Abstract
Functionally distinct Arabidopsis (Arabidopsis thaliana) genes that positively affect root or shoot growth when ectopically expressed were combined to explore the feasibility of enhanced biomass production. Enhanced root growth resulting from cytokinin deficiency was obtained by overexpressing CYTOKININ OXIDASE/DEHYDROGENASE3 (CKX3) under the control of the root-specific PYK10 promoter. Plants harboring the PYK10-CKX3 construct were crossed with four different transgenic lines showing enhanced leaf growth. For all combinations, the phenotypic traits of the individual lines could be combined, resulting in an overall growth increase. Unexpectedly, three out of four combinations had more than additive effects. Both leaf and root growth were synergistically enhanced in plants ectopically expressing CKX3 and BRASSINOSTEROID INSENSITIVE1, indicating cross talk between cytokinins and brassinosteroids. In agreement, treatment of PYK10-CKX3 plants with brassinolide resulted in a dramatic increase in lateral root growth that could not be observed in wild-type plants. Coexpression of CKX3 and the GROWTH-REGULATING FACTOR5 (GRF5) antagonized the effects of GRF5 overexpression, revealing an interplay between cytokinins and GRF5 during leaf cell proliferation. The combined overexpression of CKX3 and GIBBERELLIN 20-OXIDASE1 led to a synergistic increase in leaf growth, suggesting an antagonistic growth control by cytokinins and gibberellins. Only additive effects on root and shoot growth were visible in plants ectopically expressing both CKX3 and ARABIDOPSIS VACUOLAR PYROPHOSPHATASE1, hinting at an independent action mode. Our results show new interactions and contribute to the molecular and physiological understanding of biomass production at the whole plant level.
As the world population is estimated to grow to 9.2 billion people by 2050, the availability of plant-derived products has to increase drastically to meet the needs not only for food, feed, and fiber but also for bioenergy and other industrial applications (Borlaug, 2007). In the future, less arable land will be available while more crops will need to be produced. To lower the negative impact on the environment, increasing plant biomass production on existing agricultural land will diminish the demand for new crop acreage (Edgerton, 2009). Whereas traditional breeding combined with improved agronomical practices will not keep up with the increasing global demands, biotechnology can help serve this purpose (Borlaug, 2007). Marker-assisted breeding and introduction of transgenic traits for biotic and abiotic stress resistance have proven to increase crop gain (Eathington et al., 2007; Edgerton, 2009). Furthermore, improving yield by modulating endogenous molecular pathways will be an important feature of the next generation of biotech crops.
The introduction of multiple transgenes has become widely adopted (Naqvi et al., 2010); for example, three carotenoid biosynthesis genes enable provitamin A production in transgenic rice (Oryza sativa) cv Golden Rice (Ye et al., 2000), while as many as eight genes confer insecticide and pesticide resistance to the new biotech maize (Zea mays; James, 2009). The latter case demonstrates the great potential of stacking distinct phenotypic traits, which prompted us to investigate whether enhanced root growth can be combined with enhanced shoot growth to further improve plant productivity.
Plant growth is highly regulated throughout development, and two main levels of coordination can be distinguished. First, the final plant size is determined by a global growth coordination of distinct organs. This indicates that for optimal growth, energy production in photosynthetic leaves has to be fine-tuned to the availability of water and nutrients supplied by the root, inevitably linking shoot and root growth (Paul and Foyer, 2001). Since photosynthesis and source tissue development are regulated by sink tissue through the carbon/nitrogen balance (Paul and Foyer, 2001), altering the source/sink balance by changing the root-shoot ratio will probably impact total plant growth. An enlarged root system, for instance, could act as a strong sink and prevent enhanced growth of the shoot.
Second, different organs are delineated by specific tissue patterns that are generated by coordinated proliferation and expansion of individual cells. The proliferation process is characterized by extensive cell division and cell size homeostasis. During the expansion phase, cells become much larger than meristematic cells and start to differentiate (Beemster et al., 2005). Roots exhibit an indeterminate growth pattern. Meristematic cells in the root tip continuously produce new cells, while older cells are subsequently displaced away from the root tip. As a result, the proliferation and expansion phases coexist in roots throughout the life cycle of the plant. In contrast, the shoot consists of lateral organs of determinate size. In the leaf of Arabidopsis (Arabidopsis thaliana), cell proliferation gradually decreases from the tip toward the base, while cell expansion progressively increases in the opposite direction (Donnelly et al., 1999). The rate and duration of both cell proliferation and expansion ultimately determine the size at the organ level.
Despite this elaborate knowledge of the physiological aspect of biomass production, molecular understanding is limited. Over the past decades, many genes have been identified that improve organ size when ectopically expressed or mutated (Gonzalez et al., 2009; Krizek, 2009) and were previously referred to as “intrinsic yield genes” (IYGs). These genes belong to a diversity of functional classes and operate in different pathways, demonstrating that plant growth and yield are complex traits. However, all pathways influencing growth should eventually converge to control cell division and cell expansion, suggesting an extensive cross talk and creating the opportunity to further improve growth by combining transgenes or better-performing gene alleles that function in these pathways.
As plant hormones are involved in every aspect of plant biology, it is not surprising that a number of IYGs are part of the hormone biosynthetic and signaling pathways or exert their effect by influencing hormone metabolism. Cytokinin oxidase/dehydrogenase (CKX) enzymes catalyze the degradation of a number of biologically active cytokinins and, hence, contribute to the regulation of cytokinin levels (Mok and Mok, 2001; Werner et al., 2006). The Arabidopsis CKX family contains seven members (Schmülling et al., 2003), of which the phenotype resulting from the overexpression of six (CKX1–CKX6) members has been described (Werner et al., 2003): CKX-overexpressing plants have a reduced cytokinin content, leading to the development of the typical cytokinin deficiency syndrome characterized by an increased root growth but severely diminished shoot growth (Werner et al., 2003).
Detailed analysis of 35S-CKX1-overexpressing plants at the cellular level has revealed that cytokinins function in the process of cell proliferation in meristems (Werner et al., 2003). The shoot apical meristem of CKX-overexpressing plants is reduced in size, and cell production in the leaves is diminished, which is slightly compensated for by an enlarged cell size. In contrast, the primary root apical meristem is enlarged, additional cell files are formed, and all cell types show an increased radial expansion. These observations suggest that cytokinins have a major role in regulating meristem activity by controlling the exit of cells from the meristem and hence the duration of cell proliferation (Werner et al., 2003). Moreover, in roots, cytokinins most probably act specifically at the transition zone between dividing and expanding cells to promote cell differentiation (Dello Ioio et al., 2007).
Besides a plausible local function in the same tissue where they are synthesized, cytokinins, like other plant hormones, function in long-distance signaling between root and shoot tissues (Kudo et al., 2010). Evidence supports a role in nutrient signaling to coordinate metabolic processes between sink and source tissues (Argueso et al., 2009). Moreover, cytokinins stimulate photosynthesis (Wareing et al., 1968) and are able to establish local metabolic sinks or enhance sink strength (Kuiper, 1993; Guivarc’h et al., 2002). The effect on sink strength has also been demonstrated in 35S-CKX1-overexpressing tobacco (Nicotiana tabacum) plants, in which decreased cytokinin levels reduced the sink strength of the shoot (Werner et al., 2008). Clearly, cytokinins play a major role in fine-tuning the distribution of assimilates in the whole plant and, hence, contribute to the regulation of growth and biomass production.
Here, enhanced root growth resulting from cytokinin deficiency was exploited by overexpressing CKX3 under the control of the root-specific PYK10 (hereafter designated P10) promoter. Plants expressing the P10-CKX3 construct were crossed with four IYG-overproducing lines that enhance leaf growth (Gonzalez et al., 2009) and had been selected based on the reproducibility of their leaf phenotype and their functional diversity (Gonzalez et al., 2010). These IYGs encode GROWTH-REGULATING FACTOR5 (GRF5), which is a putative transcription factor proposed to act in a pathway that promotes cell proliferation (Horiguchi et al., 2005); GIBBERELLIN 20-OXIDASE1 (GA20ox1), which catalyzes consecutive steps in GA biosynthesis (Xu et al., 1995); BRASSINOSTEROID INSENSITIVE1 (BRI1), a key component of the brassinosteroid membrane receptor complex (Wang et al., 2001); and ARABIDOPSIS VACUOLAR PYROPHOSPHATASE1 (AVP1), a H+-pyrophosphatase involved in the generation of proton gradients in endomembrane compartments (Li et al., 2005). In all four lines, an increase in cell proliferation drives, predominantly or entirely, the enhanced leaf growth (Gonzalez et al., 2010).
Our results indicate that the phenotypic traits of the lines can be combined, resulting in enhanced root and shoot growth. We provide evidence for cross talk between cytokinins and brassinosteroids during lateral root and leaf growth and between cytokinins and GAs during leaf growth. In addition, a connection between cytokinins and GRF5 during shoot growth is shown.
RESULTS
The P10 Promoter Restricts the Cytokinin-Deficient Phenotype of CKX3 Overexpression to the Root
Overexpression of CKX1 or CKX3 under the control of the cauliflower mosaic virus 35S promoter (35S) increases the total root system length of 8-d-old plants up to 3-fold but drastically delays leaf formation and reduces leaf size (Werner et al., 2003). To benefit from the positive effect on root growth of CKX overexpression while alleviating the inhibiting effect on shoot growth, the Arabidopsis P10 promoter was chosen to drive CKX3 expression. P10 encodes a myrosinase and is highly expressed in hypocotyl and roots, while it is virtually not expressed in mature rosette leaves (Nitz et al., 2001; Sherameti et al., 2008). Plants with enhanced root growth and normal shoot growth could be obtained in this manner (Werner et al., 2010).
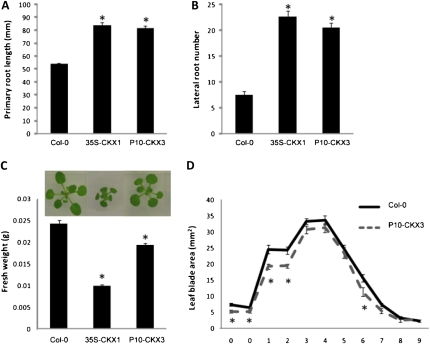
Root growth of 35S-CKX1 and P10-CKX3-GFP (hereafter referred to as P10-CKX3) was measured to evaluate the potential of the P10 promoter to confer the cytokinin-deficient root phenotype under our experimental conditions. In 35S-CKX1 plants, both primary root length (Fig. 1A) and lateral root number (Fig. 1B) had increased strongly compared with those of wild-type Columbia-0 (Col-0) 9 d after stratification (DAS). Primary root length and lateral root number of P10-CKX3 plants were increased to the same extent as for 35S-CKX1 plants (Fig. 1, A and B), demonstrating that the P10 promoter driving CKX3 expression is sufficiently strong to confer the same root phenotype as the 35S promoter driving CKX1 expression.
Figure 1.
Enhanced root growth and slightly reduced shoot growth of P10-CKX3 plants. A and B, Primary root growth and lateral root number of 35S-CKX1, P10-CKX3, and wild-type (Col-0) lines. Plants were grown on vertical plates under in vitro conditions for 9 d. Error bars indicate se (n ≥ 20). * Significantly different from the wild type (P < 0.01, Student’s t test). C and D, Shoot growth. C, Fresh weight of rosettes of 35S-CKX1, P10-CKX3, and wild-type plants grown for 21 d under in vitro conditions. Photographs were taken from 18-d-old plants grown in vitro. Error bars indicate se (n ≥ 21). D, Blade area of cotyledons (0) and leaves 1 to 9 calculated from leaf series made at 21 DAS from P10-CKX3 and wild-type plants grown in vitro. Error bars indicate se (n ≥ 14). * Significantly different from the wild type (P < 0.05, Student’s t test). [See online article for color version of this figure.]
To quantify the effect of CKX3 overexpression under the control of the P10 promoter on shoot growth, fresh weight of the rosette was measured from plants grown for 21 DAS under in vitro conditions (Fig. 1C). This revealed that P10-CKX3 plants showed only a mild reduction in rosette size compared with wild-type plants, whereas constitutive overexpression of CKX1 strongly reduced shoot growth (Werner et al., 2003; Fig. 1C). To analyze the leaf size of P10-CKX3 plants in detail, leaf parameters (blade area, length, and width) of each individual leaf were calculated from leaf series made from 21-d-old rosettes (Fig. 1D; Supplemental Fig. S1). Although the blade area of P10-CKX3 leaves was not significantly different from that of the wild type for most leaves (Fig. 1D), leaf length was significantly reduced for all leaves, while blade width remained unchanged (Supplemental Fig. S1). This polarity-dependent reduction in blade area gave P10-CKX3 plants a characteristic appearance with rounded laminas (Fig. 1C).
In conclusion, the use of the P10 promoter to drive the expression of CKX3 in Arabidopsis allows circumvention of the negative effect on shoot growth but maintenance of the positive effect of CKX overexpression on root growth.
Crossing P10-CKX3 Plants with Plants Overexpressing Genes That Enhance Leaf Size
To assess whether genes that positively affect root growth can be combined with genes that enhance leaf organ size and whether this strategy could lead to a further increase in growth and biomass production, four different genes that enhance leaf size when overexpressed (BRI1, GRF5, GA20ox1, and AVP1) were cooverexpressed with the P10-CKX3 construct.
Plants homozygous for each construct were crossed, and root and shoot growth of the resulting heterozygous F1 progeny were analyzed. Each homozygous line, in addition, was crossed with wild-type Col-0 plants, resulting in heterozygous lines that were used as appropriate controls. To evaluate the effect on growth, the expected values in case of an additive effect were calculated as the sum of the measurements for each individual heterozygous parent minus the measurement for Col-0 for a certain trait. An additive phenotype corresponds to the sum of the individual phenotypes of the heterozygous parents and, therefore, is an indication that both genes might work independently to control organ size. When the combination of two transgenes results in a value for a phenotypic trait that is higher or lower than the expected value for an additive effect, a putative synergistic or antagonistic growth phenotype occurs, respectively. The latter cases are usually an indication that the combined genes function in related pathways, making it possible to further unravel the regulatory network in which these genes take part.
Introduction of pBRI-BRI1 into P10-CKX3 Plants Results in a More Than Additive Increase of Root and Shoot Growth
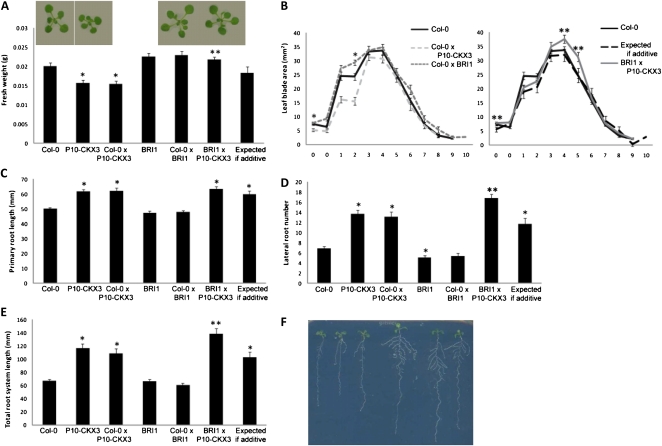
Ectopic expression of BRI1 under the control of its own promoter leads to an increased response to brassinosteroids, resulting in longer petioles and larger leaves (Wang et al., 2001). However, when the plants were grown under in vitro conditions for 21 d, enhanced BRI1 expression did not result in a significant increase in either rosette fresh weight (Fig. 2A) or blade area for most leaves (Fig. 2B) but mainly in increased blade length (Supplemental Fig. S2, A and C). Heterozygous or homozygous pBRI-BRI1-GFP (hereafter referred to as BRI1) plants were similarly affected (Fig. 2A). Also, crossing with the wild type had no effect on the extent of the reduced shoot growth in P10-CKX3 lines (Fig. 2A). Rosette fresh weight of plants that ectopically expressed both BRI1 and CKX3 was equal to that of wild-type and BRI1 plants (Fig. 2A), demonstrating that BRI1 overexpression can compensate for the reduction in fresh weight caused by CKX3 overexpression. Consequently, fresh weight of the cross was significantly higher than that of the expected value for an additive effect (Fig. 2A), as reflected in a synergistic increase in blade area for certain leaves (Fig. 2B). Detailed observation of leaf parameters showed that this was mainly attributed to an increase in leaf length that was comparable to that of Col-0×BRI1 (Supplemental Fig. S2, B and D), suggesting that ectopic expression of BRI1 overcomes the effects of P10-CKX3 in the leaf length direction. Relative transgene expression levels in shoots of BRI1×P10-CKX3 plants were similar to those of the heterozygous parents (Supplemental Fig. S3A), showing that the observed synergism in leaf blade area is not a consequence of an altered expression level of one transgene due to the presence of the other.
Figure 2.
More than additive increases in shoot and root growth by crossing P10-CKX3 plants with BRI1-overexpressing plants. A and B, Shoot phenotype of plants grown under in vitro conditions for 21 d. A, Rosette fresh weight. Photographs were taken from the rosettes of the wild type (Col-0), the heterozygous parents, and the cross grown for 18 d in vitro. B, Blade area of cotyledons (0) and leaves 1 to 10 calculated from leaf series. C to F, Root growth of wild-type plants and plants overexpressing P10-CKX3 and/or BRI1. Plants were grown on vertical plates under in vitro conditions for 9 d. C, Primary root length. D, Lateral root number. E, Total root system length. F, Root system of 9-d-old plants. From left to right: Col-0, BRI1, Col-0×BRI1, P10-CKX3×BRI1, Col-0×P10-CKX3, and P10-CKX3. Values expected if additive were calculated as the sum of Col-0×P10-CKX3 and BRI1×Col-0 minus Col-0. Asterisks indicate values significantly different from the wild type (*) and from the value expected for an additive effect (**) by Student’s t test (P < 0.05 [A and B] and P < 0.01 [C–E]). Error bars indicate se (n ≥ 16 [A and B] and n ≥ 23 [C–E]).
Whereas treatment with brassinolide (BL) has been shown to enhance lateral root formation (Bao et al., 2004), BRI1-overexpressing plants had no significantly enhanced root growth at 9 DAS (Fig. 2, C–F). No difference in root growth could be observed either between homozygous and heterozygous BRI1 lines or between homozygous and heterozygous P10-CKX3 lines (Fig. 2, C–F). Surprisingly, lateral root number and total root system length of BRI1×P10-CKX3 were significantly larger than those expected for an additive effect (Fig. 2, D and E). The expression levels of both transgenes did not differ in the cross when compared with the heterozygous controls (Supplemental Fig. S3B). Although the lateral root number in Col-0×P10-CKX3 plants was 90% higher than that in the wild type, an increase of 144% was observed for plants overexpressing both BRI1 and CKX3 (Fig. 2D). Measurements of the total root system of Col-0×P10-CKX3 revealed a 63% increase compared with control plants versus 107% in BRI1×P10-CKX3 plants (Fig. 2E). The primary root was not significantly longer than the value expected for an additive effect (Fig. 2C). Hence, combining enhanced expression of these two genes has a synergistic effect on lateral root growth. Taken together, overexpression of CKX3 and BRI1 not only results in the combination of phenotypic traits but also improves lateral root growth synergistically and the growth of particular leaves in more than an additive manner.
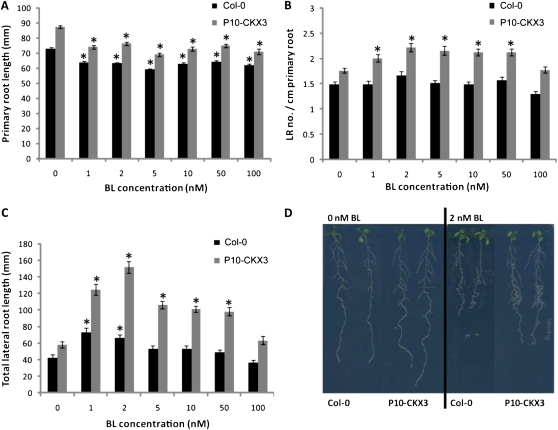
Exogenous Application of BL Enhances Lateral Root Growth of P10-CKX3 Plants
The observed positive effect of the combined overexpression of CKX3 and BRI1 suggests that reduced cytokinin levels and enhanced perception of BL act synergistically to enhance lateral root growth. To confirm the effect of brassinosteroids, P10-CKX3 plants were treated with different concentrations of BL (Fig. 3). Wild-type primary root length decreased for all BL concentrations applied (Fig. 3A), as reported previously (Müssig et al., 2003; Bao et al., 2004). The relative decrease in primary root length was similar in P10-CKX3 plants (Fig. 3A). Whereas no significant increase in lateral root number was observed when wild-type plants were treated with 1 to 100 nm BL under our conditions, lateral root number per centimeter of primary root of P10-CKX3 plants increased in response to 1 to 50 nm BL (Fig. 3B). Moreover, the total length of P10-CKX3 lateral roots increased up to 160% by treatment with 1 to 50 nm BL, whereas a significant increase up to 72% could only be detected for wild-type lateral roots for 1 and 2 nm BL (Fig. 3C). No correlation between the BL-induced lateral root phenotypes and changes in relative mRNA abundance of CKX3 and BRI1 in wild-type and P10-CKX3 seedlings could be observed (Supplemental Fig. S3B). These data show that both lateral root formation and lateral root elongation are further induced by the combined action of brassinosteroids and CKX3 overexpression and, by inference, a reduction of biologically active cytokinins.
Figure 3.
Synergistically enhanced lateral root growth of P10-CKX3 plants by exogenous application of BL. A to C, P10-CKX3 and wild-type (Col-0) seedlings grown vertically for 5 d on half-strength MS and transferred to plates containing half-strength MS + 0, 1, 2, 5, 10, 50, and 100 nm BL for 6 d. Primary root length (A), lateral root (LR) number per centimeter of primary root (B), and total lateral root length (C) were determined. * Significantly different from the mock-treated controls (P < 0.01, Student’s t test). Error bars indicate se (n ≥ 21). D, Root systems of 11-d-old plants grown for 5 d on half-strength MS and 6 d on 0 (left) or 2 nm (right) BL.
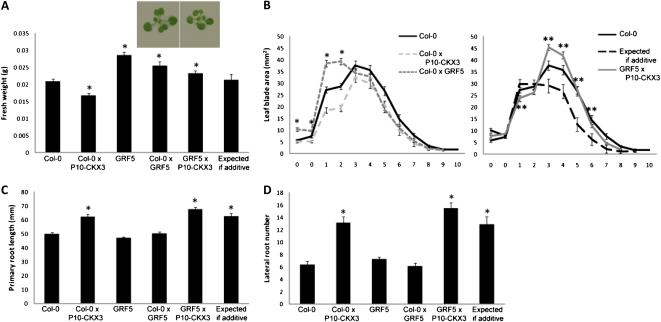
Cooverexpression of CKX3 with GRF5 Antagonizes the Effects of GRF5 during Leaf Development
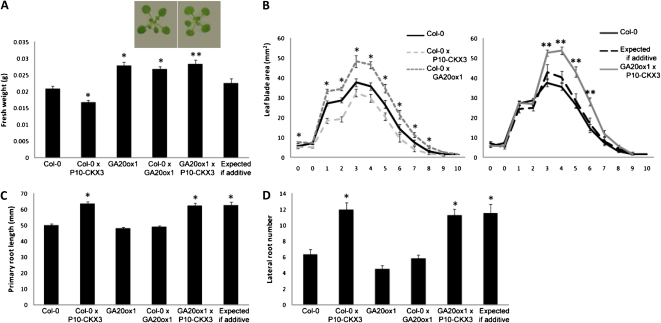
Overexpression of GRF5 leads to the development of enlarged leaves due to enhanced cell proliferation in both width and length directions (Horiguchi et al., 2005; Gonzalez et al., 2010), with an increase in rosette fresh weight of 21-d-old plants grown in vitro as a consequence (Fig. 4A). The cotyledons and first leaves were larger in size, but from leaf 3 onward, the blade area was slightly smaller than that of the wild type (Fig. 4B; Gonzalez et al., 2010). This characteristic leaf phenotype quantitatively differed between homozygous and heterozygous GRF5-overexpressing lines, being less severe for heterozygous lines, indicating the presence of a gene dosage effect for GRF5 (Fig. 4A). Crossing P10-CKX3 with GRF5 led to an average fresh weight that was slightly higher than that of the wild type but did not significantly differ from the expected value for an additive effect (Fig. 4A). Analysis of individual leaf blade areas, however, revealed that the increase in blade size caused by GRF5 overexpression was reduced more than expected for an additive effect (Fig. 4B). In addition, the younger leaves 3 to 6, which are slightly smaller in both individual overexpressing lines, surprisingly were larger than expected if additive and even larger than those of wild-type leaves 3 and 4 when overexpression of CKX3 and GRF5 was combined (Fig. 4B). Suppression of the CKX3 and GRF5 phenotypes was observed in both width and length directions (Supplemental Fig. S2, B and D). Analysis of relative mRNA levels of CKX3 and GRF5 eliminated the possibility that the effects resulted from changes in transgene expression (Supplemental Fig. S3D). A plausible explanation for the phenotype is that both genes antagonize each other: CKX3 overexpression seems to reduce the effects of GRF5 overexpression and/or vice versa, eventually converting a reduction in leaf size into an increase.
Figure 4.
Leaf and root growth phenotypes of plants cooverexpressing P10-CKX3 and GRF5. A and B, Shoot growth measurements at 21 DAS from plants grown in vitro. A, Rosette fresh weight. Photographs were taken from the rosettes of the heterozygous parent and the cross grown for 18 d in vitro. B, Blade area of cotyledons (0) and leaves 1 to 10 calculated from leaf series. C and D, Root growth measurements of plants grown on vertical plates under in vitro conditions for 9 d. C, Primary root length. D, Lateral root number. Values expected if additive were calculated as the sum of the single heterozygous lines minus the wild type (Col-0). Asterisks indicate values significantly different from the wild type (*) and from the value expected for an additive effect (**) by Student’s t test (P < 0.05 [A and B] and P < 0.01 [C and D]). Error bars indicate se (n ≥ 16 [A and B] and n ≥ 23 [C and D]). [See online article for color version of this figure.]
GRF5-overexpressing plants showed no significant enhanced root growth (Fig. 4, C and D). Root length and lateral root number in GRF5×P10-CKX3 plants increased up to the same level as in P10-CKX3 plants crossed with Col-0 (Fig. 4, C and D), indicating that GRF5 overexpression does not influence the root phenotype caused by CKX3 overexpression in the root.
The Effects of GA20ox1 and CKX3 Ectopic Expression Are Synergistic with Respect to Leaf Growth
Plants overexpressing GA20ox1 under the control of the 35S promoter show an increased content of bioactive GA4 and other GAs in the vegetative rosette (Coles et al., 1999; Gonzalez et al., 2010). These plants typically have elongated hypocotyls and stems and large leaves with long petioles (Coles et al., 1999). At 21 DAS, 35S-GA20ox1 (hereafter referred to as GA20ox1) plants showed an increase in rosette fresh weight when compared with wild-type plants, and the phenotype of heterozygous and homozygous plants was indistinguishable (Fig. 5A). The blade area of all rosette leaves had increased (Fig. 5B) as a consequence of enhanced growth in both blade width and length directions (Supplemental Fig. S2, A and C). Coexpression of GA20ox1 and P10-CKX3 resulted in a more than additive effect on shoot growth (Fig. 5, A and B). GA20ox1 overexpression not only compensated for the reduction in blade area due to CKX3 overexpression, but the blade area of GA20ox1×P10-CKX3 plants additionally increased from leaf 3 onward compared with that of Col-0×GA20ox1 plants (Fig. 5B), and this resulted from a further growth increase in the lateral direction (Supplemental Fig. S2, B and D). As the blade width of P10-CKX3 plants did not significantly change, CKX3 overexpression might enhance the growth-promoting effect of GA20ox1 overexpression in the width direction. This strong synergistic effect yielded up to a 36% increase in total rosette fresh weight compared with wild-type plants (Fig. 5A). Similar transgene mRNA levels in the cross and the heterozygous parents excluded the notion that the observed synergism is caused by mutually induced changes in expression level (Supplemental Fig. S3E).
Figure 5.
Synergistic and additive increases in leaf and root growth, respectively, by combined overexpression of P10-CKX3 and GA20ox1. A and B, Shoot phenotype of 21-d-old plants grown in vitro. A, Rosette fresh weight. Photographs were taken from the rosettes of the heterozygous parent and the cross grown for 18 d in vitro. B, Blade area of cotyledons (0) and leaves 1 to 10 calculated from leaf series. C and D, Root phenotypes of plants grown on vertical plates under in vitro conditions for 9 d. C, Primary root length. D, Lateral root number. Values expected if additive were calculated as the sum of the single heterozygous lines minus the wild type (Col-0). Asterisks indicate values significantly different from the wild type (*) and from the value expected for an additive effect (**) by Student’s t test (P < 0.05 [A and B] and P < 0.01 [C and D]). Error bars indicate se (n ≥ 16 [A and B] and n ≥ 23 [C and D]). [See online article for color version of this figure.]
Overexpression of GA20ox1 did not alter root growth either in homozygous or heterozygous conditions (Fig. 5, C and D). Combination of GA20ox1 with P10-CKX3 led to an increase in total root system length characteristic for CKX3 overexpression alone (Fig. 5, C and D), demonstrating that GA20ox1 overexpression does not interfere with the effects of enhanced cytokinin breakdown in the root.
Combining CKX3 and AVP1 Overexpression Has an Additive Effect on Root and Shoot Growth
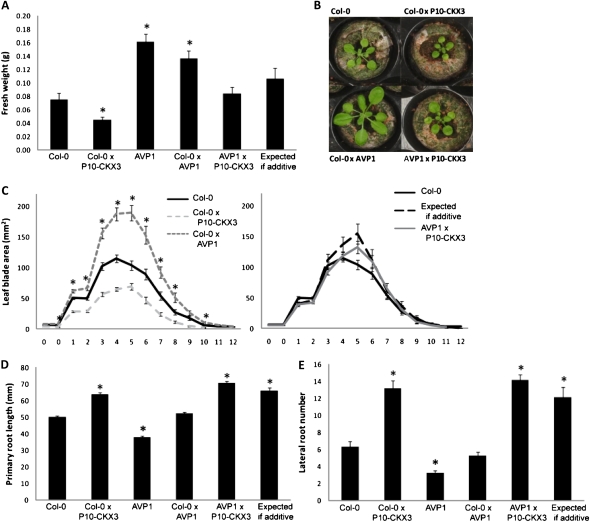
AVP1 overexpression dramatically enhances leaf organ size by a polarity-independent increase in cell number (Li et al., 2005). However, homozygous 35S-AVP1 (hereafter referred to as AVP1) plants exhibit only a slight increase in leaf organ size when grown under in vitro conditions on half-strength Murashige and Skoog (MS) medium for 21 d (Gonzalez et al., 2010). Therefore, fresh weight and leaf parameters were analyzed from plants grown in soil (Fig. 6, A–C). All rosette leaves increased in size (Fig. 6C), leading to a doubling in rosette fresh weight when compared with wild-type plants, both for homozygous and heterozygous AVP1 plants (Fig. 6A). Coexpressing AVP1 and P10-CKX3 led to an intermediate rosette growth, with measurements that equaled values expected for additive effects for fresh weight and all leaf parameters (Fig. 6, A and C; Supplemental Fig. S2, B and D). This results in a shoot phenotype at 22 DAS that more closely resembled wild-type shoot growth.
Figure 6.
Additive effects on leaf and root growth by crossing of P10-CKX3- and AVP1-overexpressing plants. A to C, Shoot growth of 22-d-old plants grown in soil. A, Rosette fresh weight. B, Photographs from the rosettes of the wild type (Col-0), the heterozygous parents, and the cross. C, Blade area of cotyledons (0) and leaves 1 to 12 calculated from leaf series. D and E, Root phenotypes of plants grown on vertical plates under in vitro conditions for 9 d. D, Primary root length. E, Lateral root number. Values expected for an additive effect were calculated as the sum of Col-0×P10-CKX3 and Col-0×AVP1 minus Col-0. Asterisks indicate values significantly different from the wild type (*) and from the value expected for an additive effect (**) by Student’s t test (P < 0.05 [A–C] and P <0.01 [D and E]). Error bars indicate se (n ≥ 12 [A–C] and n ≥ 23 [D and E]).
Also, an increase in root growth was reported when plants were grown hydroponically for 45 d (Li et al., 2005). At 9 DAS, however, root growth of homozygous AVP1 plants was drastically reduced when grown in vitro on half-strength MS medium (Fig. 6, D and E). A striking difference in growth behavior between homozygous and heterozygous AVP1-overproducing plants could be observed, because primary root length and lateral root number of heterozygous AVP1 plants were comparable to those of the wild type (Fig. 6, D and E). Taken together, the analysis of in vitro root and shoot (data not shown) growth of heterozygous and homozygous AVP1 plants suggests that the effect of ectopic expression of AVP1 depends on the gene dosage. Root growth of AVP1 plants crossed with CKX3 plants was similar to that of Col-0×P10-CKX3 plants, indicating that the combination of both genes results in the phenotype of P10-CKX3 plants (Fig. 6, D and E). In conclusion, these observations imply an independent mode of action of AVP1 and CKX3 through distinct pathways during root and shoot growth.
DISCUSSION
Here, we show that the combined introduction of two transgenes, one enhancing root growth and the other enhancing shoot growth, is a powerful strategy to increase overall plant growth. The P10 promoter driving CKX3 expression proves to be efficient to restrict cytokinin degradation to the root, thereby strongly reducing the negative effect of constitutive CKX overexpression on vegetative shoot growth (Werner et al., 2003). At 21 DAS, the leaf size of P10-CKX3 plants is smaller than that of the wild type, indicating a slightly slower development characteristic for plants with reduced cytokinin responses (Werner et al., 2003; Miyawaki et al., 2006; Riefler et al., 2006). However, rosette growth catches up with wild-type plants after bolting when plants are grown in soil (Werner et al., 2010), and expression of P10-CKX3 has no significant influence on the leaf phenotypes obtained by simultaneous ectopic expression of BRI1, GRF5, GA20ox1, or AVP1 after bolting (data not shown). This implies that in the control of whole plant size, root growth is seemingly not enhanced at the expense of enhanced shoot growth in the crosses, even in the absence of supplemented Suc. Furthermore, it demonstrates that the increased root system does not act as a strong sink, which is in agreement with observations in tobacco (Werner et al., 2008) and Arabidopsis (Werner et al., 2010). Conversely, a fast-growing root system might provide more water and nutrients to the shoot, but no additional increase in shoot fresh weight was observed in comparison with the single BRI1-, GRF5-, GA20ox1-, or AVP1-overexpressing lines, when root growth was enhanced by coexpression of P10-CKX3. Taken together, this strongly suggests that the phenotypic effects on root and shoot growth under optimal environmental conditions where assimilates are not limited are primarily delineated by changes in genetic competence and not by the global sink/source regulation.
Unexpected phenotypic outcomes from crossing the transgenic lines were uncovered when leaf size and root architecture were analyzed at 21 and 9 DAS, respectively. These phenotypes, which are not additive, are usually an indication for cross talk between pathways in which the genes function (Chandler, 2009; Pérez-Pérez et al., 2009) and will be discussed below.
Brassinosteroid-Cytokinin Cross Talk during Leaf Development
The reduction in leaf length caused by CKX3 overexpression is completely overcome by simultaneous ectopic expression of BRI1. In other words, an increased sensitivity to brassinosteroids could compensate for the decreased leaf length resulting from enhanced cytokinin breakdown. Brassinosteroids were shown to affect both leaf cell proliferation and elongation, because the reduced leaf size of biosynthetic mutants is due to a decrease in cell number and size (Nakaya et al., 2002). However, overexpression of BRI1 enhances cell proliferation in the leaf, while no changes in cell size were detected (Gonzalez et al., 2010). Moreover, the changes in leaf size caused by BRI1 overexpression might be the consequence of an increase in the period rather than the rate of cell proliferation (Gonzalez et al., 2010). A role for cytokinins in the duration of leaf cell proliferation has also been suggested through the maintenance of meristematic competence to prevent the transition to differentiation and, hence, cell expansion (Nishimura et al., 2004). Taken together, these observations suggest cross talk between pathways that guide brassinosteroid and cytokinin responses toward cell division mechanisms, thereby controlling the timing of transition from cell proliferation to cell expansion.
Brassinosteroids can increase cell division, in particular when auxin and cytokinin are limiting, and it was demonstrated that BL can substitute cytokinin to promote the growth of callus and cell suspensions of Arabidopsis (Hu et al., 2000). In good agreement with our findings, an increased brassinosteroid response compensates for lower cytokinin levels in the leaves. Little is known, however, about the molecular mechanisms that integrate the responses from both hormones. Hormonal cross talk can be accomplished by the regulation of expression of common target genes or modulation of the activity of a common protein, which is considered as direct cross talk (Chandler, 2009). Expression of Cyclin D3;1 (CYCD3;1), a positive regulator of cell proliferation, is induced by treatment with brassinosteroids and cytokinins (Riou-Khamlichi et al., 1999; Hu et al., 2000), making it a good candidate. Numerous other genes have been found to be transcriptionally regulated by both hormones (Nemhauser et al., 2006; Peng et al., 2009), but their putative role as a convergence point of brassinosteroid and cytokinin response pathways needs further investigation.
Alternatively, hormonal cross talk can be indirect, as when one hormone response pathway influences the biosynthesis, degradation, sensitivity, or perception of the other hormone (Chandler, 2009). Given that enhanced BRI1 expression compensates for lower cytokinin levels in the leaf, under normal conditions, cytokinins might potentially attenuate the brassinosteroid response in one of the above described ways. Genes involved in brassinosteroid metabolism are likely targets, although changes in their expression induced by cytokinin treatment were not solely linked to decreased brassinosteroid levels (Nemhauser et al., 2006).
Brassinosteroid-Cytokinin Cross Talk during Lateral Root Development
Root growth is impaired in brassinosteroid biosynthetic mutants and in the signaling mutant bri1, demonstrating that brassinosteroids positively affect root growth (Müssig et al., 2003; Bao et al., 2004; Mouchel et al., 2006). On the other hand, neither overexpression of BRI1 nor BL treatment increases primary root growth or lateral root formation of wild-type plants. In fact, BL treatment reduces primary root growth of wild-type and P10-CKX3 plants to the same relative extent. Also, primary root growth of P10-CKX3 plants is not influenced by simultaneous BRI1 ectopic expression. Although changes in brassinosteroid as well as cytokinin levels can affect the size of the root apical meristem (Werner et al., 2003; Mouchel et al., 2006; Dello Ioio et al., 2007), both hormones seemingly accomplish that through independent, parallel pathways.
In contrast to previous observations (Bao et al., 2004), BL has no effect on the formation of lateral roots of wild-type plants. However, when cytokinin levels are reduced in the root, BL concentrations from 1 to 50 nm enhance lateral root formation and elongation. Also, combining CKX3 overexpression with BRI1 ectopic expression enhances lateral root growth synergistically, demonstrating that an enhanced sensitivity to brassinosteroids as well as exogenous application of brassinosteroids further stimulates lateral root growth of P10-CKX3 plants. In other words, cytokinin deficiency enhances the effect of brassinosteroids on lateral root growth or vice versa. Therefore, we hypothesize that under normal conditions, cytokinins antagonize brassinosteroid function in lateral root formation and/or vice versa.
Whereas virtually nothing is known about the molecular basis of cytokinin-brassinosteroid cross talk during lateral root development, ample evidence exists for the interaction of both hormones with auxin. Brassinosteroids promote the initiation of lateral root primordia, most probably by increasing acropetal auxin transport to aid local auxin accumulation necessary for primordium development (Benková et al., 2003; Bao et al., 2004). They are proposed to act through the modulation of PINFORMED (PIN) auxin efflux carriers, because brassinosteroids have been shown to regulate PIN gene expression (Nakamura et al., 2004; Nemhauser et al., 2004). Cytokinins inhibit lateral root initiation by preventing the reentry of pericycle cells into the cell cycle, thereby perturbing asymmetric cell division to establish lateral root primordia (Li et al., 2006). In addition, cytokinins disturb auxin accumulation by down-regulation of PIN gene expression, inhibiting cell fate respecification in the developing primordium (Laplaze et al., 2007), suggesting that cross talk between both hormones is accomplished through the modulation of auxin transport. Furthermore, both brassinosteroids and cytokinins regulate the transcription of genes involved in the auxin signaling pathway (Nakamura et al., 2003, 2006; Goda et al., 2004; Mouchel et al., 2006; Nemhauser et al., 2006; Hardtke, 2007; Dello Ioio et al., 2008).
Alternatively, the observed synergism can be brought about by direct cross talk between cytokinin and brassinosteroid response pathways. BREVIS RADIX is putatively involved, because the mutant, which has a root-specific brassinosteroid deficiency (Mouchel et al., 2004), is insensitive for the inhibition of lateral root formation when cytokinins are applied, due to an altered auxin response (Li et al., 2009), underlining the complexity of the highly interconnected hormonal network that governs plant growth.
GA-Cytokinin Cross Talk during Leaf Development
Overexpression of CKX3 does not interfere with the increase in blade length resulting from GA20ox1 overexpression and synergistically enhances the increase in blade width. This observation demonstrates that increased GA levels overcome the reduction in blade length caused by enhanced cytokinin breakdown and that reduced cytokinin levels enhance the growth-promoting effect of GAs during growth in the blade width direction. Thus, cytokinins and GAs interact antagonistically to promote leaf growth, consistent with their interaction during other plant developmental processes (Greenboim-Wainberg et al., 2005; Jasinski et al., 2005; Weiss and Ori, 2007).
Overexpression of GA20ox1 considerably enhances leaf cell number, whereas cell size is only moderately increased (Gonzalez et al., 2010). GAs promote growth through the proteolytic degradation of growth-repressing DELLA proteins that is likewise documented to stimulate both cell elongation and proliferation (Achard et al., 2009). Cytokinin deficiency or insensitivity affects cell number, suggesting that cross talk between cytokinins and GAs during leaf growth concerns cell division (Werner et al., 2003; Nishimura et al., 2004). Consistent with this hypothesis, CYCD3;1 plays an important role in executing cytokinin (Dewitte et al., 2007) as well as GA responses, because overexpression of CYCD3;1 can rescue the growth of GA-deficient ga1-3 plants (Achard et al., 2009).
Evidence for cross talk between cytokinin and GA response pathways exists at both the level of biosynthesis regulation and signal transduction. Two GA biosynthesis genes, GA20ox and GA4, are down-regulated, while two DELLA genes, GAI and RGA, are up-regulated upon cytokinin treatment of seedlings (Brenner et al., 2005). The GA response inhibitor SPINDLY (SPY) is proposed to mediate cross talk between cytokinin and GA responses (Greenboim-Wainberg et al., 2005; Weiss and Ori, 2007). Whereas SPY promotes cytokinin responses, it represses GA responses, while GAs inhibit cytokinin signaling by suppression of SPY activity through a DELLA-independent pathway (Maymon et al., 2009). Whether SPY integrates GA and cytokinin responses to control cell division and/or cell expansion mechanisms during leaf growth remains to be elucidated.
GRF5-Cytokinin Cross Talk during Leaf Development
Coexpression of CKX3 and GRF5 suppresses leaf phenotypes of the individual overexpressing parent plants, giving rise to blade areas that more closely resemble those of the wild type. This implies that when cytokinin levels are reduced, the effects of GRF5 overexpression disappear and vice versa, suggesting that cytokinins and GRF5 work in concert to stimulate leaf cell proliferation. Like cytokinins, GRF5 is postulated to regulate the transition between cell proliferation and expansion (Gonzalez et al., 2010). Expression of this putative transcription factor coincides with actively dividing cells and rapidly decreases when expansion starts (Horiguchi et al., 2005; M. Andriankaja, S. Dhondt, S. De Bodt, F. Coppens, A. Skirycz, N. Gonzalez, G.T.S. Beemster, and D. Inzé, unpublished data).
Transcript profiling of GRF5-overexpressing seedlings did not reveal any significant enrichment for genes involved in cytokinin responses (Gonzalez et al., 2010), and GRF5 was not transcriptionally regulated by cytokinin treatment or CKX1 overexpression (Brenner et al., 2005; Nemhauser et al., 2006; Lee et al., 2007; Argyros et al., 2008), suggesting that GRF5 does not function directly in the cytokinin signaling pathway and does not influence cytokinin metabolism. Therefore, cross talk between GRF5 and CKX3, and by inference cytokinins, might rather be accomplished by the regulation of (a) common target gene(s) or protein(s). Genes found to be differentially regulated by overexpression of GRF5 (Gonzalez et al., 2010) and in the same direction by cytokinin treatment (Argyros et al., 2008) and/or in the opposite direction by CKX1 overexpression (Brenner et al., 2005) are putative candidates for the convergence of both response pathways.
Interestingly, the leaves of GRF5 plants are dark green due to an increased chlorophyll content (V.B. Tognetti, L. Vercruyssen, N. Gonzalez, D. Inzé, and F. Van Breusegem, unpublished data). Cytokinins are known to stimulate chlorophyll synthesis and to increase the photosynthetic rate (Wareing et al., 1968; Mok, 1994; Riefler et al., 2006; Argyros et al., 2008), illustrating that GRF5 and cytokinin functions are most probably interconnected during leaf development.
AVP1
AVP1 is implicated in polar auxin transport (Li et al., 2005), and overexpression of AVP1 increases auxin content in seedlings (Gonzalez et al., 2010). Although several lines of evidence exist for cytokinin-auxin cross talk (Bögre et al., 2008; Benková and Hejátko, 2009; Chandler, 2009; Veit, 2009), coexpression of AVP1 and CKX3 results in root and leaf phenotypes corresponding to the sum of the phenotypes of the individual heterozygous parent plants, at least under our growth conditions. Therefore, AVP1 and CKX3 seem to function in independent pathways. Alternatively, because the leaf size of AVP1×P10-CKX3 plants closely resembles that of the wild type, the effects of increased auxin levels can be counteracted by those of reduced cytokinin levels, resulting in a restored auxin/cytokinin balance that favors wild-type cell proliferation timing.
CONCLUSION
Diverse hormonal inputs are guided through different, but interconnected, response pathways that form complex networks and determine the final extent of cell proliferation and cell expansion in roots and leaves. By simultaneously altering the expression of two functionally distinct components in the network, we reveal cross talk and bring insight into the hierarchies among the distinct pathways. Further study is needed to identify the “nodes” of the network, the molecular players that integrate the information from these different pathways to control final organ size. On the other hand, root and shoot growth appear to be less tightly linked, and it seems likely from our experiments that the effects on cell proliferation and/or expansion are superimposed to changes in the sink/source balance in the whole plant, at least under optimal conditions.
Enhancing root growth can provide several advantages, such as a better exploitation of soil nutrients and water when environmental conditions are less favorable (Nibau et al., 2008), and is becoming a key feature of ameliorating crop productivity (Gewin, 2010). Simultaneously increasing the size of leaves, which provide the major energy for the plant, might alter the photosynthetic efficiency and create the potential for further biomass improvement under diverse environmental conditions (Zhu et al., 2010). Furthermore, many examples illustrate that the above-described mechanisms that control plant growth and, additionally, tolerance to abiotic stress in Arabidopsis can be readily translated to improve crop productivity. For instance, overexpression of OsGA20ox1 greatly enhances the plant stature of rice (Oikawa et al., 2004), and homologs of BRI1 and CKX genes are present in many crop species, where they perform similar functions (Yamamuro et al., 2000; Chono et al., 2003; Schmülling et al., 2003). Increased tolerance to salt or drought stresses, as observed in AVP1-overexpressing plants (Gaxiola et al., 2001), has been seen in tobacco, creeping bentgrass (Agrostis stolonifera), and tomato (Solanum lycopersicum) by heterologous expression (Park et al., 2005; Duan et al., 2007; Li et al., 2010). This suggests that combining these transgenes will most probably result in the combination of beneficial phenotypic traits in crops.
MATERIALS AND METHODS
Plant Material
Seeds of Arabidopsis (Arabidopsis thaliana) were from the 35S-CKX1 (CKX1; At2G41510) and P10-CKX3 (CKX3; At5G56970) lines (Werner et al., 2003, 2010). Seeds of the AVP1-overexpressing (At1G15690), GRF5-overexpressing (At3G13960), BRI1-overexpressing (At4G39400), and GA20ox1-overexpressing (At4G25420) lines were kindly provided by Dr. Roberto A. Gaxiola (University of Connecticut; Li et al., 2005), Dr. Hirokazu Tsukaya (University of Tokyo; Horiguchi et al., 2005), Dr. Joanne Chory (Salk Institute; Wang et al., 2001), and Dr. Peter Hedden (Rothamsted Research; Coles et al., 1999), respectively. Seeds from all the transgenic lines were in the Col-0 background.
Growth Conditions
As environmental conditions during seed formation and seed storage duration can affect seed vigor, all experiments were conducted with wild-type and transgenic seeds that had been harvested from plants grown side by side on the same tray. For in vitro experiments, seeds were sown on sterile plates containing half-strength MS medium (Murashige and Skoog, 1962) supplemented with 1% Suc and 0.8% agar. The plates were sealed and put in a tissue culture room at 21°C under a 16-h-day/8-h-night regime for shoot growth analysis and under continuous light (110 μmol m−2 s−1) for root growth analysis. AVP1-overexpressing plants were grown in soil for shoot growth analysis at 22°C under 16-h-day/8-h-night conditions (50 μmol m−2 s−1). BL was purchased from Fuji Chemical Industries, dissolved in dimethyl sulfoxide, and added to the agar medium after autoclaving. Plants were grown under 16-h-day/8-h-night conditions for 5 d on half-strength MS prior to transfer to half-strength MS medium containing different concentrations of BL for another 6 d.
Growth Analysis
To measure rosette leaf parameters, seedlings were grown under in vitro conditions for 21 d (AVP1-overexpressing plants in soil for 22 d). Individual leaves (cotyledons and rosette leaves) were dissected and photographed. Their blade area, blade width, and blade length were measured with the ImageJ software (http://rsb.info.nih.gov/ij/).
For root system analysis, seedlings were grown on vertical plates for 9 or 11 d (BL treatments), after which the plates were scanned (Epson). Primary root length, lateral root length, and total root system length were measured with the ImageJ software (http://rsb.info.nih.gov/ij/).
Quantitative Reverse Transcription-PCR
RNA was extracted according to a combined protocol of TRI Reagent RT (Molecular Research Center) and the RNeasy Kit (Qiagen) with on-column DNase (Qiagen) digestion. cDNA was prepared from 250 ng to 1 μg of RNA with the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. The following primer pairs were used: 5′-TCACTAGCGGTCCTGTTCTTG-3′ and 5′-TCAAAAGCCTCCCAATTGTC-3′ for CKX3, 5′-AAAGTTGCGGTTGCGTGTTTG-3′ and 5′-GTTGACTGTGAATCTATCCCTGACC-3′ for BRI1, 5′-TCAGTTCAATGTCTTAGCCTCTGC-3′ and 5′-CCCAACTCCTCCAACTCTCTCC-3′ for GRF5, and 5′-CATCAACGTTCTCGAGCTTGATGTTC-3′ and 5′-GCGGCTCGTGTATTCATGAGCG-3′ for GA20ox1. Quantitative reverse transcription-PCR was done on a LightCycler 480 (Roche Diagnostics) in 384-well plates with LightCycler 480 SYBR Green I Master (Roche) according to the manufacturer’s recommendations. All individual reactions were done in triplicate. Expression levels were normalized against the average of the housekeeping genes UBQ10, CKA2, and CDKA1;1, and relative expression levels of the crosses were determined with the method of Livak and Schmittgen (2001) or qBase (Hellemans et al., 2007). Normalized values for the biological replicates of BL-treated samples were used for statistical analysis according to the proc mixed procedure from SAS (SAS 9.2; SAS Institute), including plate as a random factor. Least square mean differences with mixed-model-based t tests and P values were calculated between 0 nm BL and each BL concentration and that for each line.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf blade length and width for wild-type and P10-CKX3 plants.
Supplemental Figure S2. Leaf blade length and width for P10-CKX3 plants crossed with plants overexpressing BRI1, GRF5, GA20ox1, and AVP1 and their respective controls.
Supplemental Figure S3. Relative expression levels of CKX3 and BRI1 in shoots and roots of the crosses and in roots of wild-type and P10-CKX3 plants upon BL treatment, and CKX3, GRF5, and GA20ox1 expression levels in crossed seedlings.
Supplementary Material
Acknowledgments
We sincerely thank our colleagues in the Systems Biology of Yield group for fruitful discussions, Véronique Storme for the statistical analysis of the quantitative reverse transcription-PCR data, and Martine De Cock for help in preparing the manuscript.
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GTS, Genschik P. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Argueso CT, Ferreira FJ, Kieber JJ. (2009) Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ 32: 1147–1160 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z. (2004) Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134: 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Hejátko J. (2009) Hormone interactions at the root apical meristem. Plant Mol Biol 69: 383–396 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bögre L, Magyar Z, López-Juez E. (2008) New clues to organ size control in plants. Genome Biol 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlaug N. (2007) Feeding a hungry world. Science 318: 359. [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Chandler JW. (2009) Auxin as compère in plant hormone crosstalk. Planta 231: 1–12 [DOI] [PubMed] [Google Scholar]
- Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y. (2003) A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol 133: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, García-Lepe R, Lewis MJ, Hedden P. (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Scaglia Linhares F, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al. (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Duan XG, Yang AF, Gao F, Zhang SL, Zhang JR. (2007) Heterologous expression of vacuolar H+-PPase enhances the electrochemical gradient across the vacuolar membrane and improves tobacco cell salt tolerance. Protoplasma 232: 87–95 [DOI] [PubMed] [Google Scholar]
- Eathington SR, Crosbie TM, Edwards MD, Reiter RS, Bull JK. (2007) Molecular markers in a commercial breeding program. Crop Sci 47: S154–S163 [Google Scholar]
- Edgerton MD. (2009) Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol 149: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR. (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewin V. (2010) Food: an underground revolution. Nature 466: 552–553 [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Beemster GTS, Inzé D. (2009) David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr Opin Plant Biol 12: 157–164 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guivarc’h A, Rembur J, Goetz M, Roitsch T, Noin M, Schmülling T, Chriqui D. (2002) Local expression of the ipt gene in transgenic tobacco (Nicotiana tabacum L. cv. SR1) axillary buds establishes a role for cytokinins in tuberization and sink formation. J Exp Bot 53: 621–629 [DOI] [PubMed] [Google Scholar]
- Hardtke CS. (2007) Transcriptional auxin-brassinosteroid crosstalk: who’s talking? Bioessays 29: 1115–1123 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19.1–R19.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Hu Y, Bao F, Li J. (2000) Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J 24: 693–701 [DOI] [PubMed] [Google Scholar]
- James C. (2009) Global Status of Commercialized Biotech/GM Crops: 2009. ISAAA Brief Number 41. International Service for the Acquisition of Agri-biotech Applications, Ithaca, NY [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M. (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Krizek BA. (2009) Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Kudo T, Kiba T, Sakakibara H. (2010) Metabolism and long-distance translocation of cytokinins. J Integr Plant Biol 52: 53–60 [DOI] [PubMed] [Google Scholar]
- Kuiper D. (1993) Sink strength: established and regulated by plant growth regulators. Plant Cell Environ 16: 1025–1026 [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Park JY, Ku SJ, Ha YM, Kim S, Kim MD, Oh M-H, Kim J. (2007) Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7) overexpression in cytokinin response. Mol Genet Genomics 277: 115–137 [DOI] [PubMed] [Google Scholar]
- Li J, Mo X, Wang J, Chen N, Fan H, Dai C, Wu P. (2009) BREVIS RADIX is involved in cytokinin-mediated inhibition of lateral root initiation in Arabidopsis. Planta 229: 593–603 [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Li X, Mo X, Shou H, Wu P. (2006) Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Li Z, Baldwin CM, Hu Q, Liu H, Luo H. (2010) Heterologous expression of Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ 33: 272–289 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Maymon I, Greenboim-Wainberg Y, Sagiv S, Kieber JJ, Moshelion M, Olszewski N, Weiss D. (2009) Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J 58: 979–988 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T. (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DWS, Mok MC. (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Mok MC. (1994) Cytokinins and plant development: an overview. DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 155–166 [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS. (2006) BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443: 458–461 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Müssig C, Shin GH, Altmann T. (2003) Brassinosteroids promote root growth in Arabidopsis. Plant Physiol 133: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Goda H, Shimada Y, Yoshida S. (2004) Brassinosteroid selectively regulates PIN gene expression in Arabidopsis. Biosci Biotechnol Biochem 68: 952–954 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S. (2003) Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K-i, Nozaki H, Asami T, Yoshida S, Fujioka S. (2006) Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J 45: 193–205 [DOI] [PubMed] [Google Scholar]
- Nakaya M, Tsukaya H, Murakami N, Kato M. (2002) Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol 43: 239–244 [DOI] [PubMed] [Google Scholar]
- Naqvi S, Farré G, Sanahuja G, Capell T, Zhu C, Christou P. (2010) When more is better: multigene engineering in plants. Trends Plant Sci 15: 48–56 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz I, Berkefeld H, Puzio PS, Grundler FMW. (2001) Pyk10, a seedling and root specific gene and promoter from Arabidopsis thaliana. Plant Sci 161: 337–346 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M. (2004) A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol 55: 687–700 [DOI] [PubMed] [Google Scholar]
- Park S, Li J, Pittman JK, Berkowitz GA, Yang H, Undurraga S, Morris J, Hirschi KD, Gaxiola RA. (2005) Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci USA 102: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Foyer CH. (2001) Sink regulation of photosynthesis. J Exp Bot 52: 1383–1400 [DOI] [PubMed] [Google Scholar]
- Peng ZY, Zhou X, Li L, Yu X, Li H, Jiang Z, Cao G, Bai M, Wang X, Jiang C, et al. (2009) Arabidopsis Hormone Database: a comprehensive genetic and phenotypic information database for plant hormone research in Arabidopsis. Nucleic Acids Res (Database issue) 37: D975–D982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Candela H, Micol JL. (2009) Understanding synergy in genetic interactions. Trends Genet 25: 368–376 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH. (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Schmülling T, Werner T, Riefler M, Krupková E, Bartrina y Manns I. (2003) Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116: 241–252 [DOI] [PubMed] [Google Scholar]
- Sherameti I, Venus Y, Drzewiecki C, Tripathi S, Dan VM, Nitz I, Varma A, Grundler FM, Oelmüller R. (2008) PYK10, a β-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J 54: 428–439 [DOI] [PubMed] [Google Scholar]
- Veit B. (2009) Hormone mediated regulation of the shoot apical meristem. Plant Mol Biol 69: 397–408 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wareing PF, Khalifa MM, Treharne KJ. (1968) Rate-limiting processes in photosynthesis at saturating light intensities. Nature 220: 453–457 [DOI] [PubMed] [Google Scholar]
- Weiss D, Ori N. (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144: 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Holst K, Pörs Y, Guivarc’h A, Mustroph A, Chriqui D, Grimm B, Schmülling T. (2008) Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J Exp Bot 59: 2659–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. (2006) New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8: 371–381 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. (2010) Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD. (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I. (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR. (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.