE-miRNAd, including abscisic acid, gibberellin, and auxin-related genes (Supplemental Table S2). Notably, a Populus ortholog of PIN3 is down-regulated in 35S::PRE-miRNAd. This gene has been shown to be expressed preferentially in the cortex of Populus stems (Schrader et al., 2003) and is directly or indirectly regulated by the class III HD ZIP antagonist KANADI1 (Ilegems et al., 2010). A large number of cell wall-related genes are misregulated in 35S::PRE-miRNAd (Supplemental Table S3), including genes involved in cellulose biosynthesis (cellulose synthases) and lignification (e.g. laccases, ferulate-5-hydroxylase, and cinnamoyl CoA reductase). Interestingly, fasciclin-like arabinogalactan proteins are also misregulated in 35S::PRE-miRNAd. The proteins have recently been shown to affect cellulose deposition and cell wall matrices in Arabidopsis and Eucalyptus (MacMillan et al., 2010). Additionally, genes encoding proteins involved in cell signaling and protein phosphorylation (Supplemental Table S1) and cytoskeleton functions (Supplemental Table S2) are misexpressed in 35S::PRE-miRNAd.
DISCUSSION
The functional analyses presented here demonstrate that the Populus class III HD ZIP, PRE, plays a role in cambium initiation and patterning of secondary vascular tissues. PRE is an ortholog of the Arabidopsis gene REV and is expressed during both primary and secondary growth. Unexpectedly, PRE expression during secondary growth is not uniform around the circumference of the stem, with expression sometimes restricted to only some regions of the cambial zone, secondary xylem, or phloem fibers. One possible dynamic is that once PRE expression is established in the cambial region, expression is maintained in the derived cambial daughter cells as they differentiate. This nonuniform expression could reflect a response to environmental cues, physiological or hormonal changes, or epigenetic regulation. Differential gene expression around the circumference of stems is known to be associated with the production of reaction wood, which acts to counteract the force of gravity on leaning stems. However, examination of published gene expression profiles (Déjardin et al., 2004; Sterky et al., 2004) did not reveal any expression of PRE associated with reaction wood, and there is no significant overlap of genes expressed during reaction wood and in response to PRE misexpression. Alternatively, the nonuniform expression could reflect the dynamic physiological or hormonal changes occurring around the circumference of the stem. For example, positions of primary vascular bundles around the stem (Larson, 1980) may be associated with differences in auxin concentrations that affect gene expression, and class III HD ZIPs are known to affect auxin transport (Zhong and Ye, 2001; Izhaki and Bowman, 2007; Ilegems et al., 2010). A third possibility is that epigenetic mechanisms affect PRE expression. The instability of PRE misexpression phenotypes (see below) and previous observations of epigenetic regulation of the Arabidopsis class III HD ZIPs, PHB and PHV (Bao et al., 2004), support this possibility.
We made transgenic Populus with up- or down-regulated PRE to assess PRE function during secondary growth. Transgenic Populus expressing artificial miRNA or RNA interference transgenes targeting PRE transcripts resulted in plants with either no change or slightly up-regulated PRE transcript levels; thus, we were not able to ascertain the effects of PRE down-regulation. Similar to reports for class III HD ZIP transgenics in Arabidopsis, overexpression of the wild-type PRE did not result in mutant phenotypes, but overexpression of PRE with an altered miRNA-binding site did (35S::PRE-miRNAd mutants), indicating that negative regulation by miR165/166 can effectively counteract the effects of misexpression of a wild-type allele. 35S::PRE-miRNAd mutants display leaf phenotypes ranging from upward curling of leaf margins to partially radialized leaves, which is consistent with the promotion of adaxial growth, as has been described for REV gain-of-function mutants in Arabidopsis (Talbert et al., 1995; Emery et al., 2003). Surprisingly, the 35S::PRE-miRNAd mutant phenotypes were unstable, with the phenotype of genetically identical clones or even individual plants gaining or waning in severity over time or within different parts of the same plant. This result is somewhat similar to Arabidopsis REV mutants, which were originally named VARIABLE MERISTEM (Pogany et al., 1998; Otsuga et al., 2001), which show variation in severity within individual plants for phenotypes such as the presence of axillary meristems, and to the observation that rev mutant alleles show differing severity of phenotype in different genetic backgrounds (Prigge et al., 2005). Physiological or hormonal explanations are possible, although mutant and normal leaves can be found in close proximity on the same 35S::PRE-miRNAd plant and nearly wild-type root suckers can emerge from plants with relatively strong phenotypes. An alternative explanation would be that epigenetic factors may influence PRE expression. Variable transgene silencing could play a role, although it is less likely to explain the transitions observed for plants with weak phenotypes to stronger phenotypes. Normal epigenetic regulation affecting expression could also play a causative role, as has been described for methylation of PHB/PHV loci (Bao et al., 2004).
The most striking aspect of 35S::PRE-miRNAd mutants is the abnormal initiation of cambia in cortical parenchyma and the changes in patterning and polarity of the derived secondary vascular tissues. Like most woody plants, vascular cambia in Populus arise between xylem and phloem in vascular bundles (fascicular cambia) and are joined by interfascicular cambia to create a cylinder of cambium (Larson and Isebrand, 1974). 35S::PRE-miRNAd mutants displayed a range of patterning defects with regard to cambium position, ranging from normal initiation to initiation in various parts of the cortical parenchyma. These abnormal cambia produced secondary vascular tissues with reversed polarity (i.e. secondary xylem was produced toward the epidermis and secondary phloem toward the pith). That these cambia were vascular cambia and not phellogen (cork) cambia was evidenced by parallel files of secondary xylem emanating from the cambial initials. Initiation of cambial activity in cortical parenchyma does occur in nature, for example during the growth of stems with successive cambia (Carlquist, 2007). However, we are not aware of examples of polarity reversals of secondary growth, perhaps because of the physical restrictions an externally oriented ring of lignified secondary xylem would impose on growth. Interestingly, positions of abnormal cambial growth and polarity reversal were correlated with leaves and axillary buds. This correlation could reflect alterations in PRE function in response to hormonal differences in these fields; alternatively, they could reflect that polarity and patterning relationships in stems must be reestablished after interruption by leaf gaps and require proper expression of PRE. Additional abnormal growth was also present on some 35S::PRE-miRNAd stems in the form of callus growth from the epidermis or outer cortical cell layers. Because the cells derived from this meristematic activity were not arranged in files, their origin cannot be unequivocally attributed to cambial activity. However, classical studies have demonstrated that exposed cambial tissues (i.e. through wounding) can respond by producing callus tissue (Brown and Sax, 1962; Noel, 1968), and pressure has been identified as a factor influencing the function of cambia (Brown and Sax, 1962). It is thus possible that the observed callus production was from additional cambial activity from outer cell layers, growing without the physical restraint of encasing tissue.
Microarray profiling of 35S::PRE-miRNAd mutant stems revealed portions of the biological networks in which PRE functions. A surprising overrepresentation of transcriptional regulators was found among the genes misregulated in 35S::PRE-miRNAd, potentially indicating the PRE is a major hub in this regulatory network, and could account in part for pleiotropic phenotypes of REV and PRE mutants. While direct targets of the PRE ortholog, REV, have not been identified, multiple genes and mechanisms have been identified that interact at various levels with REV function. Several of these key regulators are misregulated in 35S::PRE-miRNAd, indicating that our microarray profiling was successful in identifying relevant genes involved in PRE function and that there are important points of overlap of the function of REV during primary growth in Arabidopsis and PRE during secondary growth in Populus. One important point of overlap involves antagonists and interaction partners of class III HD ZIPs and auxin transport. Several lines of evidence point to the influence of REV and hormones in meristem functions. PIN-FORMED (PIN) genes encode auxin efflux carriers that have been shown to be expressed in leaf procambial strands and cells of apical meristems in Arabidopsis (Reinhardt et al., 2003; Scarpella et al., 2006) and during secondary vascular growth in Populus (Schrader et al., 2003). Misexpression of KANADI1, which acts antagonistically with class III HD ZIPs (Emery et al., 2003), results in down-regulation of PIN1, PIN3, PIN4, and AS2 (Izhaki and Bowman, 2007; Wu et al., 2008; Ilegems et al., 2010). Additionally, DRNL physically interacts with REV and acts in part to regulate PIN expression (Chandler et al., 2007). Orthologs of AS2, DRNL, and PIN3 are all misregulated in 35S::PRE-miRNAd stems, suggesting that these auxin-related pathways are also important for secondary vascular growth. On the other hand, PRE may have unique binding partners, target genes, or functions that differ between species, tissues, or growth stages. For example, a previously proposed regulatory network (Wenkel et al., 2007) includes a negative feedback loop in which REV promotes the expression of LITTLE ZIPPER genes, which encode small Leu zipper-containing proteins that physically interact with and negatively regulate class III HD ZIP proteins. In contrast, we found no evidence for up-regulation of Populus LITTLE ZIPPERs in 35S::PRE-miRNAd. Additionally, we found a large overrepresentation of cell differentiation and cell wall-related genes and genes involved in cell signaling processes that have not previously been identified in class III HD ZIP regulatory networks. However, it is important to note that our experiments were not only in a different species and tissue type than those of Wenkel et al. (2007) but also used a constitutive promoter versus an inducible promoter to drive transgene expression. Future experiments will be required to determine which genes are directly regulated by PRE and to fully describe the regulatory networks in which PRE participates.
Class III HD ZIPs are interesting in that they play central roles in several fundamental aspects of plant development, including meristem functions in both shoot and root apical meristems, polarity of lateral organs, and development of primary vascular tissues (Eshed et al., 2001; McConnell et al., 2001; Emery et al., 2003; Hawker and Bowman, 2004; Ilegems et al., 2010). Because class III HD ZIPs are evolutionarily ancient and predate the evolution of leaves, flowers, and vascular tissues, many of the functions attributed to class III HD ZIPs in angiosperms are derived. We demonstrated here another role for class III HD ZIPs, in regulating secondary vascular growth. While REV and other class III HD ZIPs may have unique functions in different tissues and developmental events (e.g. through interactions with different binding partners), it is also possible that there are commonalities for class III HD ZIP functions in diverse developmental processes. Comprehensive comparative studies among plant meristems could reveal such commonalities.
MATERIALS AND METHODS
Phylogenetic Analysis
Sequences used for phylogenetic analysis were retrieved from the Phytozome database (http://www.phytozome.net/) on October 6, 2010, and are included as Supplemental File S1. The supplemental file also includes aliases for the different genes analyzed. MUSCLE software (Edgar, 2004) was used for the alignment of entire protein sequences using default settings. MUSCLE results were used to produce tree diagrams using TreeView (Page, 1996).
In Situ Hybridization
Whole mount in situ hybridizations were performed as described previously (Du et al., 2009). Digoxigenin-labeled RNA probes were generated by in vitro transcription from purified PCR products. An template for generating an antisense PRE probe was generated using primers P9isF-a-sense (5′-CAGCACGGGATATGTGGACTCTG-3′) and P9is-Ra-sense (5′-AATTAATACGACTCACTATAGGGCTGGGCAGCGGCAGCTGCATC-3′; containing a T7 primer sequence). A template for generating a negative control sense PRE probe was made using primers P9is-Fsense (5′-AATTAATACGACTCACTATAGGGCAGCACGGGATATGTGGACTCTG-3′; containing a T7 primer sequence) and P9is-Rsense (5′-CTGGGCAGCGGCAGCTGCATCGG-3′). A positive control probe complementary to the Populus 50S gene was generated using primers Pop50s-sense-R2 (5′-AATTAATACGACTCACTATAGGGCGGGTGAAAAAGGCACTAGA-3′; containing a T7 primer sequence) and Pop50s-sense-F2 (5′-ATATGGTTCAGGGGGTCTCC-3′).
Cloning and DNA Manipulations
Total RNA from leaves and young shoots was isolated from greenhouse-grown Populus trichocarpa plants (clone Nisqually1) using the RNeasy PlantMiniprep RNA extraction kit (Qiagen) following the manufacturer’s protocol. cDNA was generated from total RNA using the iScript cDNA synthesis kit from Bio-Rad, according to the manufacturer’s protocol. The full-length REV ortholog, PRE, was amplified from target estExt_Genewise1_v1.C_660759 using primers P9cDNA1-L (5′-CACCATGGAAGTGGCCCCTCTG-3′) and P9cDNA1-R (5′-TCACAAAAAAGACCAGTTTACAAAAGAG-3′), with the forward primer containing a CACC element to allow for directional cloning into a Gateway-compatible entry vector. The PCR product was cloned into vectors pENTR-D-TOPO, pCR2.1, and pCR blunt (Invitrogen) and confirmed by sequencing. The Gateway cassette containing the gene of interest was subcloned into the binary Gateway expression vector Pk2GW7, yielding binary expression vector 35S:42.
A miRNA-resistant form of PRE was created from two DNA fragments amplified from vector 35S:42 using overlapping primers that introduced mismatches to the original miRNA-binding site. The resulting sequence rendered the miRNA recognition site resistant to miRNA without changing the protein-coding sequence. Primers P9cDNA1-L (5′-CACCATGGAAGTGGCCCCTCTG-3′) and mutP9rR (5′-CCCAATCGAATCTGGTCCAGGC-3′) and mutP9L (5′-CTGGAATGAAGCCTGGACCAGATTC-3′) and P9cDNA1-R (5′-TCACAAAAAAGACCAGTTTACAAAAGAG-3′) were used to amplify the 5′ and 3′ portions of the cDNA, respectively. Primers mutP9rR and mutP9L contain internal mismatches that alter the miRNA recognition site in the cDNA, thus rendering the resulting transcript resistant to miRNA degradation. The two fragments were subsequently joined using overlapping PCR. The complete mutated sequence was cloned into vectors pCR2.1 and pCR blunt (Invitrogen). For generation of miRNA-resistant overexpression lines, the mutated cDNA was cloned into vector pArt7 (Gleave, 1992) behind the cauliflower mosaic virus 35S promoter and then shuttled to binary expression vector pArt27 (Gleave, 1992).
The 35S::miRNA-PRE construct was assembled to drive the expression of a synthetic miRNA by the 35S promoter. A 21-nucleotide sequence (5′-TGCCTGAAGATTCAGGTGATC-3′) specific to PRE and its paralog, PtHB1.8, was targeted based on published targeting parameters (Alvarez et al., 2006) and uniqueness to PRE and its paralog, PtHB1.8. The exact complementary sequence (miRNA) is 5′-GATCACCTGAATCTTCAGGCA-3′. Mismatches were introduced at positions four, nine, and 10, and at positions 20 and 21 in the complementary strand, to produce the following miRNA and miRNA* sequences: miRNA, 5′-GATCACCTGAATCTTCAGGCA-3′; miRNA*, 5′-CTTCACCTGAAAGTTCACGCA-3′.
A DNA was synthesized with these sequences replacing the normal miRNA and miRNA* sequences within MIR164b to produce the following sequence containing XhoI and XbaI restriction sites at the 5′ and 3′ ends, respectively. The sequence CACC was added at the 5′ end for directional cloning: Xho-Xba/MIR-PRE (5′-caccCTCGAGGAGAATGATGAAGGTGTGTGATGAGCAAGATGCCTGAAGATTCAGGTGATCTTACTAGCTCATATATACACTCTCACCACAAATGCGTGTATATATGCGGAATTTTGTGATATAGATGTGTGTGTGTGTTGAGTGTGATGATATGGATGAGTTAGTTCCTTCACCTGAAAGTTCACGCATCATGACCACTCCACCTTGGTGACGATGACGACGAGGGTTCAAGTGTTACGCACGTGGGAATATACTTATATCGATAAACACACACGTGCGTCTAGA-3′).
To create RNAi vector RNAi26, two PCR products were amplified from PRE cDNA (primer pair 1: rnaIP9-1-L [5′-caccGTTTCCAGATGATGCTCCACTG-3′] and rnaIP9-1-R [5′-GCAAACTATGACACGATGAGCCATCC-3′]; primer pair 2: rnaiP8-2-L [5′-caccAGCTTTGGGACCAAAGC-3′] and rnaiP9-2-R [5′-CTGCTCCTAAATGGTAATTAAGCAGAAC-3′]). The two products were directional cloned into Gateway entry vectors, and then fragments were recombined into expression vector Pk7gwiwg2(ii).
Plant Transformation
Hybrid aspen clone INRA 717-IB4 (Populus tremula × Populus alba) was micropropagated and transformed using the protocols of Han et al. (1996) by cocultivation with Agrobacterium tumefaciens strain GV3101 containing the transformation vector. A single transformed shoot was selected from each independent callus to ensure that each transgenic line created represented an independent, unique transformation event.
Histology
For light microscopy of wild-type and mutant stem, leaf, and root tissue, samples were embedded into 5% agarose and affixed onto a metal block using cyanoacrylate. Samples were sectioned using a Vibratome Series 1000 tissue-sectioning system to a thickness of 25 to 50 μm. Sections were cleared with 70% ethanol before being stained with toluidine blue, safrinin, or safrinin with alcian blue as a counterstain (Ruzin, 1999). For serial sectioning, samples were fixed in formaldehyde-acetic acid-alcohol and processed for embedding in Paraplast (Fisher) as described (Ruzin, 1999) prior to sectioning with a Leitz 1512 microtome. After dewaxing and rehydration, slides were stained with alcian blue followed by safranin.
Microarray Analysis
Total RNA was isolated from defoliated stems from 1-month-old 35S::PRE-miRNAd (line 3-8-1) plants and matched wild-type controls using the Qiagen Plant RNA extraction kit following the manufacturer’s instructions. RNA quantity and quality were assayed using the Agilent 2100 bioanalyzer (Agilent Technologies). cDNA synthesis and amplification were carried out using the GeneChip Whole Transcript cDNA synthesis and amplification kit, followed by labeling using the GeneChip Whole Transcript terminal labeling kit (Affymetrix). Labeled and fragmented RNA was hybridized onto Affymetrix GeneChip Poplar Genome Array chips according to the manufacturer’s instructions. Details of biological samples, experimental procedures, and hybridization data are available as National Center for Biotechnology Information Gene Expression Omnibus record GSE19467.
Data were analyzed using dChip (http://biosun1.harvard.edu/complab/dchip/) software. Data were normalized using median probe intensity of the baseline array. Model-based expression data were filtered by removing genes whose representative probes did not exceed 40% (presence call percentage) on a given array and 50% among arrays. Filtered genes were then compared based on fold expression difference and t test (P = 0.05). Gene Ontology analysis was through Provart and Zhu (2010) using Arabidopsis (Arabidopsis thaliana) best BLAST hits as input.
Q-RT-PCR
cDNA was generated from total RNA extracted from transgenic and wild-type lines grown to the same age and under identical conditions. RNA was subjected to DNase treatment prior to RT using the iScript cDNA synthesis kit (Bio-Rad). cDNA was amplified using Bio-Rad real-time PCR reagents following the manufacturer’s instructions on a Bio-Rad Mycycler.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XM_002328121.1 (PRE) and AY919616.1 (PtHB1.8).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PRE transcript levels in Populus transgenic stems relative to wild-type control stems, as determined by Q-RT-PCR using a tubulin gene transcript level as a reference.
Supplemental Figure S2. Stem phenotype of a plant expressing a miRNA-resistant version of PRE (35s::RBL-miRNAd).
Supplemental Table S1. Genes involved in cell signaling misregulated in popREVOLUTA transgenics.
Supplemental Table S2. Cytoskeleton-related genes misregulated in popREVOLUTA transgenics.
Supplemental Table S3. Microarray analysis results.
Supplemental File S1. Supplemental table sequences.
Supplementary Material
Acknowledgments
We thank Gayle Dupper for Populus tissue culture and cultivation, Annie Mix for greenhouse cultivation of plants, and Maichi Phan for photography of plants.
References
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G. (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK. (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7: 653–662 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK. (2008) Patterning and polarity in seed plant shoots. Annu Rev Plant Biol 59: 67–88 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K. (2007) Green genes: comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Brown CL, Sax K. (1962) The influence of pressure on the differentiation of secondary tissues. Am J Bot 49: 683–691 [Google Scholar]
- Carlquist S. (2001) Cambial variants (anomalous secondary growth). Carlquist S, , Comparative Wood Anatomy: Systematic, Ecological and Evolutionary Aspects of Dicotyledon Wood. Springer-Verlag, Berlin, pp 271–295 [Google Scholar]
- Carlquist S. (2007) Successive cambia revisited: ontogeny, histology, diversity, and functional significance. J Torrey Bot Soc 134: 301–332 [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Grewe B, Werr W. (2007) The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Déjardin A, Leplé JC, Lesage-Descauses MC, Costa G, Pilate G. (2004) Expressed sequence tags from poplar wood tissues: a comparative analysis from multiple libraries. Plant Biol (Stuttg) 6: 55–64 [DOI] [PubMed] [Google Scholar]
- Du J, Mansfield SD, Groover AT. (2009) The Populus homeobox gene ARBORKNOX2 regulates cell differentiation during secondary growth. Plant J 60: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Esau K. (1960) The Anatomy of Seed Plants. John Wiley & Sons, New York [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL. (2007) The ancestral developmental tool kit of land plants. Int J Plant Sci 168: 1–35 [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL. (2006) Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173: 373–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Groover AT, Nieminen K, Helariutta Y, Mansfield SD. (2010) Wood formation in Populus. Jansson S, Bhalerao RP, Groover AT, , Plant Genetics and Genomics: Crops and Models, Vol 8: Genetics and Genomics of Populus. Springer, New York, pp 201–224 [Google Scholar]
- Han KH, Gordon MP, Strauss SH. (1996) High frequency transformation of cotonwoods (genus Populus) by Agrobacterium rhizogenes. Can J For Res 27: 464–470 [Google Scholar]
- Hawker NP, Bowman JL. (2004) Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol 135: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, Stieger PA. (2010) Interplay of auxin, KANADI and class III HD-ZIP transcription factors in vascular tissue formation. Development 137: 975–984 [DOI] [PubMed] [Google Scholar]
- Izhaki A, Bowman JL. (2007) KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Prassinos C, Han KH. (2006) Developmental and seasonal expression of PtaHB1, a Populus gene encoding a class III HD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166). New Phytol 169: 469–478 [DOI] [PubMed] [Google Scholar]
- Larson PR. (1975) Development and organization of primary vascular system in Populus deltoides according to phyllotaxy. Am J Bot 62: 1084–1099 [Google Scholar]
- Larson PR. (1976) Procambium vs cambium and protoxylem vs metaxylem in Populus deltoides seedlings. Am J Bot 63: 1332–1348 [Google Scholar]
- Larson PR. (1980) Interrelations between phyllotaxis, leaf development and the primary-secondary vascular transition in Populus deltoides. Ann Bot (Lond) 46: 757–769 [Google Scholar]
- Larson PR. (1994) The Vascular Cambium. Springer-Verlag, Berlin [Google Scholar]
- Larson PR, Isebrand JG. (1974) Anatomy of primary-secondary transition zone in stems of Populus deltoides. Wood Sci Technol 8: 11–26 [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG. (2010) Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J 62: 689–703 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Bürglin TR. (2006) MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol 140: 1142–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel A. (1968) Callus formation and differentiation at an exposed cambial surface. Ann Bot (Lond) 32: 347–359 [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Page RD. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pogany J, Simon E, Katzman R, De Guzman B, Yu L, Trotochaud A, Clark S. (1998) Identifying novel regulators of shoot meristem development. J Plant Res 111: 307–313 [Google Scholar]
- Prigge MJ, Clark SE. (2006) Evolution of the class III HD-Zip gene family in land plants. Evol Dev 8: 350–361 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart N, Zhu T. (2010) Classification SuperViewer Tool w/Bootstrap: The Bio-Array Resource for Plant Functional Genomics. http://bar.utoronto.ca/ (January 10, 2010)
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Ruzin S. (1999) Plant Microtechnique and Microscopy. Oxford University Press, New York [Google Scholar]
- Scarpella E, Marcos D, Friml JÃ, Berleth T. (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J, Baba K, May ST, Palme K, Bennett M, Bhalerao RP, Sandberg G. (2003) Polar auxin transport in the wood-forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proc Natl Acad Sci USA 100: 10096–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. (2004) A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Morelli G, Ruberti I. (1997) DNA-binding specificity of the homeodomain-leucine zipper domain. J Mol Biol 274: 303–309 [DOI] [PubMed] [Google Scholar]
- Spicer R, Groover A. (2010) Evolution of development of vascular cambia and secondary growth. New Phytol 186: 577–592 [DOI] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, et al. (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101: 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. (1995) The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou BH, Evans MMS, Barton MK. (2007) A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19: 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. (2008) KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc Natl Acad Sci USA 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (1999) IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11: 2139–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (2001) Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol 126: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.