Abstract
In the Rhizobium-legume symbiosis, calcium/calmodulin-dependent protein kinase (CCaMK) is a key regulator for both rhizobial infection and nodule organogenesis. Deregulation of CCaMK by either a point mutation in the autophosphorylation site or the deletion of the carboxyl-terminal regulatory domain results in spontaneous nodule formation without rhizobia. However, the underlying biochemical mechanisms are poorly understood. Here, using the kinase domain of CCaMK as a bait in yeast two-hybrid screening, we identify a novel protein, CIP73 (for CCaMK-interacting protein of approximately 73 kD), that interacts with CCaMK. CIP73 contains a Scythe_N ubiquitin-like domain and belongs to the large ubiquitin superfamily. Deletion and mutagenesis analysis demonstrate that CIP73 could only interact with CCaMK when the calmodulin-binding domain and three EF-hand motifs are removed from the kinase domain. The amino-terminal 80 amino acid residues (80–160) of CCaMK are required for interacting with CIP73 in yeast cells. On the other hand, protein pull-down assay and bimolecular fluorescence complementation assay in Nicotiana benthamiana show that the full-length CCaMK could interact with CIP73 in vitro and in planta. Importantly, CCaMK phosphorylates the amino terminus of CIP73 in a Ca2+/calmodulin-dependent manner in vitro. CIP73 transcripts are preferentially expressed in roots, and very low expression is detected in leaves, stems, and nodules. The expression in roots is significantly decreased after inoculation of Mesorhizobium loti. RNA interference knockdown of CIP73 expression by hairy root transformation in Lotus japonicus led to decreased nodule formation, suggesting that CIP73 performed an essential role in nodulation.
In the Rhizobium-legume symbiosis, a new organ, the nodule, is formed to provide a niche for bacterial nitrogen fixation. Initiation and development of nodules relies on a continuous molecular dialogue between the host plant and its rhizobial partner. The legume roots can release metabolites, such as flavonoids or isoflavonoids, which interact with rhizobial NodD transcription factor and activate the expression of rhizobial nodulation (nod) genes that are required for the synthesis and secretion of bacterial lipochitooligosaccharide signaling molecules named Nod factors (NFs; Long, 1989; Lerouge et al., 1990). The structural features of NFs define the specificity of the interaction between the rhizobia and their plant hosts (Lerouge et al., 1990). Purified NFs, at concentrations as low as 10−12 m, are sufficient to trigger many of the early host plant symbiotic responses, including alkalinization of the cytosol, depolarization of the plasma membrane, calcium (Ca2+) influx and spiking, and activation of early nodulin genes (Ehrhardt et al., 1996; Felle et al., 1999; Stacey et al., 2006). NFs also induce altered root hair growth and the mitotic activation of inner cortical cells that leads to the formation of nodule primordium (Foucher and Kondorosi, 2000).
In recent years, forward genetics and map-based cloning approaches in the model legumes Lotus japonicus and Medicago truncatula have led to the identification of a set of host genes essential for NF perception and signal transduction (Oldroyd et al., 2005). Receptor-like kinases, such as NFR1 and NFR5 of L. japonicus (Madsen et al., 2003; Radutoiu et al., 2003, 2007) or LYK3 and NFP of M. truncatula (Amor et al., 2003; Arrighi et al., 2006; Smit et al., 2007), with chitin-binding LysM motifs in their extracellular domain, have been postulated as receptors for the chitin-like NFs. Following perception, a series of genes including putative ion channels (DOESN'T MAKE INFECTIONS1 [DMI1]/POLLUX and CASTOR; Ané et al., 2004; Imaizumi-Anraku et al., 2005), Leu-rich repeat receptor-like kinases (SYMRK/DMI2/NORK; Endre et al., 2002; Stracke et al., 2002), nucleoporins (NUP133 and NUP85; Kanamori et al., 2006; Saito et al., 2007), a Ca2+/calmodulin (CaM)-dependent protein kinase (CCaMK/DMI3; Lévy et al., 2004; Mitra et al., 2004), and a protein of unknown function (CYCLOPS/IPD3; Messinese et al., 2007; Yano et al., 2008) have been identified as the components of the “common symbiosis pathway” that is required for both rhizobial and mycorrhizal endosymbioses (Kistner and Parniske, 2002; Banba et al., 2008). Two GRAS family transcription regulators, NSP1 and NSP2 (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006), NIN (Schauser et al., 1999; Marsh et al., 2007), and an ERF transcription factor, ERN1 (Middleton et al., 2007), participate in the nodulation-specific pathway following the common symbiosis pathway.
Ca2+ spiking consists of periodic peaks and valleys of Ca2+ concentrations in the perinuclear and nuclear regions stimulated by microbial signals. Ca2+ spiking is an important part of the symbiotic signaling pathway (Ehrhardt et al., 1996). Among the seven common symbiosis genes (Kistner and Parniske, 2002) in L. japonicus, SYMRK CASTOR and POLLUX NUP133 and NUP85 act upstream of Ca2+ spiking while CCaMK and CYCLOPS are allocated downstream of Ca2+ spiking (Yano et al., 2008). Proteins that are able to decode Ca2+ spiking have not yet been described in plants. But in animal cells, CaMK II can be activated in a Ca2+ spiking frequency-dependent manner (Hudmon and Schulman, 2002). CCaMK is composed of a Ser/Thr kinase domain, a CaM-binding domain, and three visinin-like EF-hand motifs. Its kinase domain and CaM-binding domain are highly similar to those of mammalian CaMK II. The lily (Lilium longiflorum) CCaMK (Patil et al., 1995) has been studied in great detail at the biochemical level. These studies have shown that the binding of free Ca2+ induces autophosphorylation to enhance the binding of Ca2+/CaM, which in turn promotes substrate phosphorylation (Takezawa et al., 1996; Ramachandiran et al., 1997; Sathyanarayanan et al., 2000). This dual regulation of CCaMK by calcium indicates that CCaMK might recognize complex calcium signatures and serve as a strong candidate for the decoder of Ca2+ spiking in the symbiotic signaling pathway.
Functional analysis has indicated that the CCaMK is a central regulator of nodule organogenesis and rhizobial infection. CCaMK localizes to the nucleus of root cells, corresponding to the site with the highest Ca2+ spiking intensity (Kaló et al., 2005; Smit et al., 2005; Oldroyd and Downie, 2006). In L. japonicus, snf1, a gain-of-function CCaMK point mutation that resides in the autophosphorylation site of the kinase domain, develops spontaneous nodules in the absence of rhizobia (Tirichine et al., 2006). In addition, the deletion of the C-terminal CaM-binding and EF-hand regulatory domains generates the constitutively active CCaMK, resulting in the formation of spontaneous nodules in M. truncatula. However, unlike the mutation in the autophosphorylation site, the deletion mutants are not sufficient to allow bacterial entry. However, both types of mutations can induce nodules when inoculated with rhizobia (Gleason et al., 2006). These results indicate that both the CaM-binding and EF-hand domains are essential for rhizobial infection but are independent for nodule organogenesis. Recently, the epistatic relationships between symbiotic genes show that a gain-of-function CCaMKT265D mutation suppresses loss-of-function common symbiotic genes required for the generation of Ca2+ spiking not only for nodule organogenesis but also for successful infection of rhizobia and arbuscular mycorrhiza (AM) fungi. These findings demonstrate a key role of CCaMK in the coordination of nodule organogenesis and infection (Hayashi et al., 2010; Madsen et al., 2010). However, one question still remains: how does CCaMK differentially activate organogenesis and the infection-related pathway? Further genetic studies and yeast two-hybrid (Y2H) interaction screening have identified a protein, CYCLOPS/IPD3, in both L. japonicus and M. truncatula. CYCLOPS is a phosphorylation target of CCaMK. The CaM-binding and EF-hand domains are required for the CCaMK-CYCLOPS interaction in yeast. The cyclops mutants block symbiotic infection but are dispensable for nodule organogenesis. It has been proposed that CYCLOPS forms an ancient, preassembled signal transduction complex with CCaMK that is specifically required for infection, whereas organogenesis likely requires additional yet-to-be identified CCaMK-interacting partners or substrates (Yano et al., 2008).
In this work, using the sole kinase domain of CCaMK as bait by the Y2H interaction screening approach, we identified a novel protein named CIP73 (for CCaMK-interacting protein of approximately 73 kD) that contains a Scythe_N ubiquitin-like domain and belongs to the large ubiquitin superfamily. Contrary to CCaMK-CYCLOPS interaction, CIP73 can only interact with the kinase domain of CCaMK. The CaM-binding and EF-hand domains inhibit the CCaMK-CIP73 interaction in yeast. Most importantly, like CYCLOPS, CIP73 is a phosphorylation substrate of CCaMK in vitro. Our study suggested that CIP73 may be a new regulator of nodule organogenesis.
RESULTS
Characterization of a CCaMK-Associated Protein from L. japonicus
CCaMK is characterized by an N-terminal Ser-Thr kinase domain, an autoinhibitory domain, a CaM-binding domain, and a C-terminal neural visinin-like domain with three EF hands. Using the full-length CCaMK as a bait, the Y2H system identified a homology of eukaryotic elongation factor 1α in lily (Wang and Poovaiah, 1999) and a novel nuclear protein named IPD3 in Medicago (Messinese et al. 2007). To identify new interacting partners for CCaMK in this study, the kinase domain of CCaMK was used as a bait to screen a root cDNA Y2H library of L. japonicus constructed in the prey vector pGADT7-Rec (Zhu et al., 2008). Interactions were tested in repeated experiments on stringent selective medium (synthetic dextrose [SD]-Trp-Leu-His-Ade). Two independent positive yeast colonies were revealed, and the two cDNAs showed identical nucleotide sequence coding the C-terminal region (413–691 amino acids) of a gene.
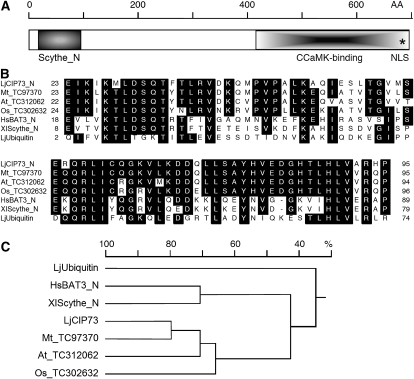
Using 5′ RACE, a full-length cDNA (GenBank accession no. GU980966) was identified. It contained an open reading frame of 2,076 nucleotides encoding a protein of 691 amino acids with a predicted molecular mass of approximately 73 kD (Supplemental Fig. S1). This protein was designated as CIP73 (Fig. 1A). The protein sequence analysis revealed that CIP73 contained an N-terminal ubiquitin homology region that is similar to the N terminus of Scythe in Xenopus and Bat3 (HLA-B-associated transcript 3) in human (Banerji et al., 1990; Thress et al., 1998). This domain was named the Scythe_N domain (Fig. 1A). The proteins containing the Scythe_N domain are widely present in animals and regulate apoptosis in a variety of settings (Thress et al., 1998; Desmots et al., 2005). Besides the Scythe_N ubiquitin-like domain, CIP73 bears only limited resemblance to identified proteins with well-known functions. PSORT (Horton et al., 2007) analysis revealed a potential nuclear localization sequence (NLS; 686–689, KRQK) located in the C terminus.
Figure 1.
CIP73 contains a Scythe_N ubiquitin-like domain and belongs to the large ubiquitin superfamily. A, Schematic illustration of the CIP73 protein. The deduced amino acid sequence of CIP73 contains 691 amino acid residues with a calculated molecular mass of approximately 73 kD. Notable features include the Scythe_N (Scythe is also known as BAT3) ubiquitin-like domain at the N terminus (21–93) and a putative NLS (686–689) shown by the asterisk. The CCaMK-binding region identified in the original Y2H screening (414–691) is in the CIP73 C-terminal region. B, Multiple sequence alignment of the N-terminal ubiquitin-like domain of CIP73 and the homologous sequence from Medicago (TC97370), Arabidopsis (TC312062), rice (TC302632), human BAT3_N (NP_542433), Xenopus Scythe_N (NP_001080008), and L. japonicus ubiquitin (AW720576). The numbers on the left and right indicate the positions of amino acids. C, Homology tree of the N-terminal ubiquitin-like domain of CIP73 homologs, Scythe_N, and ubiquitin. The tree was constructed using the DNAMAN software (version 5.2.2; Lynnon Biosoft). Note that the N-terminal ubiquitin-like domain of CIP73 is more closely related to the animal Scythe_N domain than to LjUbiquitin.
A search for CIP73 sequences in plant databases identified CIP73-like proteins from M. truncatula, Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), Ricinus communis, and Populus trichocarpa (Supplemental Fig. S2). Proteins containing the Scythe_N-like domain are also widely present in plant genomes, but their functions remain unknown. Homology analysis showed that the N-terminal ubiquitin-like domain was more closely related to the Scythe_N domain in animals than to LjUbiquitin (Fig. 1C).
CIP73 Interacts with CCaMK in the Y2H System
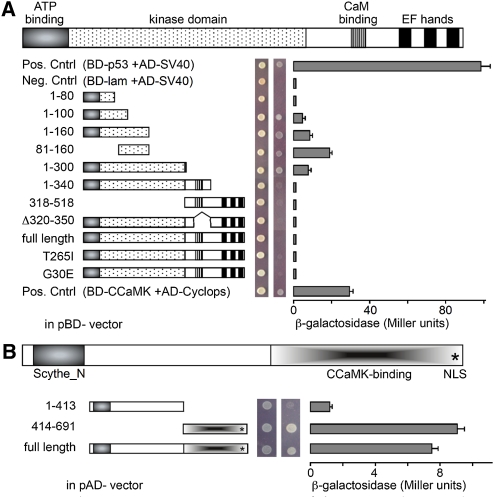
To confirm the interaction between CIP73 and CCaMK, a series of CCaMK deletions and point mutants were fused to the GAL4 DNA-binding domain (BD). A full-length CIP73, CIP73-N (residues 1–413), and CIP73-C (residues 414–691) were fused to the GAL4 activation domain (AD; Fig. 2). Using the Y2H approach, we detected that CIP73 only interacted with the N terminus of CCaMK (residues 1–300) but not with the full-length CCaMK or the fragment containing the CaM-binding domain (residues 1–340). Moreover CIP73 did not interact with several CCaMK mutants: a point mutation in the autophosphorylation site (T265I) that causes nodule development in the absence of rhizobia (Tirichine et al., 2006), a kinase-defective mutant (G30E), and a CaM-binding deleted mutant (Fig. 2A). The further analysis of deletion constructs revealed that the N-terminal 80 amino acid residues (80–160) of CCaMK was sufficient to interact with CIP73 in the Y2H system (Fig. 2B). However, CCaMK1-300 interacted with CIP73-C and did not interact with CIP73-N (Fig. 2B). The full-length CIP73 and the N terminus of CIP73 fused to the GAL4 BD showed autoactivation in yeast (Supplemental Fig. S3A). The results of the Y2H analysis revealed that CIP73 could form a homodimer and that CIP73-C (414–691) is necessary for self-interaction (Supplemental Fig. S3B).
Figure 2.
CIP73 interacts with CCaMK in yeast. A, Y2H assays for interaction between CCaMK and CIP73. The kinase domain (1–300) was used in the initial isolation of CIP73 from the Y2H Library. SD-Leu-Trp medium was used for testing successful mating, and SD-Leu-Trp-His-Ade medium was used for testing the interaction. The strength of interaction was measured through the β-galactosidase activity. The combination of BD-53/AD-SV40 or BD-CCaMK/AD-CYCLOPS (Yano et al., 2008) was used as a positive control, and BD-Lam/AD-SV40 was used as a negative control (Clontech). CIP73 could only interact with CCaMK when the CaM-binding domain and three EF-hand motifs were removed from the kinase domain. The N-terminal 80 amino acid residues (80–160) of CCaMK are sufficient for interacting with CIP73 in yeast cells. B, The kinase domain of CCaMK (1–300) can interact with full-length CIP73 and CIP73-C (414–691) but cannot interact with CIP73-N (1–413). [See online article for color version of this figure.]
CIP73 Interacts with CCaMK in Vitro and in Planta
We have demonstrated that the full-length CCaMK could not interact with CIP73 in yeast cells (Fig. 2A). Next, we tested whether the full-length CCaMK can interact with CIP73 by in vitro protein pull-down assay and bimolecular fluorescence complementation (BiFC) assay. In most cases, the interaction between a protein kinase and its substrate is transient and difficult to detect in the Y2H system unless a mutated kinase is used (Rodriguez Milla et al., 2006; Uno et al., 2009). However, protein pull-down assay or immunoprecipitation could detect such interactions (Patharkar and Cushman, 2000; Lee et al., 2003).
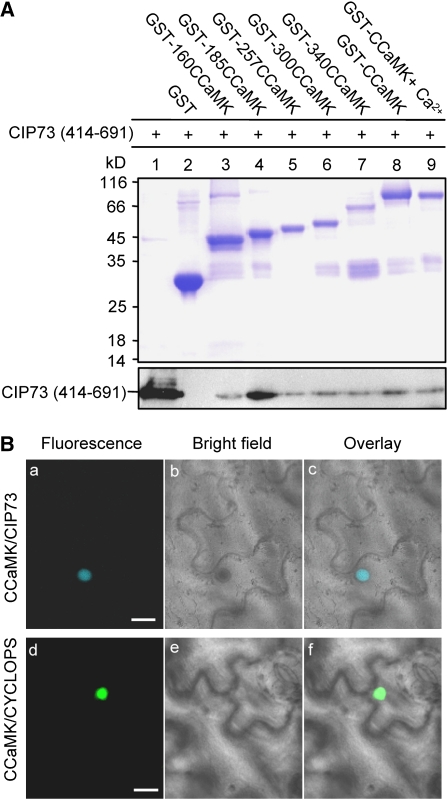
The full-length CCaMK and a series of CCaMK deletion mutants were expressed as glutathione S-transferase (GST) fusion proteins. CIP73-C (414–691) was expressed with 6× His tag in Escherichia coli for the interaction assays in vitro. The His-tagged CIP73-C protein was incubated with different bead-bound GST-CCaMK truncated proteins or GST alone. After washing with buffer, proteins retained to the beads were separated by SDS-PAGE. Immunoblot analysis showed that His-CIP73-C associated with the beads, demonstrating that CIP73-C was able to interact with all deletion mutants and the full-length CCaMK (Fig. 3A).
Figure 3.
CIP73 interacts with CCaMK in vitro and in planta. A, In vitro protein pull-down assay for testing the interaction between CCaMK and CIP73. His-tagged CIP73 (414–691) was incubated with the immobilized GST-CCaMK fusion protein or GST alone in the presence of EGTA (lanes 2–8) or Ca2+ (lane 9). After washing, the proteins were separated by SDS-PAGE and visualized by staining with Coomassie Brilliant Blue R250 (top). A similar gel was used for immunoblot using anti-His tag antibody (bottom). B, Interaction of CCaMK and CIP73 in planta. N. benthamiana leaves were cotransformed with SCFPC::CCaMK and CIP73::SCFPN (a–c) or with SCFPC::CCaMK and CYCLOPS::VenusN (d–f). Left images show fluorescence signal of the cells (a and d); middle images show the cell architecture (b and e); right images show the overlays of fluorescence and bright-field images (c and f). Bars = 20 μm.
It has been shown that CCaMK interacted with CYCLOPS in the nucleus of Nicotiana benthamiana using the BiFC assay (Yano et al., 2008). Taking advantage of a multicolor BiFC system (Waadt et al., 2008), we can simultaneously visualize the multiple protein interactions in the same cell. The constructs SCFPC::CCaMK and CIP73::SCFPN were cotransformed and transiently expressed in N. benthamiana epidermis cells to confirm CIP73 protein interactions in planta. Strong cyan fluorescence was observed in the nucleus when CIP73 and CCaMK fused to the C-terminal and N-terminal halves of cyan fluorescent protein (CFP), respectively (Fig. 3B, a–c). The SCFPC::CCaMK and CYCLOPS::VenusN constructs were coexpressed in N. benthamiana epidermis cells, and strong green fluorescence was observed in the nucleus (Fig. 3B, d–f). However, only weak marginal background fluorescence was detected when all three constructs were infiltrated at the same time (data not shown). These results indicated that CCaMK could interact with either CIP73 or CYCLOPS in living plant cells. However, whether they could form a ternary complex remained to be tested.
CIP73 Expression and Subcellular Localization
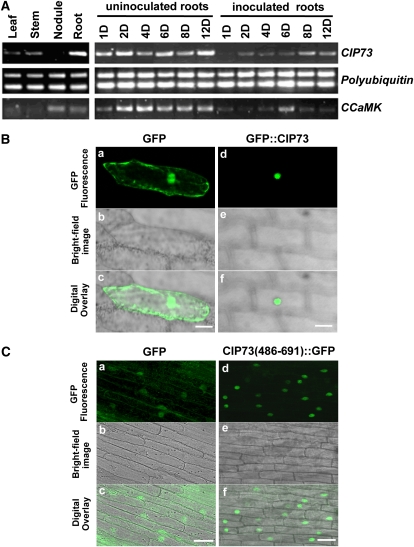
Using semiquantitative reverse transcription (RT)-PCR, we examined the CIP73 and CCaMK mRNA levels in different organs of L. japonicus. Both CIP73 and CCaMK mRNA were preferentially expressed in roots as compared with leaves and stems. But unlike CCaMK, which had a high mRNA level in nodules, the CIP73 mRNA level was very low in nodules (Fig. 4A, left). After inoculation with Mesorhizobium loti, CIP73 exhibited an expression pattern very similar to that of CCaMK, with a transient decrease in expression after inoculation (Fig. 4A, right). This postinoculation decrease was also observed in IPD3 (Messinese et al., 2007). These results indicated that CIP73 mRNA was constitutively expressed in uninoculated roots and down-regulated by inoculation of M. loti.
Figure 4.
CIP73 expression and subcellular localization. A, CIP73 and CCaMK expression was investigated by semiquantitative RT-PCR before and after M. loti inoculation using polyubiquitin as an internal control. Uninoculated roots (1D–12D) are shown as controls for the same periods post inoculation with M. loti. B, Nuclear localization of GFP::CIP73 in onion epidermal cells. The control GFP gene (a–c) and GFP::CIP73 fusion gene (d–f), delivered by particle bombardment, were transiently expressed in onion epidermal cells and observed with a laser-scanning confocal microscope 24 h after bombardment. Note that the control GFP is distributed both in the cytoplasmic and nuclear compartments, whereas GFP::CIP73 only localizes in the nucleus. Top images show GFP fluorescence of the cells (a and d); middle images show the cell architecture (b and e); bottom images show the overlays of GFP and bright-field images (c and f). C, The C terminus of CIP73 (486–691) shows strong nuclear localization in epidermal and cortex cells of transformed hairy roots. Bars = 50 μm for B and 20 μm for C. [See online article for color version of this figure.]
Sequence analysis showed that there was a putative NLS segment located in the C terminus of CIP73 (Fig. 1A). This suggested that CIP73 might be located in the nucleus of living cells. To determine the subcellular localization of CIP73, a construct containing CIP73 fused in-frame with GFP (GFP::CIP73) was made under the control of the cauliflower mosaic virus (CaMV) 35S promoter. This construct was transiently expressed in onion (Allium cepa) epidermal cells via particle bombardment. Confocal laser-scanning microscopic examination revealed that cells expressing the control 35S::GFP exhibited strong fluorescence both in the cytoplasmic and nuclear compartments (Fig. 4B, a–c). In contrast, when fused with the full-length CIP73 protein, the GFP signal was mainly concentrated in the nuclear compartment (Fig. 4B, d–f). These data indicated that CIP73 localized to the nucleus of onion epidermal cells.
To examine the intracellular distribution of CIP73 in L. japonicus roots, we transformed the 35S::GFP and CIP73 (486–691)::GFP constructs into L. japonicus by means of Agrobacterium rhizogenes. GFP localization was scored on 3-week-old transformed hairy roots. Similar to the results obtained with onion epidermal cells, 35S::GFP hairy roots exhibited GFP fluorescence in both the cytoplasmic and nuclear compartments (Fig. 4C, a–c), while GFP signal of the CIP73 (486–691)::GFP hairy roots was mainly concentrated in nucleus in L. japonicus (Fig. 4C, d–f).
CIP73 Is a New Phosphorylation Substrate of CCaMK in Vitro
It has been demonstrated that CCaMK was able to phosphorylate CYCLOPS in vitro (Yano et al., 2008). This indicated that CIP73 might be another potential phosphorylation substrate for CCaMK. To further investigate the biochemical functions of the interaction between CCaMK and CIP73, we performed the phosphorylation and autophosphorylation assay between CCaMK and CIP73 in vitro. The nonspecific substrate casein was used as a control. The N-terminal (1–413 residues) and C-terminal (414–691 residues) regions of CIP73 were expressed and purified as 6× His-tagged fusion proteins. Our result showed that the N terminus of CIP73, which did not interact with CCaMK in the Y2H assay, was phosphorylated by CCaMK. On the other hand, the C terminus of CIP73, which showed interaction with CCaMK in the Y2H assay, was not phosphorylated by CCaMK (Fig. 5A). These results were quite different from what was observed with CYCLOPS. The N terminus of CYCLOPS, which interacted with CCaMK in the Y2H assay, was phosphorylated by CCaMK. The C terminus of CYCLOPS, which did not interact with CCaMK in the Y2H assay, was not phosphorylated by CCaMK (Yano et al., 2008). Thus, our data suggested that the biological function of the interaction between CCaMK and CIP73 was different from that of CCaMK and CYCLOPS both in vitro and probably also in planta, not only in yeast cells.
Figure 5.
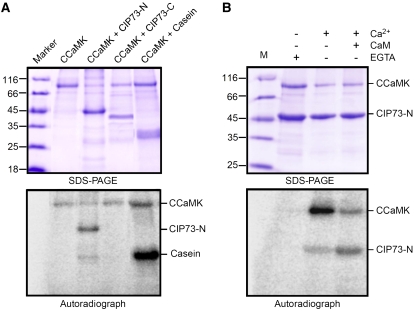
Phosphorylation of CIP73 by CCaMK in vitro. A, Autophosphorylation and phosphorylation assay of CCaMK in the presence of Ca2+ and CaM. CCaMK can phosphorylate CIP73-N (1–413) but not CIP73-C (414–691). Casein served as a positive control. B, Phosphorylation of CIP73-N (1–413) in the presence (+) or absence (−) of Ca2+ EGTA and CaM. Bottom images show autoradiographs of kinase assays, and top images show Coomassie Brilliant Blue staining of the same gels. The autophosphorylation activity of CCaMK was increased in the presence of Ca2+, and substrate (CIP73) phosphorylation was accelerated by the addition of Ca2+/CaM. [See online article for color version of this figure.]
The autophosphorylation activity of CCaMK was increased in the presence of Ca2+ alone, whereas substrate (CIP73) phosphorylation was accelerated by the addition of CaM (Fig. 5B). These phosphorylation effects were very similar to CYCLOPS and myelin basic protein, which were phosphorylated by CCaMK. Thus, these data suggested that CIP73 is a new phosphorylation substrate of CCaMK in vitro.
CIP73 RNA Interference Roots Are Impaired in Nodulation
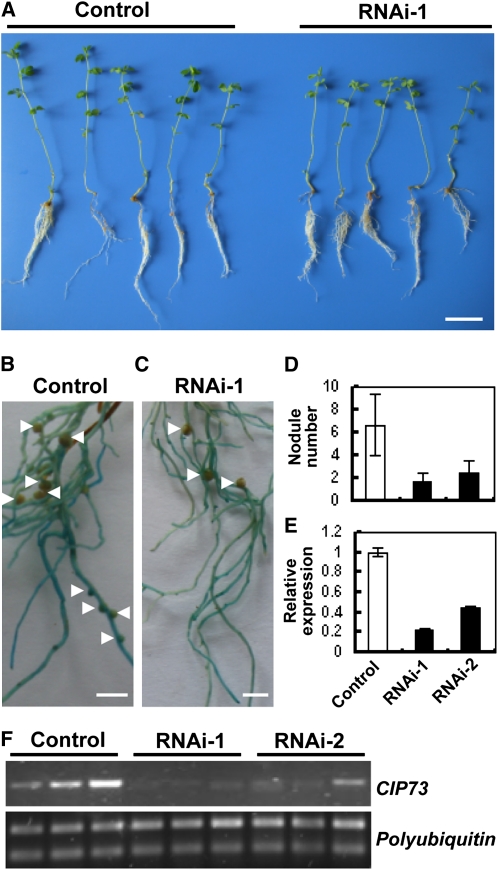
In order to explore the possible function of CIP73 during the nodulation process, RNA interference (RNAi) experiments were carried out. We transformed the L. japonicus hairy roots with two CIP73-specific RNAi constructs under the control of the CaMV 35S promoter by A. rhizogenes LBA1334, as described in “Materials and Methods.” The transformed hairy roots were selected by GUS staining, and nontransformed roots were excised from the seedling. The identified plants were then transferred to pots filled with vermiculite and sand (1:1) and grown in a growth chamber. After 5 d, the transgenic hairy roots were inoculated with M. loti MAFF303099 and grown for 4 weeks for observation of growth and nodulation phenotypes (Fig. 6, A–C). Expression of CIP73 in transgenic hairy roots was examined by real-time RT-PCR. The level of CIP73 transcripts in the transgenic roots was 20% to 50% of that of the control roots (Fig. 6E). The average number of nodules formed on control hairy roots was 6.7, while that on the two RNAi-transformed plants RNAi-1 and RNAi-2 was 1.7 and 2.5, respectively. Statistical analysis showed that the nodule number with the two RNAi constructs was significantly different from the controls (P < 0.03, n = 17 and P < 0.01, n = 19, respectively; Fig. 6D). Also, we observed that approximately 30% of the CIP73 RNAi roots did not develop any nodule, whereas 100% of the control roots contained nodules.
Figure 6.
Nodulation phenotype of CIP73-specific RNAi-transformed hairy roots. A, Five representative images of plants transformed with the CIP73 RNAi-1 construct compared with plants that were transformed with an empty vector (control). The plants were inoculated with M. loti MAFF303099 and grown in the absence of nitrogen, and the photographs were taken at 4 weeks after inoculation. B and C, Nodulation phenotype of control vector transgenic roots (B) and RNAi-1 transgenic roots (C). The transgenic hairy roots were further confirmed by GUS staining before photographing. Arrowheads indicate nodule positions on the roots. D, Total nodule number of vector control and two RNAi construct transgenic roots determined 4 weeks after inoculation with M. loti MAFF303099. E and F, Estimation of CIP73 transcripts in transgenic hairy roots by quantitative real-time RT-PCR (E) and RT-PCR (F). Bars = 2 cm for A and 2 mm for B and C. [See online article for color version of this figure.]
As root growth defects may influence nodule number, we examined the growth phenotype of CIP73 RNAi roots that had not been inoculated with M. loti under the same conditions as the nodulation assay. As shown in Supplemental Figure S5, the growth of CIP73 RNAi roots was not significantly different with the vector control. We also examined the growth of the CIP73 RNAi and control roots on half-strength Murashige and Skoog (MS) and Broughton and Dilworth agar plates, and no obvious difference was observed (data not shown).
Rhizobial and Mycorrhizal Infection Phenotypes of CIP73 RNAi Roots
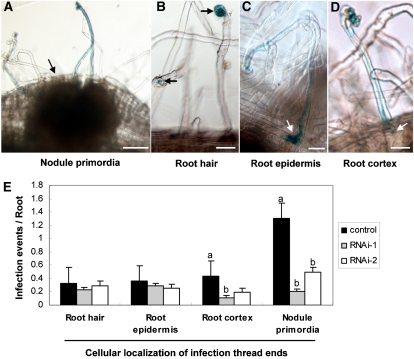
CCaMK is suggested to be involved not only in the rhizobial infection through infection threads (ITs) but also in nodule organogenesis. CYCLOPS is also known to be required for the initiation of ITs but not nodule organogenesis (Yano et al., 2008). To determine how the decreased nodule formation was mediated by CIP73 RNAi, we individually analyzed the rhizobial infection process by using lacZ-labeled M. loti to visualize IT formation at 7 d post inoculation. In CIP73 RNAi roots, ITs could initiate from curled root hair tips (Fig. 7B), grew through well-elongated root hair to root epidermis (Fig. 7C), and further penetrated into the root cortex (Fig. 7D). Also, some ITs could reach into the nodule primordia tissues and rhizobia could release from ITs into the nodule cells. However, the number of ITs that reached the cortex and nodule primordia was significantly reduced compared with the control roots (Fig. 7E). These data indicated that the impaired nodulation in CIP73 RNAi roots is probably because of cortical events associated with nodule organogenesis but not rhizobial infection.
Figure 7.
Analysis of rhizobia infection of CIP73 RNAi hairy roots. A to D, Histochemical staining of lacZ-labeled M. loti strain to follow the infection events at 7 d post inoculation. Representative ITs observed in vector control roots (A) and CIP73 RNAi-1 roots (B–D) are shown (blue). Arrows indicate the cellular localization of IT ends. E, Frequencies of the infection events per root. The data are presented as 15 individual transgenic plants for each construct and randomly scored in four roots between 4 to 6 cm per plant. Different letters above the bars indicate significant differences (P < 0.05, t test) between pairwise comparisons. Bars = 50 μm for A and 25 μm for B to D. [See online article for color version of this figure.]
As both CCaMK and CYCLOPS also control mycorrhization, we next determined the mycorrhization phenotype of CIP73 RNAi roots. In the CIP73 RNAi-1 and RNAi-2 roots, 2 weeks after inoculation with Glomus intraradices, the AM fungi penetrated into the outer cell layers, colonized the root cortex, and formed arbuscules and vesicles, and the hyphal, arbuscular, and vesicular colonizations did not differ from the vector control (Supplemental Fig. S6). These results suggested that RNAi knockdown of CIP73 expression by hairy root transformation had no obvious effect on AM fungal colonization.
DISCUSSION
In the Rhizobium-legume symbiosis, CCaMK is a central regulator of nodule organogenesis and rhizobia infection (Gleason et al., 2006; Tirichine et al., 2006). CYCLOPS, an interacting partner with CCaMK, is a phosphorylation substrate of CCaMK (Yano et al., 2008). In this report, we identified a novel CCaMK-interacting protein, CIP73, in L. japonicus. CIP73 contained a Scythe_N ubiquitin-like domain. Similar to CYCLOPS, CIP73 was also a phosphorylation target of CCaMK in vitro (Fig. 5A). CaM could enhance the level of substrate phosphorylation in the presence of Ca2+ (Fig. 5B). However, additional work is needed to determine the biologically relevant effect of this phosphorylation event. The CCaMK autophosphorylation defect mutant (T265I) led to spontaneous nodule development. This mutant can also phosphorylate myelin basic protein in a Ca2+/CaM-dependent manner, although the activity was lower as compared with the wild type (Tirichine et al., 2006). The deletion mutants DMI3/CCaMK (1–311) and DMI3/CCaMK (1–326), which also led to spontaneous nodulation, phosphorylated glycogen synthase peptide constitutively independent of Ca2+ and CaM (Gleason et al., 2006). Although both T265I and the deletion mutant can activate nodule morphogenesis, only the T265I mutant is fully functional to allow bacterial entry (Gleason et al., 2006). These observations prompted us to test whether these mutants were functionally different in the phosphorylation of CYCLOPS or CIP73. Determining the differences in the biochemical mechanisms will help to illustrate the functional significance of the phosphorylation of CIP73 and CYCLOPS by CCaMK in the nodulation signaling pathway.
Unlike the MtIPD3 screening, which used the full-length DMI3 CCaMK as bait, CIP73 was isolated with the kinase domain as bait. Using a series of CCaMK deletions and point mutants, we demonstrated that the interaction between CCaMK and CIP73 was quite different from the interaction between CCaMK and CYCLOPS in yeast cells. The site between the CaM-binding domain and the second EF hand was required for the CCaMK-CYCLOPS interaction in yeast. In contrast, CIP73 could interact with CCaMK only when both the CaM-binding domain and three EF-hand motifs were removed from the kinase domain. This was equivalent to a constitutively active kinase resulting in spontaneous nodule development in the absence of rhizobia (Gleason et al., 2006). CCaMK was required for both nodule organogenesis and rhizobial infection. It has been shown that the CaM-binding and EF-hand domains are essential for rhizobial infection but not for nodule organogenesis. CYCLOPS was also identified to be specifically required for rhizobial infection but not for nodule organogenesis (Yano et al., 2008). These biologic functions of CCaMK and CYCLOPS might be partially explained by their interaction identified in yeast cells. To this extent, CIP73 could only interact with the kinase domain of CCaMK, indicating that CIP73 is possibly required for nodule organogenesis.
Although the full-length CCaMK did not interact with CIP73 in yeast cells, their interaction was detected using in vitro protein pull-down assay and BiFC assay in N. benthamiana (Fig. 3). Both CIP73 and CYCLOPS could interact with CCaMK within the nucleus of heterozygous N. benthamiana cells. However, whether both the CCaMK-CYCLOPS and CCaMK-CIP73 interactions could occur simultaneously in a single cell remained to be tested. To address this question, the multicolor BiFC approach was used (Waadt et al., 2008). Coinfiltration of A. tumefaciens strains carrying SCFPC::CCaMK and CIP73::SCFPN constructs into N. benthamiana leaves resulted in the formation of cyan BiFC complexes in the nucleus, whereas coexpression of SCFPC::CCaMK and CYCLOPS::VenusN resulted in the formation of green BiFC complexes that were also localized in the nucleus. However, when all three constructs were infiltrated at the same time, only weak marginal background fluorescence was detected (data not shown). It is possible that CIP73 and CYCLOPS were competing for CCaMK. This results in a reduced fluorescence emission that could not be detected by the system. Alternatively, the undetectable signal might be due to the low transformation efficiency for this triple infiltration.
In M. truncatula, CCaMK (DMI3) with both C-terminal and N-terminal GFP fusions, driven by the 35S promoter, show strong nuclear localization in epidermal root cells (Kaló et al., 2005), and a similar localization was observed in root hair cells when the N-terminal GFP fusion was driven by the DMI3 promoter (Smit et al., 2005). CIP73 has a putative NLS segment located in the C terminus (Fig. 1A), suggesting that CIP73 might also be located in the nucleus. We first attempted to detect CIP73 with C-terminal GFP fusion under the control of the 35S promoter in L. japonicus hairy roots and onion epidermal cells. However, no fluorescence was detected in these cells. In contrast to the full-length CIP73, the C terminus (486–691) of CIP73 with C-terminal GFP fusion showed strong fluorescence in nuclei both in L. japonicus hairy roots (Fig. 4C, d–f) and onion epidermal cells (Supplemental Fig. S4, D–F). Murakami et al. (2006) also reported that no fluorescence could be detected in L. japonicus hairy roots for NSP2 with a C-terminal GFP fusion. On the other hand, CIP73 with an N-terminal GFP fusion in pMON30060 vector showed strong nuclear localization in onion epidermal cells (Fig. 4B, d–f). At this stage, we cannot explain this strange phenomenon. Continued work to test CIP73 with C- and N-terminal GFP fusions in L. japonicus root or other organs by regenerating plants via somatic embryogenesis is likely to solve the problem.
mRNA examination in different organs showed that the expression of CIP73 was organ regulated. The mRNA level was much higher in uninoculated roots as compared with leaves and stems. This suggested a potential role of CIP73 in root-related processes. However, the expression of CIP73 was very low in nodules as compared with roots. This was in contrast with CCaMK, which showed similar expression in both uninoculated roots and nodules. Interestingly, the expression level was significantly decreased after inoculation with M. loti for both CIP73 and CCaMK (Fig. 4A). This implied that CIP73 might be functional downstream of CCaMK and probably in the negative feedback in the NF signal transduction pathway (Oldroyd et al., 2001). RNAi knockdown of CIP73 expression by hairy root transformation in L. japonicus led to decreased nodule formation (Fig. 6). These data suggested that CIP73 performed an essential role in nodulation. On the other hand, except for regulating rhizobial and mycorrhizal symbioses, little is known about the biological role of CCaMK in plants. But the preferential expression of CCaMK in developing anthers and root tips (Poovaiah et al., 1999) suggested that CCaMK may play a role in mitosis and meiosis (Yang and Poovaiah, 2003). As an interacting partner, CIP73 may also be involved in these processes in addition to its role in the development of the nodule. We are currently generating stable transformants with the CIP73 RNAi constructs by conventional A. tumefaciens-mediated hypocotyl transformation to study the biological functions of CIP73 in more detail.
A thorough search of the available protein databases revealed that CIP73 bears limited resemblance to proteins with well-known functions. Analysis of the CIP73 sequence identified an N-terminal region with substantial homology to the Scythe_N domain belonging to the UBQ superfamily. Scythe protein (also known as Bat3) is an apoptotic regulator that is highly conserved in eukaryotes and contains a ubiquitin-like domain near its N terminus. Scythe binds Reaper, a potent apoptotic inducer, and Scythe/Reaper are thought to signal apoptosis, in part through regulating the folding and activity of apoptotic signaling molecules (Thress et al., 1998; Desmots et al., 2005). However, except for the Scythe_N-like domain, CIP73 contained the downstream sequence with no homology to Scythe protein (Fig. 1A). In addition, the biochemical mechanisms of the Scythe_N domain alone in apoptosis have not been elucidated. So there are very limited clues for us to test whether CIP73 serves as an apoptotic regulator like Scythe.
The Scythe_N ubiquitin-like domain indicates that CIP73 is a member of the large ubiquitin family proteins. The ubiquitin family proteins are generally divided into two groups: type Ι ubiquitin-like modifiers and type II ubiquitin-like domain proteins (UDPs). CIP73 can be classified into type II UDPs embedding ubiquitin-like domains, since it lacks the type Ι-specific conserved C terminus diglycine motif, which can form conjugates with other proteins. There is emerging evidence indicating an exciting link between the functions of UDPs, proteasome, chaperones, and the apoptotic pathway (Jentsch and Pyrowolakis, 2000). At least three UDPs, the DSK2 relative PLIC2/Chap1 BAG-1, and BAT3/Scythe/BAG6 have shown affinities for chaperones of the Hsp70 family (Kaye et al., 2000; Thress et al., 2001; Sasaki et al., 2008; Corduan et al., 2009). We have identified Hsp70-interacting protein as an interacting partner for CIP73 using Y2H interaction screening (data not shown). Nowadays, there is increasing evidence about the ubiquitin-mediated proteolysis during nodule development (Kondorosi et al., 2005; Kiss et al., 2009; Yano et al., 2009). All of these findings implied that CIP73 might mediate protein folding/degradation pathways in the regulation of nodulation signal transduction cascades.
Although proteins containing the Scythe_N domain are widely present in all sequenced eukaryotic organisms, the overall structure of CIP73 appears to be unique to plants, since genes coding for proteins with similar structures could not be found in other organisms in the databases. Genes homologous to CIP73 are clearly present in the moss Physcomitrella patens and in higher plants like R. communis, P. trichocarpa, Vitis vinifera, Arabidopsis, and rice. But amino acid sequences with high overall similarity to CIP73 were only found in legume plants (Supplemental Fig. S2). The Medicago CIP73 orthologs have overall 72% sequence identity with CIP73 and have at least two isoforms (J.M. Ané, personal communication). The rice CIP73 orthologs, with the same exon and intron structure to the Medicago ortholog, only have overall 30% sequence identity with CIP73 and also have two isoforms (data not shown).
MATERIALS AND METHODS
Plant Materials
Seeds of Lotus japonicus MG-20 were surface sterilized in 75% ethanol for 2 min and followed by 8 min in 2% sodium hypochlorite and six washes with sterile water. The seeds were left to germinate for 48 h at 22°C on sterile water-soaked filter paper in petri dishes in the dark. Seedlings were subsequently planted in pots with sterile sands supplemented with nitrogen-free Fahraeus medium. The seeds were grown in a growth chamber maintained at 22°C with a 16/8-h day/night cycle. Five-day-old seedlings were inoculated with approximately 107 colony-forming units mL−1 Mesorhizobium loti NZP2037 or MAFF303099. Various organs were collected from plants 2, 4, 6, 8, and 12 d after rhizobial inoculation and were immediately frozen in liquid nitrogen. Nicotiana benthamiana plants were grown in a growth chamber at 22°C with a 16/8-h day/night cycle.
Y2H Library Construction
Total RNA was isolated from the equally mixed roots collected 2, 4, 6, 8, and 12 d after rhizobial inoculation using the TRIzol reagent (Invitrogen). Total RNA (2 μg) was reverse transcribed into single-stranded cDNAs using oligo(dT) as the primer. The cDNAs were size fractionated using BD Matchmaker Library Construction and Screening Kits (BD Biosciences-Clontech). cDNA fragments longer than 500 bp were cotransformed with linearized pGADT7-Rec into yeast AH109 cells. The transformants were selected on SD-Leu medium according to the manufacturer’s instruction. The transformation efficiency was approximately 2 × 106 colony-forming units per 3 μg of pGADT7-Rec.
Y2H Library Screening
For the Y2H screenings, bait constructs were cloned into the vector pGBKT7. The partial cDNA corresponding to the CCaMK (GenBank accession no. AM230793) kinase domain was fused in-frame with the GAL4 BD into the pGBKT7 vector. Bait constructs were transformed into yeast strain Y187 by the lithium acetate method. Screening of interaction clones was carried out via mating according to the manufacturer’s instructions (Clontech). A total of 10 million transformants from the cDNA library were screened for growth on the stringent SD-Leu-Trp-His-Ade dropout medium. Positive clones were confirmed by assaying for another yeast reporter, LacZ, via β-galactosidase assays. To validate observed interactions, prey plasmids were rescued, analyzed by restriction digestion, and transformed again into AH109. Yeast cells with preys were mated one-on-one in parallel against the yeast Y187 expressing the target baits or the negative control plasmid pGBKT7-Lam (Clontech).
β-Galactosidase Assay
Yeast cells grown in liquid SD selection medium were pelleted and washed twice with Z-buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1.0 mm MgSO4, pH 7.0). The cells were resuspended in 100 μL of Z-buffer and permeabilized by three freeze-thaw cycles with liquid nitrogen and a 37°C water bath. Cell extracts were added to 0.7 mL of Z-buffer containing 50 mm β-mercaptoethanol and 160 μL of O-nitrophenyl β-d-galactopyranoside (4 mg mL−1 in Z-buffer). After incubation at 30°C for 30 min or until the yellow color appeared, the reaction was terminated by the addition of 0.4 mL of 1.0 m Na2CO3. The reaction mixture was centrifuged for 10 min at 13,000 rpm to remove cell debris. β-Galactosidase activity in the supernatant was measured at an optical density at 420 nm and expressed in Miller units.
Purification of CCaMK and CIP73 Protein
The full-length and truncated CCaMK cDNA fragments were PCR amplified and inserted in-frame at the SmaΙ/XhoΙ sites of pGEX-6P1 vector (Amersham Pharmacia Biotech) for GST fusion. The full-length CCaMK cDNA fragment was inserted into the XbaΙ/HindΙΙΙ sites of pMAL-c2X vector (New England Biolabs) for maltose-binding protein (MBP) fusion. The two truncated CIP73 cDNA fragments (1–413 and 414–691) were PCR amplified and cloned into pET-28a vector (Novagen) using EcoRΙ and SalΙ sites. For protein expression, Escherichia coli BL21-Codon Plus (DE3)-RIL (Stratagene) harboring the plasmids were induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside in Luria-Bertani broth for 5 h at 22°C. The GST fusion proteins were purified using glutathione-Sepharose 4B columns (GE Healthcare) using standard procedures. The MBP fusion proteins were purified using amylose resin (New England Biolabs). The His-tagged proteins were purified using nickel-agarose beads (Qiagen) under native conditions according to the manufacturer’s instructions and eluted with a buffer solution (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, and 200 mm imidazole, pH 8.0). Purified proteins were desalted by dialysis in phosphate-buffered saline and concentrated with polyethylene glycol-8000 powder.
In Vitro Protein-Protein Interaction
To assay the interaction between CCaMK and CIP73, GST-tagged CCaMK or GST alone was bound to glutathione-Sepharose 4B beads. The beads were incubated with 2 μg of purified His-CIP73 protein in 1 mL of interaction buffer (20 mm Tris-HCl, 100 mm KCl, 2 mm MgCl2, and 5% glycerol, pH 8.0) in the presence of 0.5 mm CaCl2 or 2.5 mm EGTA. The reaction was incubated for 1 h on ice with gentle shaking. The Sepharose beads were collected and washed three times in 1 mL of 20 mm Tris-HCl buffer containing 300 mm NaCl and either 0.5 mm CaCl2 or 2.5 mm EGTA. The retained proteins were eluted by boiling for 5 min in 1× SDS sample buffer (2% SDS, 29.1 mmol L−1 Tris, pH 6.8, 10% glycerol, and 0.01% bromphenol blue). Samples were analyzed by 12% SDS-PAGE followed by immunoblotting with the anti-His tag antibody or visualized by staining with Coomassie Brilliant Blue R250.
In Vitro Kinase Assay
A full-length cDNA of CaM (TC8422) for L. japonicus was amplified by RT-PCR using the primers 5′-GGACTGCCATATGGCAGATCAACTCAC-3′ and 5′-CGGAATTCTCACTTGGCCATCATGACC-3′. The amplified fragment was cloned into the NdeI/EcoRΙ site of pET28a vector and expressed as N-terminal 6x His-tagged fusion protein in E. coli strain BL21. Expression protein was affinity purified via nickel-agarose beads (Qiagen) under native conditions and eluted with a buffer solution (50 mm Tris-HCl, 200 mm NaCl, and 200 mm imidazole, pH 8.0). Purified protein was desalted and concentrated by Microcon Ultracel YM-3 centrifugal filter devices (Millipore). Protein concentrations were determined by SDS-PAGE using bovine serum albumin as a standard (Fermentas). The kinase assays were performed at 25°C for 30 min in 40 μL of reaction buffer containing 50 mm HEPES (pH 7.5), 10 mm MgCl2, 1 mm dithiothreitol, 10 μm ATP, and 10 μCi of [γ-32P]ATP. Either 4 mm EGTA or 0.5 mm Ca2+ with or without 0.5 μg of purified CaM was added as indicated. Each reaction was carried out by using approximately 1 μg of MBP-CCaMK protein and 2 μg of substrate. Kinase reactions were stopped by mixing with SDS-PAGE sample buffer and boiling for 5 min. Samples were separated by 12% SDS-PAGE, and the gel was subsequently stained by Coomassie Brilliant Blue R250. Signal was scanned with a Fujifilm FLA-5100 phophorimager.
BiFC Analysis
Full-length cDNA of CCaMK was PCR amplified and cloned into the SpeI/XhoΙ site of pSCYCE-R vector (Waadt et al., 2008) to obtain CCaMK::SCFP3AC fusion. Full-length cDNA of CIP73 and CYCLOPS without stop codon were cloned into the BamHI/SalΙ site or the SalΙ/SmaΙ site of pSCYNE or pVYNE, respectively, to obtain CIP73::SCFP3AN or CYCLOPS::VenusN fusion. The constructs were transferred into Agrobacterium tumefaciens strain GV3101/PMP90 by electroporation for future N. benthamiana transformation. The Agrobacterium cells were harvested by centrifugation and were resuspended in agroinfiltration buffer (10 mm MES, pH 5.7, 10 mm MgCl2, and 150 μm acetosyringone). The strains were mixed to a final optical density at 600 nm of 0.5 for each Agrobacterium and incubated on the bench for 2 to 4 h at room temperature. The bacterial mixture was infiltrated into the top leaves of 6-week-old N. benthamiana with a 1- to 2-mL syringe. The fluorescence was assayed 3 to 5 d after infiltration using a Zeiss LSM510 laser-scanning microscope with CFP (excitation wavelength, 405 nm; emission wavelength, 477 nm) for SCFP3AC/SCFP3AN complexes and GFP (excitation wavelength, 488 nm; emission wavelength, 515 nm) for SCFP3AC/VenusN complexes.
Expression Analysis
The expression profile of the CIP73 gene was analyzed by semiquantitative RT-PCR in leaves, shoots, nodules, and roots of L. japonicus MG-20 following inoculation with M. loti MAFF303099. Roots were collected at each time point, and total RNA was isolated using the TRIzol reagent (Invitrogen) and treated with DNase Ι (Promega) to eliminate genomic DNA contamination. The amount of total RNA was normalized by measuring the RNA concentration at 260 nm, and 1 μg of total RNA was added to individual tubes to synthesize first-strand cDNA using oligo(dT) primer. PCR was carried out on an S1000 Thermal Cycler system (Bio-Rad; 5-min soak at 95°C, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s, followed by a 6-min soak at 72°C). The amplified products were separated on 1.2% agarose gels and visualized by ethidium bromide staining. The experiment was repeated three times to confirm the expression profile of CIP73.
Real-time RT-PCR was performed using the one-step SYBR PrimeScript RT-PCR Kit II (Takara) on a Roche LightCycler apparatus according to the manufacturer’s instructions. CIP73 transcripts were amplified using the forward 5′-ACACAACAAGAGCCATCAAGTTC-3′ and reverse 5′-CACTGCTTGGTACCTGTCTCTC-3′ primers. The forward 5′-TTCACCTTGTGCTCCGTCTTC-3′ and reverse 5′-AACAACAGCACACACAGACAATC-3′ primers were used to amplify the Ubiquitin (AW720576) transcript as an internal constitutive control. The thermal cycle was set as follows: 95°C for 10 s; 40 cycles of 95°C for 5 s and 60°C for 30 s.
Subcellular Localization of CIP73 in Onion Epidermal Cells and Hairy Root Cells
The full-length cDNA of CIP73 was cloned into the EcoRΙ/BamHI site of pMON30060 vector. The resulting construct consisted of CIP73 fused to the C terminus of the GFP under the control of the CaMV 35S promoter. The plasmid was used for transient expression in the onion (Allium cepa) epidermal cells by particle bombardment using a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad). After incubation for 12 to 24 h at 23°C in the dark, the epidermal cell layers were examined using a confocal laser-scanning microscope (Zeiss LSM510; excitation wavelength, 488 nm by argon laser; emission wavelength, 550 nm).
The C-terminal part of CIP73 (486–691 amino acids) was PCR amplified by the primers 5′-CATGCCATGGTAGCAGCAGTACCAGTTC-3′ and 5′-GACTAGTCTCCATCTTTTGACGC-3′. The amplified fragment was cloned into the NcoI/SpeΙ site of pCAMBIA1302 vector. For the selection of transformed hairy roots, the hygromycin resistance gene in pCAMBIA1302 was replaced by the GUS gene. The construct was transferred into Agrobacterium rhizogenes LBA1334 by electroporation. The transformed hairy roots selected by GUS staining were examined using the Zeiss LSM510 confocal microscope.
Construction of the RNAi Plasmid
A 225-bp fragment of the 5′ untranslated region with a short part of the coding region of CIP73 (RNAi-1) and a 261-bp fragment of the 3′ untranslated region (RNAi-2) were amplified by RT-PCR using the following primers: 5′-TGGGTGATTATAATAATCCAC-3′, containing a SmaI or PstΙ site at the 5′ end, and 5′-GTCTGCATTTCCAGTGCTG-3′, containing a BamHI or SalΙ site at the 5′ end, for RNAi-1; and 5′-CAGGTTGAAATTGGTGCTG-3′, containing a SmaI or PstΙ site at the 5′ end, and 5′-CTCGGTTGAAGAACAGTC-3′, containing a BamHI or SalΙ site at the 5′ end, for RNAi-2. The amplification products were digested with SmaI-BamHI or with PstΙ-SalΙ and ligated into the pCAMBIA1301-35S-int-T7 plasmid vector (a gift from Dr. D. Luo), in which the sense and antisense CIP73 RNA sequences were located in tandem with the Arabidopsis (Arabidopsis thaliana) actin-11 intron between them, and this intron-containing hairpin RNA construct was placed behind the CaMV 35S promoter. The constructs were transferred into A. rhizogenes LBA1334 by electroporation.
Plants harboring transformed hairy roots were transferred to pots filled with vermiculite and sand (1:1) with half-strength Broughton and Dilworth medium and grown in a chamber in a 16/8-h day/night cycle at 22°C. After 5 to 7 d, plants were inoculated with M. loti MAFF303099 and allowed to continue growing with the same medium.
Hairy Root Transformation and Identification
Hairy root transformation of L. japonicus MG20 using A. rhizogenes strain LBA1334 was performed as described previously (Kumagai and Kouchi, 2003) with some minor modifications. In brief, seeds were sterilized and germinated in water for 4 d in the dark at 22°C, followed by 2 d in a 16/8-h light/dark photocycle in a growth chamber. The seedlings were cut at the base of the hypocotyls and placed in the A. rhizogenes suspension in a petri dish for several minutes. The seedlings with cotyledons were transferred onto MS (Sigma) plates with 1.5% Suc and cocultivated for 5 d in a growth chamber. Then, the plants were transferred onto fresh MS medium containing 100 μg mL−1 cefotaxime and grown for 10 d until the hairy roots were developed from the section of hypocotyls.
For the selection of transformed hairy roots, transformed roots were immersed in GUS staining solution [100 mm sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 0.1% N-laurylsarcosine, 10 mm Na2EDTA, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid] and incubated overnight at 37°C in the dark.
Rhizobial and Mycorrhizal Infection Assay
For rhizobial infection assay, transgenic hairy roots were inoculated with M. loti strain MAFF303099 constitutively expressing the lacZ reporter gene and grown in pots containing sand:vermiculite (1:1 volume). After 5 to 7 d of inoculation, ITs were visualized by staining for β-galactosidase activity as described by Tansengco et al. (2003) and were observed using an Olympus BX51 microscope under bright-field illumination.
For mycorrhizal infection assay, transgenic seedlings were cultivated in pots containing Glomus intraradices, and sand:vermiculite (1:1 volume) was used as a substrate. After 2 weeks, roots were harvested and cleared with 10% KOH at 90°C for 10 min. AM fungal structures were stained with 0.05% trypan blue. Hyphal, arbuscular, or vesicular colonization was determined as the percentage of root length colonization using a magnified intersection method (McGonigle et al., 1990). Trypan blue-stained structures were observed with a bright-field microscope (CX41; Olympus), and images were acquired using a CCD camera (Penguin 150CL; Pixera).
Sequence data from this article can be found in the GenBank data libraries under accession number GU980966.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Predicted amino acid sequence of CIP73.
Supplemental Figure S2. Alignment of CIP73 protein homologs from plants.
Supplemental Figure S3. Transcription activation and dimerization of CIP73 in yeast cells.
Supplemental Figure S4. Nuclear localization of the C terminus of CIP73 (486–691) in onion epidermal cells.
Supplemental Figure S5. Growth phenotype of CIP73 RNAi roots under nonsymbiotic conditions.
Supplemental Figure S6. Mycorrhization phenotype of CIP73 RNAi roots.
Supplementary Material
Acknowledgments
We thank Dr. Guojiang Wu for providing M. loti MAFF303099 and Dr. D. Luo for providing RNAi vector pCAMBIA1301-35S-int-T7. We are grateful to Dr. Bin Zhao for technical advice on mycorrhizal infection and for kindly providing G. intraradices inocula. We also thank Yao Hang for advice on transient expression and confocal laser-scanning microscopy.
References
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banba M, Gutjahr C, Miyao A, Hirochika H, Paszkowski U, Kouchi H, Imaizumi-Anraku H. (2008) Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol 49: 1659–1671 [DOI] [PubMed] [Google Scholar]
- Banerji J, Sands J, Strominger JL, Spies T. (1990) A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc Natl Acad Sci USA 87: 2374–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corduan A, Lecomte S, Martin C, Michel D, Desmots F. (2009) Sequential interplay between BAG6 and HSP70 upon heat shock. Cell Mol Life Sci 66: 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmots F, Russell HR, Lee Y, Boyd K, McKinnon PJ. (2005) The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol Cell Biol 25: 10329–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB. (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M. (1999) Elevation of the cytosolic free [Ca2+] is indispensable for the transduction of the Nod factor signal in alfalfa. Plant Physiol 121: 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher F, Kondorosi E. (2000) Cell cycle regulation in the course of nodule organogenesis in Medicago. Plant Mol Biol 43: 773–786 [DOI] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GE. (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Banba M, Shimoda Y, Kouchi H, Hayashi M, Imaizumi-Anraku H. (2010) A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J 63: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA. (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudmon A, Schulman H. (2002) Neuronal Ca2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu Rev Biochem 71: 473–510 [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al. (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Jentsch S, Pyrowolakis G. (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol 10: 335–342 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al. (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye FJ, Modi S, Ivanovska I, Koonin EV, Thress K, Kubo A, Kornbluth S, Rose MD. (2000) A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett 467: 348–355 [DOI] [PubMed] [Google Scholar]
- Kiss E, Oláh B, Kaló P, Morales M, Heckmann AB, Borbola A, Lózsa A, Kontár K, Middleton P, Downie JA, et al. (2009) LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol 151: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C, Parniske M. (2002) Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci 7: 511–518 [DOI] [PubMed] [Google Scholar]
- Kondorosi E, Redondo-Nieto M, Kondorosi A. (2005) Ubiquitin-mediated proteolysis: to be in the right place at the right moment during nodule development. Plant Physiol 137: 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H, Kouchi H. (2003) Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol Plant Microbe Interact 16: 663–668 [DOI] [PubMed] [Google Scholar]
- Lee SS, Cho HS, Yoon GM, Ahn JW, Kim HH, Pai HS. (2003) Interaction of NtCDPK1 calcium-dependent protein kinase with NtRpn3 regulatory subunit of the 26S proteasome in Nicotiana tabacum. Plant J 33: 825–840 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Long SR. (1989) Rhizobium-legume nodulation: life together in the underground. Cell 56: 203–214 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle T, Miller M, Evans D, Fairchild G, Swan J. (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S. (2006) Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res 13: 255–265 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol 9: 351–357 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Harrison MJ, Udvardi M. (2005) Peace talks and trade deals: keys to long-term harmony in legume-microbe symbioses. Plant Physiol 137: 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Mitra RM, Wais RJ, Long SR. (2001) Evidence for structurally specific negative feedback in the Nod factor signal transduction pathway. Plant J 28: 191–199 [DOI] [PubMed] [Google Scholar]
- Patharkar OR, Cushman JC. (2000) A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J 24: 679–691 [DOI] [PubMed] [Google Scholar]
- Patil S, Takezawa D, Poovaiah BW. (1995) Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc Natl Acad Sci USA 92: 4897–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BW, Xia M, Liu Z, Wang W, Yang T, Sathyanarayanan PV, Franceschi VR. (1999) Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 209: 161–171 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandiran S, Takezawa D, Wang W, Poovaiah BW. (1997) Functional domains of plant chimeric calcium/calmodulin-dependent protein kinase: regulation by autoinhibitory and visinin-like domains. J Biochem 121: 984–990 [DOI] [PubMed] [Google Scholar]
- Rodriguez Milla MA, Uno Y, Chang IF, Townsend J, Maher EA, Quilici D, Cushman JC. (2006) A novel yeast two-hybrid approach to identify CDPK substrates: characterization of the interaction between AtCPK11 and AtDi19, a nuclear zinc finger protein. FEBS Lett 580: 904–911 [DOI] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, et al. (2007) NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Marcon E, McQuire T, Arai Y, Moens PB, Okada H. (2008) Bat3 deficiency accelerates the degradation of Hsp70-2/HspA2 during spermatogenesis. J Cell Biol 182: 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan PV, Cremo CR, Poovaiah BW. (2000) Plant chimeric Ca2+/calmodulin-dependent protein kinase: role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J Biol Chem 275: 30417–30422 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Stacey G, Libault M, Brechenmacher L, Wan J, May GD. (2006) Genetics and functional genomics of legume nodulation. Curr Opin Plant Biol 9: 110–121 [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Takezawa D, Ramachandiran S, Paranjape V, Poovaiah BW. (1996) Dual regulation of a chimeric plant serine/threonine kinase by calcium and calcium/calmodulin. J Biol Chem 271: 8126–8132 [DOI] [PubMed] [Google Scholar]
- Tansengco ML, Hayashi M, Kawaguchi M, Imaizumi-Anraku H, Murooka Y. (2003) crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol 131: 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K, Henzel W, Shillinglaw W, Kornbluth S. (1998) Scythe: a novel reaper-binding apoptotic regulator. EMBO J 17: 6135–6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K, Song J, Morimoto RI, Kornbluth S. (2001) Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J 20: 1033–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Uno Y, Rodriguez Milla MA, Maher E, Cushman JC. (2009) Identification of proteins that interact with catalytically active calcium-dependent protein kinases from Arabidopsis. Mol Genet Genomics 281: 375–390 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang W, Poovaiah BW. (1999) Interaction of plant chimeric calcium/calmodulin-dependent protein kinase with a homolog of eukaryotic elongation factor-1alpha. J Biol Chem 274: 12001–12008 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8: 505–512 [DOI] [PubMed] [Google Scholar]
- Yano K, Shibata S, Chen WL, Sato S, Kaneko T, Jurkiewicz A, Sandal N, Banba M, Imaizumi-Anraku H, Kojima T, et al. (2009) CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis. Plant J 60: 168–180 [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Chen T, Zhu M, Fang Q, Kang H, Hong Z, Zhang Z. (2008) A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol 148: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.