Abstract
Phosphate (Pi) deficiency triggers the differential expression of a large set of genes, which communally adapt the plant to low Pi bioavailability. To infer functional modules in early transcriptional responses to Pi deficiency, we conducted time-course microarray experiments and subsequent coexpression-based clustering of Pi-responsive genes by pairwise comparison of genes against a customized database. Three major clusters, enriched in genes putatively functioning in transcriptional regulation, root hair formation, and developmental adaptations, were predicted from this analysis. Validation of gene expression by quantitative reverse transcription-PCR revealed that transcripts from randomly selected genes were robustly induced within the first hour after transfer to Pi-deplete medium. Pectin-related processes were among the earliest and most robust responses to Pi deficiency, indicating that cell wall alterations are critical in the early transcriptional response to Pi deficiency. Phenotypical analysis of homozygous mutants defective in the expression of genes from the root hair cluster revealed eight novel genes involved in Pi deficiency-induced root hair elongation. The plants responded rapidly to Pi deficiency by the induction of a subset of transcription factors, followed by a repression of genes involved in cell wall alterations. The combined results provide a novel, integrated view at a systems level of the root responses that acclimate Arabidopsis (Arabidopsis thaliana) to suboptimal Pi levels.
As an integral constituent of nucleic acids and phospholipids, phosphate (Pi) is an essential nutrient vital to virtually all organisms. Beside its function as a structural component, Pi plays key roles in signal transduction cascades and in the regulation of metabolic pathways. Due to its low mobility in soils, Pi is limiting for plant growth in almost all agricultural soils, and Pi availability determines the distribution of plants in natural habitats. Combating Pi deficiency by fertilizers is expensive and can be detrimental to the environment when leached out of the soil and further depletes the dwindling source of available Pi rock. Understanding how plants regulate Pi acquisition and distribution within cells, tissues, and organs may help to identify routes to improve crop performance and reduce the detrimental side effects caused by fertilization. In addition, deciphering the environmental factors that are determinants in the adaptation of plants to a given Pi supply is of utmost importance for maintaining and restoring natural ecosystems.
The mechanisms of Pi homeostasis are complex. A large subset of genes responds to the availability of Pi; most of these transcripts have been identified but not placed within a network controlling cellular Pi homeostasis (Misson et al., 2005; Bari et al., 2006; Morcuende et al., 2007). The Myb-type transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1) is a conserved key regulator, directly controlling a subset of Pi deficiency genes by binding to an imperfect palindromic sequence motif and thereby activating these genes (Rubio et al., 2001). Consistent with a critical regulatory role, overexpression of PHR1 led to increased Pi accumulation (Nilsson et al., 2007). The activity of PHR1 is controlled by the SUMO E3 protein ligase SIZ1 (Miura et al., 2005), representing the most upstream component of the Pi deficiency signaling cascade identified thus far. Another subset of Pi deficiency genes is regulated by the E2 ubiquitin conjugase PHOSPHATE2 (PHO2). A connection between these two central switches is established by MICRORNA399 (MiR399), which systemically controls PHO2 through transcript cleavage (Aung et al., 2006; Pant et al., 2008). Expression of MiR399 is strongly induced by Pi deficiency, possibly controlled by PHR1 (Bari et al., 2006).
Numerous biochemical processes aimed at reducing the requirement for Pi, remobilization of internal Pi, or improving its acquisition from the soil are altered by Pi deficiency. These global processes include changes in membrane lipid composition (Kobayashi et al., 2006), secretion of phosphatases (Bozzo et al., 2002), and expression of Pi transporters (Poirier and Bucher, 2002; Guo et al., 2008; Cubero et al., 2009). In addition, the acquisition of Pi is facilitated by alterations in root architecture and by the formation of root hairs with increased length and density (Bates and Lynch, 1996; Ma et al., 2001; Williamson et al., 2001; Müller and Schmidt, 2004). The latter trait is of particular importance when depletion zones, caused by the low mobility of Pi via mass flow and diffusion, develop around the root surface (Ma et al., 2001). UBIQUITIN-SPECIFIC PROTEASE14 (UBP14) has been shown to be crucial for root development under Pi-deficient conditions (Li et al., 2010). A mutant harboring a synonymous substitution in the UBC14 gene, presumably reducing the translation efficiency of the UBC14 transcript, forms short hairs under Pi-deficient conditions, while root hair development was indistinguishable from the wild type under control conditions. These results indicate that ubiquitin-dependent protein degradation plays an important role both in Pi deficiency signaling and in adapting root development to the prevailing Pi availability. Other genes specifically required for inducing the root hair phenotype typical of Pi-deficient plants have not yet been identified.
Microarray experiments using the ATH1 Affymetrix gene chip indicate that a subset of about 800 to 1,000 genes are responsive to Pi starvation (Misson et al., 2005; Morcuende et al., 2007; this study). Only a surprisingly small percentage of the differentially expressed genes overlap among these studies, indicating that the majority of these genes does not represent a robust and conserved Pi deficiency response but rather reflects more complex, secondary interactions between Pi supply and differences in growth and assay conditions. A biologically meaningful interpretation of high-throughput experiments, in particular for experiments addressing rapidly induced changes in gene expression, necessitates a distinction between genes that are responsive primarily to the treatment and secondary responses that are induced or repressed by mechanical handling of the plants or by other events not directly associated with the treatment. Moreover, concerted processes comprising multiple, coregulated components are often obscured by temporal kinetics (i.e. by the fact that only a minority of these genes are induced or repressed at a given time point at a given cutoff value).
Large microarray data sets for the model plant Arabidopsis (Arabidopsis thaliana) are available in public databases. These experimental data are collected from various tissues, developmental stages, and following various stimuli. Assuming that groups of genes encode synchronized processes that control or optimize the plant performance, correlation of their coexpression may help to discover new genes that function in this same process. In this study, we sought to identify genes that are involved in early Pi deficiency signaling processes. In order to gain insights into the complete orchestration of the transcriptional response, as opposed to tracking down single genes, we clustered genes that are induced or repressed by Pi starvation into groups of closely correlated modules based on their coexpression under various sets of experimental conditions by mining public databases. To this end, we have developed an easy-to-use software package that allows fast coexpression-based clustering of large data sets into modules that help to validate hypotheses and to direct follow-up research. The Multi-Array Correlation Computation Utility (MACCU) is a toolbox that computes coexpression gene clusters using a personal computer. In addition, this utility allows deducing organ- and tissue-specific coexpression networks that further help to reduce biological complexity. Using this approach, we discovered potentially critical calcium-, hormone-, and cell wall-related signaling events as the earliest traceable changes to Pi deficiency at the transcriptional level. In addition, our data analysis revealed a cluster of genes most of which have predicted or verified functions in cell wall alterations. A comprehensive reverse genetic screening of mutants harboring defects in these genes suggested a function of some of these genes in root hair elongation, including known and novel players in root hair development. The results indicate that root hair elongation in response to Pi deficiency requires the concerted action of a surprisingly large subset of genes and presents a proof-of concept for the use of coexpression networks to direct follow-up experimentation.
RESULTS
Identification of Early Pi-Responsive Genes
With the goal of identification of early Pi-responsive genes, we conducted time-course microarray experiments using Arabidopsis roots subjected to short (1 h) and intermediate (6 and 24 h) periods of Pi starvation. To avoid the drawbacks inherent to the transfer of plants grown in agar-based medium (e.g. physical injuries and the transfer of Pi-containing medium adhering to the roots), we choose a hydroponic system that allows rapid transfer of plants practically without damage (Buckhout et al., 2009). A further advantage of this approach over agar-based medium is that transcriptional changes can be monitored with a delay-free, clear-cut onset of Pi-deficient conditions. Transcriptional profiling was conducted using Affymetrix ATH1 gene chips.
A total of 797 genes were found to be responsive to Pi deficiency with a fold change of greater than 2-fold for at least one time point using a cutoff value of P < 0.05 (Supplemental Table S1). The Pi status of the plants was documented by the induction of several genes that had previously been described as being responsive to the Pi supply (Table I). Transcripts of several genes were found to be approximately 3-fold higher in abundance 24 h after transfer to Pi-free medium when compared with control plants that had been transferred to fresh Pi-containing medium. Thus, the plants were Pi deficient at this time point. The glutathione transferase ATGSTU11, for example, was highly induced 1 h after transfer. We concluded that a rapid change in the Pi status of the plants was reflected in equally rapid changes in transcript abundance.
Table I. Induction of Pi-responsive genes in Arabidopsis roots.
Data represent means from three biological replicates. Fold changes are based on the differences in expression level between Pi-deficient and Pi-sufficient plants. The numbers in parentheses are P values adjusted for multiple testing by the method of Benjamini and Hochberg (1995).
| Locus | Annotation | Fold Expression (Log2) |
||
| 1 h | 2 h | 24 h | ||
| At3g17790 | ACP5, acid phosphatase | −0.37 (0.92) | 1.10 (0.15) | 3.12 (0.002) |
| At5g20150 | SPX1, SPX DOMAIN GENE1 | 0.22 (0.95) | 1.63 (0.007) | 3.17 (0.0003) |
| AT2G45130 | SPX3, SPX DOMAIN GENE3 | −0.10 (0.97) | 0.06 (0.92) | 1.18 (0.003) |
| At2g38940 | Pht1;4, Pi transporter | −1.27 (0.14) | −0.19 (0.87) | 1.37 (0.16) |
| At3g02040 | SRG3, glycerophosphodiester phosphodiesterase | 0.11 (0.97) | 0.56 (0.12) | 1.82 (0.0004) |
| At1g73220 | OCT1, ORGANIC CATION/CARNITINE TRANSPORTER1 | 0.74 (0.76) | 0.47 (0.71) | 4.18 (0.0008) |
| At5g20790 | Unknown protein | 0.01 (1.00) | 0.08 (0.94) | 1.57 (0.009) |
| At1g69930 | ATGSTU11 | 2.34 (0.09) | 3.82 (0.0009) | 0.43 (0.94) |
| At2g02990 | RNS1, ribonuclease | 0.21 (0.96) | 4.16 (7.39E-06) | 2.93 (0.001) |
Computing Root-Specific Coexpression Networks with the MACCU Toolbox
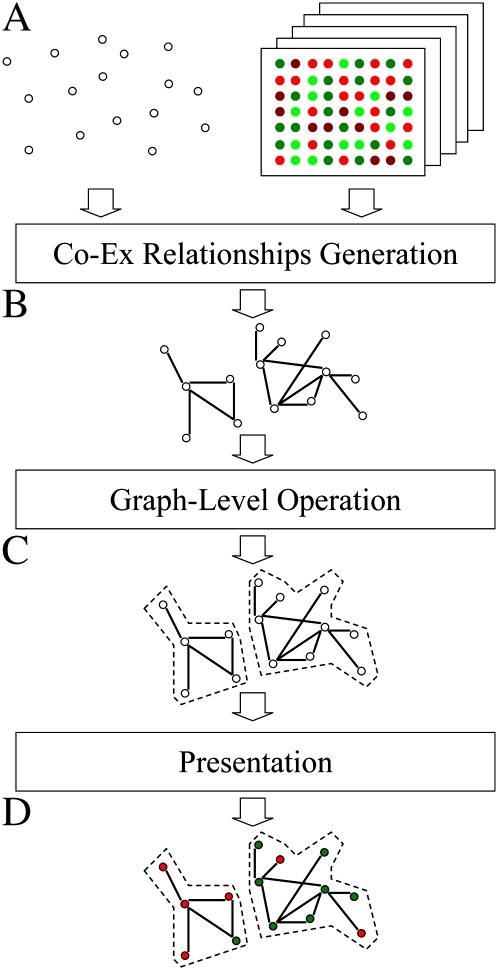
To construct root-specific gene networks, we developed the MACCU toolbox, which is outlined in Figure 1. The software package is available with a thorough description at http://maccu.sourceforge.net/. MACCU is composed of programs in three layers: the generation layer, the operation layer, and the presentation layer. The generation layer consists of two programs, clustering and fishing, both of which identify coexpression relationships of gene pairs with Pearson correlation above a chosen cutoff value. Based on a given list of genes, the clustering module computes coexpression relationships between genes within the list, and the fishing module computes coexpression relationships between the bait genes and the whole genome. Computational results of this layer are saved in a simple graph format. The operation layer executes graph-level operations (i.e. computing intersections or subtraction of multiple graphs). Finally, the presentation layer creates image files of the graphs using the Graphviz program (http://www.graphviz.org/), allowing for inclusion of additional information (e.g. gene symbols and/or colors of fold changes).
Figure 1.
Outline of the MACCU toolkit. A, Input data: a gene list (white nodes) and normalized microarray data from public databases. B, Coexpression relationships are identified by pairwise comparison of genes. Nodes without any relationships are removed. C, Generation of gene networks. D, Biologically meaningful information, such as gene symbols and colors indicating fold changes, can be added to the image. [See online article for color version of this figure.]
To identify functional modules involved in the adaptation of plants to Pi deficiency, we clustered the 797 Pi-responsive genes on the basis of their coexpression determined in experiments taken from public microarray databases. In order to reliably identify coexpression relationships in Arabidopsis roots, the following procedure was carried out. First, 2,671 microarray sample hybridizations using the ATH1 gene chip were collected from the NASCarrays database, and 300 microarrays were manually identified as reporting data from root-related experiments. Second, the data were normalized by the robust multiarray average (RMA) method using the R software package, and the 300 root-related arrays were extracted from the normalized data. Third, coexpression relationships between two genes were identified by calculating the pairwise Pearson correlation with a cutoff value of 0.8 based on the root-related arrays. It was found that 273 genes were associated with at least one coexpression relationship, and a network was generated in which these 273 genes were represented as nodes and the coexpression relationships between them as edges (Supplemental Fig. S1). The largest connected subnetwork (cluster) contained 220 genes and was composed of two subclusters uniformly containing either up-regulated or down-regulated genes, each with about 100 genes. To identify root-specific networks, we removed coexpression relationships with a Pearson correlation of 0.7 or greater that were also apparent when the total set of 2,671 arrays was used as a database (Supplemental Fig. S2). We did not attempt to further subdivide these clusters.
The cutoff value of 0.8 was selected as follows. We initially clustered the 797 Pi-responsive genes based on root-related arrays with a threshold between 0.7 and 0.9. It was found that a cutoff value of 0.9 was too stringent (i.e. the number of clustered genes was too small), while a cutoff of 0.7 generated a very large cluster. We then applied a series of Gene Ontology (GO) enrichment computations for thresholds from 0.78 to 0.85 and looked for the cutoff with the best enrichments of GO categories among the 797 Pi-responsive genes (P < 5e-5). A cutoff of 0.8 resulted in the best GO enrichment. An example for the subcluster of up-regulated genes (Supplemental Fig. S1) is shown in Supplemental Table S2. Then, root-specific edges were selected with regard to their stronger correlations based on root-related arrays (0.8 or greater) and weaker correlations based on all array experiments (less than 0.7). Following this, we experimentally confirmed the validity of the procedure.
Coexpression-Based Gene Clustering Predicts Modules with Functions in the Pi Deficiency Response
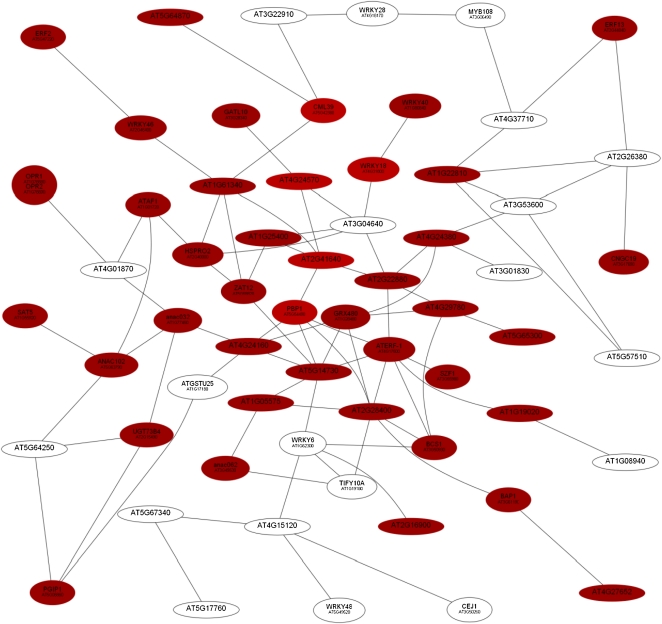
Using the criteria described above, the computed network contained 187 genes that are connected by at least one edge after subtraction of non-root-related edges, comprising three large clusters of 62, 52, and 17 genes, respectively. The first cluster, referred to as Root Module 1 (RM1), was exclusively composed of genes that were up-regulated upon Pi deficiency (Table II; Fig. 2). Most of the genes showed highest expression 1 h (16 genes) or 6 h (35 genes) after exposure to Pi-free medium. Generally, genes in this cluster responded faster to Pi deficiency than those in the other two clusters. GO enrichment (P values ranging between 1e-6 and 1e-4) and the number of genes were highest for “response to salicylic acid stimulus” (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb2/). A subset of the genes in this cluster (17 genes) encode transcription factors, mainly of the NAC, WRKY, and ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR (ERF) type, or genes with predicted or confirmed roles in signal transduction (Table II). The majority of transcription factors are reportedly involved in jasmonic acid, salicylic acid, and ethylene signaling, indicating that these hormones play critical roles in initial Pi deficiency-signaling events. Another group of genes, including two calmodulin-like or calmodulin-related genes, the calcium-transporting ATPase At3g22910, and the calcium-binding protein PINOID-BINDING PROTEIN1 (PBP1), is related to calcium signaling. Some of these genes showed high induction levels 1 h after transfer to Pi-deficient conditions. Three genes, OXOPHYTODIENOATE REDUCTASE1 (OPR1), BON ASSOCIATION PROTEIN1 (BAP1), and the lysophosphatidic acid acyltransferase At4g24160, are related to lipid binding, metabolism, or homeostasis. The strongest expression after 1 h was observed for WRKY18 and WRKY40, which have partially redundant roles (Xu et al., 2006), and for PBP1, CALMODULIN LIKE39 (CML39), the transferase AT2g41640, and the dicarboxylate carrier DIC2, which could be involved in the transport of Pi across the inner mitochondrial membrane (Table II; Palmieri et al., 2008).
Table II. Genes comprising RM1.
Boldface values indicate genes with the strongest repression upon Pi deficiency. Fold changes are based on the differences in expression level between Pi-deficient and Pi-sufficient plants. Dashes indicate not differentially expressed at this time point. Data represent means from three biological replicates. The numbers in parentheses are P values adjusted for multiple testing by the method of Benjamini and Hochberg (1995).
| Locus | Annotation | Molecular Function | Fold (Log2) |
||
| 1 h | 6 h | 24 h | |||
| At1g01720 | ANAC002 (ATAF1) | Sequence-specific DNA-binding transcription factor activity, transcription activator activity | 1.36 (0.34) | 1.33 (0.13) | 1.43 (0.37) |
| At5g63790 | ANAC102 | Sequence-specific DNA-binding transcription factor activity | 1.81 (0.07) | 1.47 (0.05) | 1.06 (0.50) |
| At3g49530 | ANAC062 | Sequence-specific DNA-binding transcription factor activity | 1.23 (0.0007) | 1.16 (0.0005) | – |
| At1g77450 | ANAC032 | Sequence-specific DNA-binding transcription factor activity | 1.14 (0.58) | 1.81 (0.08) | 1.25 (0.59) |
| At1g62300 | WRKY6 | Sequence-specific DNA-binding transcription factor activity | – | 1.63 (0.12) | – |
| At4g31800 | WRKY18 | Sequence-specific DNA-binding transcription factor activity | 2.51 (1.74E-05) | 1.03 (0.01) | – |
| At4g18170 | WRKY28 | Sequence-specific DNA-binding transcription factor activity | – | 1.51 (0.09) | 1.72 (0.25) |
| At1g80840 | WRKY40 | Sequence-specific DNA-binding transcription factor activity | 1.97 (0.001) | 2.16 (0.0003) | – |
| At2g46400 | WRKY46 | Sequence-specific DNA-binding transcription factor activity | 1.35 (0.19) | 2.41 (0.002) | – |
| At5g49520 | WRYY48 | Sequence-specific DNA-binding transcription factor activity | – | 1.50 (0.007) | 1.01 (0.26) |
| At4g17500 | ATERF-1 | Sequence-specific DNA-binding transcription factor activity | 1.10 (0.11) | 1.54 (0.005) | – |
| At5g47220 | ERF2 | DNA binding | 1.52 (0.16) | 2.02 (0.01) | – |
| At3g06490 | MYB108 | Sequence-specific DNA-binding transcription factor activity | – | 1.84 (1.79E-05) | 1.38 (0.001) |
| At3g50260 | CEJ1 | Sequence-specific DNA-binding transcription factor activity | – | 1.31 (0.10) | 1.26 (0.40) |
| At1g19180 | JAZ1, TIFY10A | Protein binding | – | 1.29 (0.01) | – |
| At1g22810 | ERF/AP2 transcription factor family | DNA binding | 1.44 (0.004) | – | – |
| At2g44840 | ERF13 | Sequence-specific DNA-binding transcription factor activity | 1.72 (0.01) | 1.66 (0.005) | – |
| At3g53600 | Zinc finger (C2H2 type) family protein | Nucleic acid binding, zinc ion binding | – | 1.44 (0.01) | – |
| At1g76680/At1g76690 | OPR1/OPR2 | 12-Oxophytodienoate reductase activity | 1.67 (0.19) | 1.14 (0.22) | – |
| At5g54490 | PBP1 | Calcium ion binding, protein binding | 2.25 (0.18) | 1.93 (0.10) | – |
| At5g42380 | CML39 | Calcium ion binding | 2.42 (0.05) | 1.80 (0.06) | – |
| At2g41640 | Transferase | Transferase activity | 2.54 (0.0002) | 1.16 (0.03) | – |
| At3g22910 | ATPase E1-E2 type family protein | Calmodulin binding | – | 3.29 (5.61E-06) | – |
| At3g61190 | BAP1 | Phospholipid binding, protein binding | 1.46 (0.12) | 1.44 (0.04) | – |
| At3g01830 | Calcium-binding EF-hand family protein | Calcium ion binding | -- | 1.65 (0.0004) | – |
| At5g59820 | ZAT12 | Nucleic acid binding, zinc ion binding | 1.30 (0.25) | 2.25 (0.005) | 1.18 (0.41) |
| At3g55980 | SZF1 | Sequence-specific DNA-binding transcription factor activity | 1.11 (0.06) | – | – |
| At5g64250 | Aldolase-type TIM barrel family protein | Nitronate monooxygenase activity | – | 1.50 (0.0006) | – |
| At1g17180 | ATGSTU25 | Glutathione transferase activity | – | 3.34 (0.001) | – |
| At4g01870 | TolB protein related | Not assigned | – | 2.80 (0.003) | – |
| At3g28340 | GATL10 | Polygalacturonate 4-α-galacturonosyltransferase activity | 1.06 (0.04) | – | – |
| At5g06860 | PGIP1 | Polygalacturonase inhibitor activity | 1.53 (0.43) | 4.46 (0.0005) | 1.78 (0.39) |
| At3g04640 | Gly-rich protein | Not assigned | – | 1.13 (0.007) | – |
| At5g67340 | ARM repeat superfamily protein | Ubiquitin-protein ligase activity | – | 1.94 (0.04) | 1.30 (0.52) |
| At5g17760 | P-loop-containing nucleoside triphosphate hydrolases superfamily protein | Nucleoside-triphosphatase activity | – | 2.34 (0.0007) | – |
| At4g37710 | VQ motif-containing protein | Not assigned | – | 2.77 (0.0005) | – |
| AT4G15120 | VQ motif-containing protein | Not assigned | – | 1.36 (0.07) | 1.13 (0.44) |
| At1g08940 | Phosphoglycerate mutase family protein | Catalytic activity | – | 1.28 (0.005) | – |
| At2g26380 | LRR family protein | Not assigned | – | 1.40 (0.002) | – |
| At1g55920 | SAT5 | Ser O-acetyltransferase activity | 1.15 (0.23) | 1.42 (0.03) | – |
| AT2G15490 | UGT73B4 | UDP-glucosyltransferase activity | 1.22 (0.51) | 2.82 (0.006) | – (0.86) |
| At4g24160 | Lysophosphatidic acid acyltransferase | Lysophosphatidic acid acyltransferase activity | 1.62 (0.02) | – | – |
| At2g40000 | HSPRO2 | Not assigned | 1.26 (0.21) | 1.68 (0.02) | 1.18 (0.33) |
| At3g50930 | BCS1 | ATPase activity | 1.88 (0.07) | 2.28 (0.006) | 1.18 (0.45) |
| At2g22880 | VQ motif-containing protein | Not assigned | 1.09 (0.55) | 2.34 (0.01) | 1.30 (0.50) |
| At4g24570 | DIC2 | Dicarboxylic acid transmembrane transporter activity | 2.58 (0.0004) | – | – |
| At1g61340 | F-box family protein | Not assigned | 1.91 (0.0005) | 1.19 (0.007) | – |
| At1g28480 | GRX480 | Protein disulfide oxidoreductase activity | 1.36 (0.37) | 1.44 (0.11) | – |
| At3g17690 | CNGC19 | Ion channel activity (calmodulin binding, cyclic nucleotide binding) | 1.08 (0.29) | 2.32 (0.001) | – |
| At5g64870 | SPFH/band 7/PHB domain-containing membrane-associated protein family | Not assigned | 1.93 (0.0005) | 2.40 (2.83E-05) | – |
| At2g16900 | Phospholipase-like protein (PEARLI4) family | Not assigned | 1.58 (9.00E-05) | 1.18 (0.0006) | – |
| At1g19020 | Unknown protein | Not assigned | 1.36 (0.50) | 1.92 (0.08) | 1.86 (0.34) |
| At1g05575 | Unknown protein | Not assigned | 1.51 (0.23) | 1.16 (0.18) | – |
| At2g28400 | Unknown protein | Not assigned | 1.74 (0.001) | 2.20 (7.30E-05) | – |
| At5g14730 | Unknown protein | Not assigned | 1.35 (0.43) | 1.80 (0.06) | – |
| At4g27652 | Unknown protein | Not assigned | 1.57 (0.03) | – | – |
| At5g65300 | Unknown protein | Not assigned | 1.30 (0.01) | 1.08 (0.01) | – |
| At4g24380 | Unknown protein | Not assigned | 1.39 (0.05) | 2.08 (0.0008) | – |
| At4g29780 | Unknown protein | Not assigned | 1.86 (0.001) | 1.61 (0.001) | – |
| At5g57510 | Unknown protein | Not assigned | – | 2.28 (3.69E-06) | – |
| At1g25400 | Unknown protein | Not assigned | 1.58 (0.03) | 2.53 (0.0003) | – |
Figure 2.
Coexpression relationships of genes in RM1. The red nodes indicate up-regulated genes, and the white nodes represent genes that are not differentially expressed at this time point. The figure shows the expression of genes 1 h after transfer to Pi-deplete medium. [See online article for color version of this figure.]
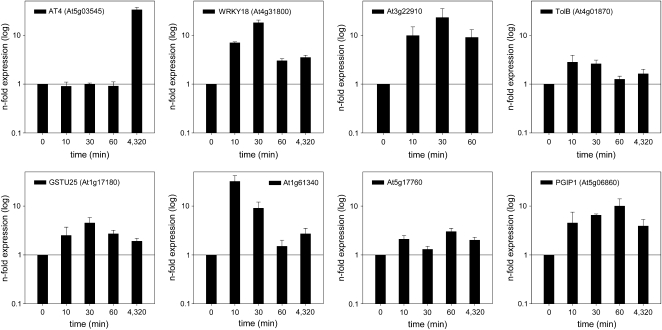
To investigate whether the early expression of genes was reliably mirrored in the microarray experiments, we analyzed the induction of a subset of these genes by quantitative reverse transcription (qRT)-PCR in roots that were subjected to Pi deficiency for 10, 30, or 60 min and 3 d. The expression of the riboregulator gene AT4, reportedly being highly responsive to Pi starvation, was used for comparison. Expression of AT4 was induced by 35-fold after 72 h of growth on Pi-deplete medium, but no induction was found after short-term exposure to Pi starvation, indicating that an increase in transcript abundance of fast-responding genes was not due to an artifact caused by the handling of the plants (Fig. 3). The genes WRKY18 and DIC2, which showed highest expression in the microarray experiments after 1 h, exhibited markedly increased transcript levels already 10 min after transfer to Pi-deplete medium. Generally, transcripts of all genes tested showed robust increases within the first 60 min of growth under Pi-deficient conditions, indicating that changes in gene expression occur very early after transfer of the plants.
Figure 3.
Short-term induction of genes with potential functions in Pi deficiency signaling derived from RM1. Relative transcript abundance was determined 10, 30, and 60 min after transfer to Pi-free medium by qRT-PCR and compared with control samples using the comparative CT method. The expression of AT4 was determined as a control.
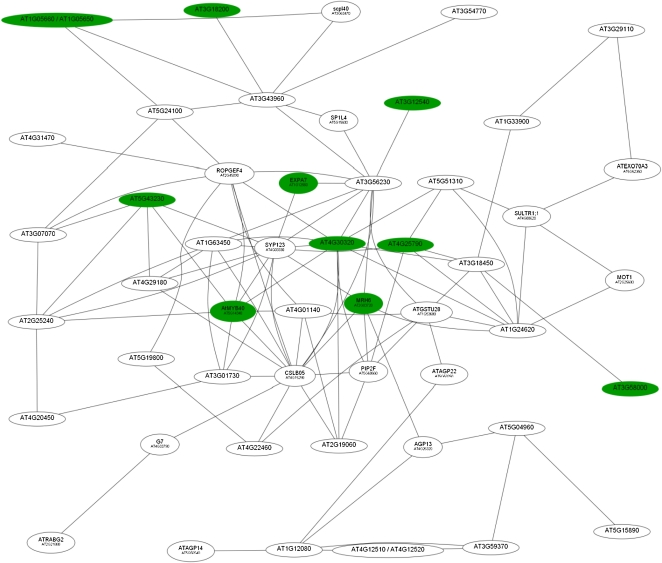
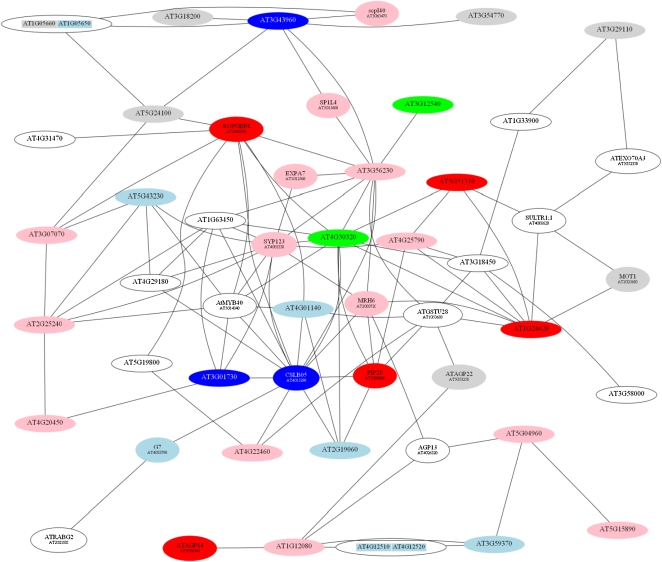
The second root module (RM2; Fig. 4) was composed of 52 genes, most of which have been related to root hair development by transcriptional profiling experiments with root hair-defective mutants (Jones et al., 2006), microarray-based deduction of root hair-specific cis-elements (Won et al., 2009), or by information-driven approaches (Breen and Grierson, 2007). Generally, genes in this cluster were expressed later in the course of the response to Pi deficiency than those in RM1. Maximal expression of most of the genes in this cluster was observed 6 h after the onset of Pi deficiency (Table III). Interestingly, all genes in this cluster were down-regulated upon Pi deficiency. Two genes involved in lipid transport (At4g12510 and At4g22460), two glycosylphosphatidylinositol (GPI)-anchored arabinogalactan proteins (AGP13 and AGP22), and an unknown protein (At3g59370) showed the greatest fold change upon Pi starvation of any of the differentially expressed genes.
Figure 4.
Coexpression relationships of genes in RM2. The red nodes indicate up-regulated genes, and white nodes represent genes not differentially expressed at this time point. The figure shows the expression of genes 1 h after transfer to Pi-deplete medium. [See online article for color version of this figure.]
Table III. Genes comprising RM2.
Boldface values indicate genes with the strongest repression upon Pi deficiency. Fold changes are based on the differences in expression level between Pi-deficient and Pi-sufficient plants. Dashes indicate not differentially expressed at this time point. Data represent means from three biological replicates. The numbers in parentheses are P values adjusted for multiple testing by the method of Benjamini and Hochberg (1995).
| Locus | Annotation | Molecular Function/Process | Fold (Log2) |
||
| 1 h | 6 h | 24 h | |||
| At1g12560a,b,c | EXP7 | Plant-type cell wall loosening | 1.83 (0.29) | 1.35 (0.25) | 1.24 (0.65) |
| At2g03720a,b,c | MRH6 | Root hair cell differentiation | 1.00 (0.58) | 1.45 (0.11) | 1.30 (0.48) |
| At1g05650/At1g05660 | Polygalacturonase/pectinase, putative | Polygalacturonase activity | 1.44 (0.07) | 2.43 (0.0005) | 1.72 (0.56) |
| At3g18200 | Nodulin MtN21 family protein | Not assigned | 1.09 (0.08) | 1.51 (0.003) | – |
| At3g12540a,b,c | Unknown protein | Not assigned | 1.28 (0.13) | – | 1.02 (0.37) |
| At3g58000a,b | VQ motif-containing protein | Not assigned | 1.83 (0.20) | −2.05 (0.04) | −1.15 (0.62) |
| At4g25790a,b,c | CAP superfamily protein | Not assigned | 1.26 (0.51) | −1.68 (0.10) | – |
| At4g30320a,b | CAP superfamily protein | Not assigned | 1.32 (0.54) | −1.90 (0.09) | −1.22 (0.66) |
| At5g14340 | MYB40 | Sequence-specific DNA-binding transcription factor activity | 1.14 (0.03) | – | – |
| At5g43230a,b | Unknown protein | Not assigned | 1.04 (1.00) | – | – |
| At4g29180a,b,c | RHS16 | Kinase activity | – | −1.00 (0.06) | – |
| At1g63450a,c | RHS8 | Not assigned | – | −1.69 (0.005) | −1.49 (0.09) |
| At2g45890a,c | RHS11, ROPGEF4 | Rho guanyl nucleotide-exchange factor activity | – | – | −1.08 (0.32) |
| At2g21880a,b | ATRAB7A | GTP binding | – | −2.01 (0.004) | −1.51 (0.14) |
| At3g07070a,b,c | Protein kinase family protein | Protein Ser/Thr kinase activity | – | −1.04 (0.11) | – |
| At4g03330a,b,c | SYP123 | SNAP receptor activity | – | −1.09 (0.08) | – |
| At3g43960a | Cys proteinase | Cys-type endopeptidase activity | – | −1.44 (0.03) | – |
| At5g15600 | SP1L4 | Not assigned | – | −1.33 (0.04) | – |
| At3g54770 | RNA-binding (RRM/RBD/RNP motifs) family protein | RNA binding | – | −2.42 (0.002) | −1.04 (0.46) |
| At5g24100 | LRR transmembrane protein kinase | Protein amino acid phosphorylation | – | −1.37 (0.004) | −1.08 (0.12) |
| At3g63470 | SCPL40 | Ser-type carboxypeptidase activity | – | −1.50 (0.002) | – |
| At4g33790 | CER4 | Fatty acyl-CoA reductase (alcohol-forming) activity | – | −2.17 (0.0002) | −1.44 (0.03) |
| At2g25240b | Ser protease inhibitor | Ser-type endopeptidase inhibitor activity | – | – | −1.14 (0.37) |
| At3g56230b | Speckle-type POZ protein related | Not assigned | – | – | −1.40 (0.22) |
| At1g24620 | EF-hand calcium-binding protein family | Calcium ion binding | – | −1.51 (0.02) | −1.36 (0.19) |
| At1g53680a,b,c | ATGSTU28 | Glutathione transferase activity | – | −1.68 (0.02) | −1.25 (0.33) |
| At2g19060a | SGNH hydrolase-type esterase superfamily protein | Hydrolase activity | – | – | −1.03 (0.37) |
| At2g25680 | MOT1 | Molybdate ion transmembrane transporter activity, sulfate transmembrane transporter activity | – | −1.47 (0.22) | −2.04 (0.31) |
| At3g01730a,b | Unknown protein | Not assigned | – | −1.18 (0.02) | – |
| At3g18450a,b | Unknown protein | Not assigned | – | −2.55 (0.007) | −1.87 (0.21) |
| At3g29110 | Terpenoid cyclase/protein prenyltransferase superfamily protein | Lyase activity | – | −1.80 (0.03) | −1.42 (0.32) |
| At4g08620a,b | SULTR1;1 | Sulfate transmembrane transporter activity | – | −1.50 (0.08) | −2.04 (0.12) |
| At4g12510/At4g12520 | Bifunctional inhibitor/lipid transfer protein/seed storage 2S albumin superfamily protein | Lipid binding | – | −4.81 (0.0005) | −1.93 (0.38) |
| At4g15290 | ATCSLB5 | Cellulose synthase activity | – | −1.22 (0.18) | −1.56 (0.32) |
| At4g20450a,b | LRR protein kinase family protein | Kinase activity | – | −1.21 (0.09) | −1.15 (0.38) |
| At4g22460a,b | Bifunctional inhibitor/lipid transfer protein/seed storage 2S albumin superfamily protein | Lipid binding | – | −2.88 (0.001) | −2.12 (0.08) |
| At4g26320 | AGP13 | Not assigned | – | −3.72 (1.00E-05) | −1.63 (0.07) |
| At5g56540 | AGP14 | Not assigned | – | −1.90 (2.79E-05) | −1.04 (0.03) |
| At5g53250 | AGP22 | Not assigned | – | −3.26 (7.94E-05) | −1.24 (0.26) |
| At4g31470 | CAP superfamily protein | Not assigned | – | −1.16 (0.008) | – |
| At5g04960a,b,c | Plant invertase/pectin methylesterase inhibitor superfamily | Enzyme inhibitor activity, pectinesterase activity | – | −1.62 (0.21) | −1.43 (0.62) |
| At5g19800a,c | Hyp-rich glycoprotein family protein | Not assigned | – | −1.39 (0.09) | – |
| At5g60660 | PIP2;4 | Water channel activity | – | −1.37 (0.31) | −2.46 (0.24) |
| At5g51310a,b | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | Oxidoreductase activity, iron ion binding | – | −1.11 (0.008) | – |
| At5g52350 | ATEXO70A3 | Exocytosis | – | −1.43 (0.06) | −1.05 (0.52) |
| At5g15890a,c | TBL21 | Not assigned | – | – | −1.15 (0.20) |
| At1g12080 | Vacuolar calcium-binding protein related | Not assigned | – | – | −1.11 (0.16) |
| At3g59370a,b | Vacuolar calcium-binding protein related | Not assigned | – | −2.96 (0.001) | −1.71 (0.23) |
| At4g01140 | Unknown protein | Not assigned | – | −1.71 (0.04) | −1.25 (0.43) |
| At1g33900 | P-loop-containing nucleoside triphosphate hydrolase superfamily protein | GTP binding | – | −1.54 (0.002) | −1.37 (0.03) |
Genes with root hair-specific cis-regulatory elements (Won et al., 2009).
Genes listed in the root hair elongation transcriptome (Jones et al., 2006).
Genes that have been attributed to root hair differentiation by bioinformatic approaches (Breen and Grierson, 2007).
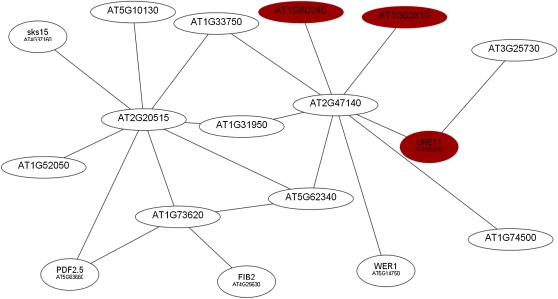
A third root module (RM3) consisted of 17 genes, some of which have been associated with root developmental processes (Fig. 5; Table IV). In contrast to the other two major clusters that contain genes that were either up- or down-regulated, RM3 is composed of genes that showed both increased and decreased mRNA abundance. The majority of genes in this cluster showed highest induction 6 h after transfer to Pi-deplete medium.
Figure 5.
Coexpression relationships of genes in RM3. The red nodes indicate up-regulated genes, and white nodes represent genes not differentially expressed at this time point. The figure shows the expression of genes 1 h after transfer to Pi-deplete medium. [See online article for color version of this figure.]
Table IV. Genes comprising RM3.
Boldface values indicate genes with the strongest repression upon Pi deficiency. Fold changes are based on the differences in expression level between Pi-deficient and Pi-sufficient plants. Dashes indicate not differentially expressed at this time point. Data represent means from three biological replicates. The numbers in parentheses are P values adjusted for multiple testing by the method of Benjamini and Hochberg (1995).
| Locus | Annotation | Function | Fold (Log2) |
||
| 1 h | 6 h | 24 h | |||
| At1g02810 | Plant invertase/pectin methylesterase inhibitor superfamily | Enzyme inhibitor activity, pectinesterase activity | 1.12 (0.03) | – | – |
| At1g31950 | Terpenoid cyclase/protein prenyltransferase superfamily protein | Lyase activity, magnesium ion binding | – | 0.77 (0.0005) | – |
| At1g33750 | Terpenoid cyclase/protein prenyltransferase superfamily protein | Lyase activity, magnesium ion binding | – | −1.29 (0.001) | – |
| At1g52050 | Man-binding lectin superfamily protein | Not assigned | – | – | 1.60 (0.17) |
| At1g73620 | Pathogenesis-related thaumatin superfamily protein | Not assigned | – | −1.99 (0.0004) | – |
| At1g74500 | TMO7 | Sequence-specific DNA-binding transcription factor activity | – | −1.74 (0.004) | – |
| At1g80240 | Unknown protein | Not assigned | 1.22 (0.20) | 1.70 (0.01) | – |
| At2g20515 | Unknown protein | Not assigned | – | −1.66 (0.0004) | – |
| At2g47140 | NAD(P)-binding Rossmann-fold superfamily protein | Oxidoreductase activity | – | −1.02 (0.03) | – |
| At3g25730 | EDF3 | Sequence-specific DNA-binding transcription factor activity | – | 1.39 (0.02) | 1.02 (0.34) |
| At4g00080 | UNE11 | Enzyme inhibitor activity, pectinesterase activity, pectinesterase inhibitor activity | 1.86 (0.003) | – | – |
| At4g25630 | FIB2 | snoRNA binding | – | −1.02 (0.0008) | – |
| At4g37160 | SKS15 | Oxidoreductase activity, copper ion binding | – | −2.54 (0.002) | 1.49 (0.25) |
| At5g10130 | Pollen Ole e 1 allergen and extensin family protein | Not assigned | – | −1.88 (0.008) | – |
| At5g14750 | WER | Sequence-specific DNA-binding transcription factor activity | – | −1.56 (0.001) | – |
| At5g62340 | Invertase/pectin methylesterase inhibitor superfamily protein | Pectinesterase activity, pectinesterase inhibitor activity | – | −2.06 (0.001) | 1.34 (0.14) |
| At5g63660 | LCR74 | Not assigned | – | −1.62 (0.0004) | – |
Gene Networks Imply a Concerted Interplay of Genes Mediating Root Hair Elongation under Pi-Deficient Conditions
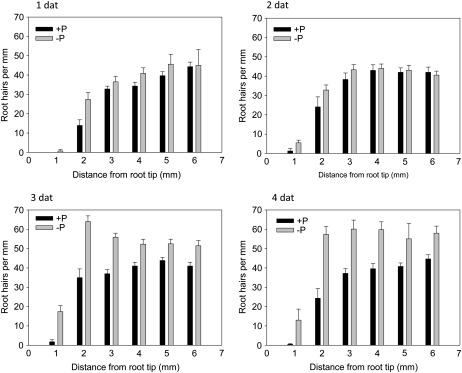
To investigate the time course of Pi deficiency-induced changes in the root hair phenotype, we monitored root hair frequency over a period of 4 d in roots of the wild type (Fig. 6). Increased root hair density at the root tip (up to 2 mm toward the base) was detected 1 d after transfer to Pi-free medium. This increase was probably due to a decrease in primary root length. No changes in the root hair pattern were observed at later developmental stages of the roots at this time point. Differences became more pronounced over time, being evident after 3 d over the investigated length of 6 mm and being most pronounced at day 4 after transfer.
Figure 6.
Changes in root hair number after transfer to Pi-deplete medium. dat, Days after transfer.
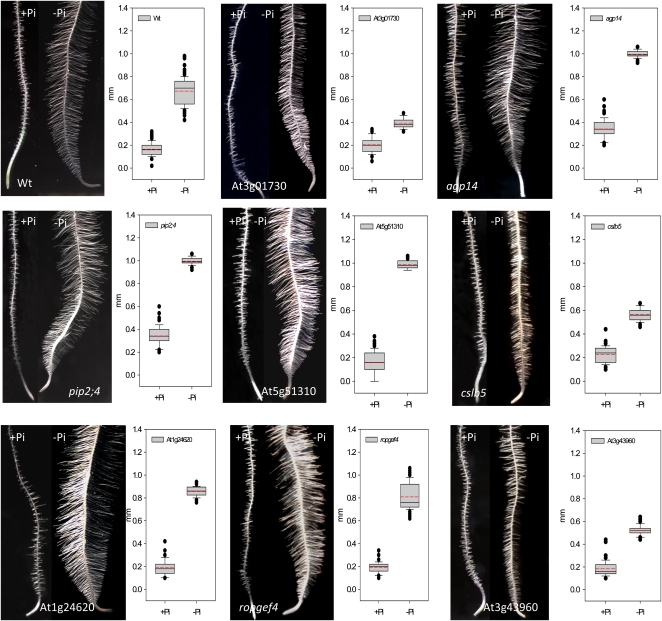
To prove whether reduced expression of the genes in RM2 has consequences for root hair pattern or elongation, we analyzed 54 homozygous mutant lines covering 31 genes in RM2 with regard to their root hair phenotype (Supplemental Table S3). Under Pi-deficient conditions, statistically significant changes in root hair length were observed in 28 of the tested lines (Supplemental Table S3). Only five of the lines under investigations did not show a deviation from the wild type. It should be noted, however, that for these five lines only one homozygous line was tested. In eight genes, two homozygous lines showed significant deviations from the wild type with the same trend, three of which, showing shorter root hairs and mutations in five genes, led to longer root hairs under Pi-deficient conditions (Figs. 7 and 8; Table V). Mutations in two genes, AGP14 and PLASMA MEMBRANE INTRINSIC PROTEIN2;4 (PIP2;4), also led to a markedly increased length of root hairs under control conditions. Four genes, namely the calcium-binding protein At1g24620, CELLULOSE SYNTHASE LIKE5 (ATCSLB5), AGP14, and PIP2;4 have not previously been associated with root hair development.
Figure 7.
Root hair length of homozygous T-DNA insertion lines harboring mutations in genes of RM2. The red nodes indicate genes that cause the formation of longer root hairs under Pi-deficient conditions, when the transcript level is reduced or absent and supported by two different T-DNA insertion lines. Pink nodes also indicate mutants with longer root hairs, but supported only by one line. Blue nodes indicate shorter root hairs (validated by two lines), light blue nodes represent mutants with shorter root hairs supported by one line. Gray nodes indicate mutant lines that did not deviate from the wild type (validated by one line), and white nodes represent genes that were not tested. Green nodes denote genes in which two mutant lines showed opposite effects. P < 0.001 (Student’s t test) indicates a significant difference from the wild type.
Figure 8.
Phenotypes and root hair lengths of mutants harboring defects in genes from RM2. Black lines in the box plots indicate the median, and red lines indicate the mean. One hundred root hairs from five roots were measured for each genotype and growth type. P < 0.001 (Student’s t test) indicates a significant difference from the wild type (Wt). [See online article for color version of this figure.]
Table V. Mutant plants with root hair length under Pi-deficient conditions that deviate from the wild type.
Mutant lines indicated in boldface are shown in Figure 8.
| Locus | Name | Mutant Lines | Phenotype under –Pi Conditions | Highest Expression |
| At3g43960 | Cys proteinase | sail_893_F01, salk_135674 | Shorter root hairs | Root hair zone |
| At4g15290 | ATCSLB5 | salk_133711, salk_016242 | Incomplete elongation | Root hair zone |
| At3g01730 | Unknown protein | salk_073740, salk_120148 | Shorter root hairs | Lateral root, elongation zone |
| At5g51310 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | sail_91_E12, salk_072163 | Longer root hairs | Lateral root, elongation zone |
| At2g45890 | RHS11, ROPGEF4 | sail_184_C08, salk_148056 | Longer root hairs | Root hair zone |
| At1g24620 | EF-hand calcium-binding protein family | salk_114440, salk_000164 | Longer root hairs | Pollen |
| At5g56540 | AGP14 | salk_107040, salk_016693 | Longer root hairs (also in +Pi plants) | Endodermis, root hair zone |
| At5g60660 | PIP2;4 | sail_1251_G12, sail_535_D05 | Longer root hairs (also in +Pi plants) | Root hair zone |
DISCUSSION
Expression of Pi-Responsive Genes Adjusts Rapidly to Changes in Pi Supply
Most biological processes depend on the coordinated interaction of a suite of genes. The microarray technology allows parallel interrogation of the expression of thousands of genes in one experiment, but a major challenge is to decipher interactions among genes that participate in signaling cascades or mediate developmental or metabolic processes. We developed a utility that computes the pairwise correlation of a large number of genes based on a customized selection of microarray experiments. The most important feature of the MACCU toolbox is the capacity to customize the database for gene clustering and to compare results of different clustering computations. Collecting array data and normalization can easily be done using bioinformatics databases and tools such as the NASCarrays database and the Bioconductor software package. Using these tools, we separated three major clusters of Pi-responsive transcripts from a time-course experiment that likely contain genes that encode critical components of processes that acclimate plants to low Pi availability. The clustering further allowed for noise-reduced identification of genes that were induced early upon the onset of Pi starvation. One of the clusters, RM1, was composed of a subset of genes that were rapidly induced by Pi starvation and enriched in transcription factors and genes with potential function in signal transduction. In addition, genes involved in hormonal response were overrepresented in this cluster. Out of the hormone-related genes, the highest expression after 1 h was observed for four salicylic acid-related genes (OPR1, WRKY18, WRKY40, and BAP1), one ethylene-related gene (ERF13), and one auxin-related gene (PBP1), indicating that a combined action of different hormones is critical in early Pi deficiency signaling. Interestingly, the expression pattern of the genes in RM1 resembled that of a defense response, linking the response to Pi deficiency and the response to plant pathogens through shared signaling components.
Verification by transcript analysis via qRT-PCR showed that all of the randomly chosen genes were induced already 10 min after transfer to Pi-free medium and remained induced when determined at later time points of up to 3 d. Clustering based on root-related coexpression relationships reliably identified candidates that were robustly induced upon Pi starvation and revealed potentially new candidate genes functioning in early Pi signaling.
Calcium Signaling and the Activity of Hydrolytic Cell Wall Enzymes May Be Critical for the Development of Root Hairs in Response to Pi Deficiency
Several lines of evidence indicate that the activity of cell wall hydrolytic enzymes (polygalacturonases, pectate lyases, pectin methylesterases) and enzymes involved in cell extension (extensins and expansins) is down-regulated upon Pi deficiency as one of the earliest and most pronounced responses observed. The highest induction in the microarray experiments was observed for POLYGALACTURONASE-INHIBITING ENZYME1 (PGIP1) in RM1. The protein is localized in the cell wall, inhibiting the activity of pectin-degrading enzymes. A putative polygalacturonase (At1g05650) and a pectinesterase (At5g04960) were down-regulated in RM2 upon Pi deficiency (Table III). In RM3, UNFERTILIZED EMBRYO SAC11 (UNE11), a pectinesterase inhibitor, and At1g02810, a pectinesterase with a pectinesterase inhibitor motif, were up-regulated, and a gene encoding a protein with pectinesterase activity was down-regulated. Application of exogenous pectin methylesterase was shown to inhibit pollen tube growth (Bosch et al., 2005); a similar effect may be inferred for root hair growth. Pectin biosynthesis requires the activity of glycosyltransferase. A glycosyltransferase (At2g41640; RM1) was found to be strongly induced upon Pi deficiency 1 h after transfer, further supporting the importance of pectin-related processes. It remains to be shown, however, if the transferase At2g41640 is mediating the synthesis of pectins. Interestingly, AGPs were suggested to bind pectins (Majewska-Sawka and Nothnagel, 2000), providing a link between these two classes of genes. While a mechanism cannot be inferred from these data, the robustness and early induction of the response suggest that direct contact of cells with the environment through the extracellular matrix is crucial for the adjustment of developmental programs to Pi-deficient conditions.
Tip-focused calcium gradients have been shown to be critical for pollen tube growth and in maintaining tip growth of root hairs (Bibikova et al., 1997; Hepler et al., 2001). Rho-related GTPases from plants (ROPs) have been associated with the regulation of calcium influx controlling tip growth both in pollen tubes and in root hairs (Molendijk et al., 2001; Jones et al., 2002; Yang and Fu, 2007). Guanine nucleotide-exchange factors (GEFs) play a critical role in ROP signaling (Berken et al., 2005; Rossman et al., 2005; Gu et al., 2006), including root hair development (Shin et al., 2010), by activating ROPs via catalyzing the dissociation of guanine nucleotides from ROP. ROPGEF4 is expressed chiefly in root hairs, and decreased transcript levels of ROPGEF4 led to longer root hairs under Pi-deficient conditions (Table V). Taken together, our data indicate that calcium signaling is important for the development of root hairs in response of Pi deficiency and that ROPGEF4 is critical in the activation of ROPs in response to extracellular stimuli.
Induction of the Pi Deficiency Root Phenotype Requires the Concerted Action of Genes
Root hair morphology is substantially altered by Pi starvation, mainly by inducing extra root hairs that are markedly longer than those formed under control conditions (Müller and Schmidt, 2004). In addition, root hairs of Pi-deficient plants are denser, due to decreased longitudinal elongation of epidermal cells and to the development of root hairs in positions normally occupied by nonhair cells. Changes in root hair patterning became apparent in the tip of the roots 1 d after transfer to Pi-free medium. The phenotype then became gradually more pronounced over the following 3 d (Fig. 6). Unexpectedly, a relatively large subset of root hair-related genes was expressed 1 to 6 h after the onset of Pi-deficient conditions, indicating that developmental alterations are induced very early after sensing Pi deficiency. Based on the coexpression analysis and subsequent screening of T-DNA insertion lines, we identified eight new players in Pi deficiency-induced root hair formation, some of which have not previously been associated with root hair development. All genes identified affected the length of Pi deficiency-induced root hairs but did not have obvious effects on the density (Table V; Fig. 8). The majority of these genes show highest expression in the root hair zone. Of particular interest is the gene At1g24620, encoding an EF-hand calcium-binding protein. This gene is chiefly expressed in pollen and links early calcium signaling inferred from RM1 to root hair development under Pi-deficient conditions.
The arabinogalactans (AGPs), a class of Hyp-rich proteoglycans, are a further interesting group producing strong phenotypes under Pi-deficient conditions. AGPs have been implicated in several developmental processes, including cell differentiation, cell-cell recognition, and cell expansion (Majewska-Sawka and Nothnagel, 2000; Seifert and Roberts, 2007; Ellis et al., 2010). Mutations in AGP14 caused the formation of very long root hairs both under control and Pi-deficient conditions. AGP14 is anchored to the membrane by a GPI anchor. GPI-anchored proteins in general and AGPs in particular have been associated with activating signaling pathways by interacting with extracellular ligands (Shi et al., 2003). Based on a bioinformatics approach, the unknown gene At3g01730 has been predicted to be GPI anchored as well (Borner et al., 2002, 2003). Homozygous mutants form short hairs under Pi-deficient conditions. The gene is closely correlated with LEUCINE-RICH REPEAT (LRR)/EXTENSIN1 (LRX1), an important regulator of root hair development, and this gene is overexpressed in a lrx1 repressor of lrx1 (rol1) double mutant, together with a set of genes involved in the modification of pectic polysaccharides (Diet et al., 2006), suggesting a function of At3g01730 in pectin-related processes. Interestingly, in lrx1 rol1 plants, the length of primary roots, root hairs, and the longitudinal length of trichoblasts are affected, traits that are typically changed in Pi-deficient plants. We suggest that cell wall-related processes are important in Pi deficiency signaling, probably including the cleavage of GPI anchors by phospholipases. Based on the phenotype of the agp14 mutant, we add a new function of GPI-anchored AGPs in root hair elongation in response to environmental stimuli. Contrary to our expectations, all of the genes with predicted or verified roles in root hair formation were down-regulated upon Pi deficiency. This indicates that repression of a suite of genes is necessary to elongate root hairs in response to Pi starvation, possibly related to the reduced elongation of epidermal cells. Interestingly, mutations in the genes in RM2 can cause either an increase or a decrease in root hair length, likely reflecting a complex network of processes involved in the Pi deficiency-induced changes in postembryonic development.
RM3 Reveals Potential Novel Players in Pi Deficiency-Induced Root Developmental Changes
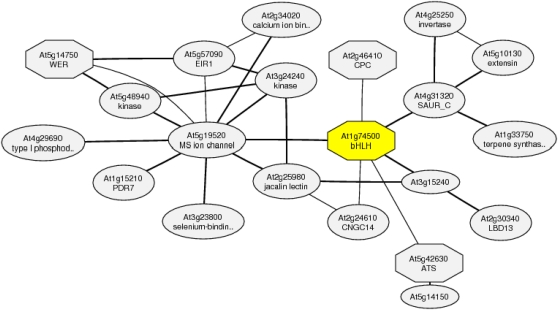
The differential expression of genes that have been associated with developmental processes represent a suite of novel players in root developmental changes induced by Pi deficiency. TARGET OF MONOPTEROS (TMO7) encodes a mobile basic helix-loop-helix (bHLH) transcription factor modulating brassinosteroid signaling that has been shown to be involved in cell elongation and embryonic root initiation (Wang et al., 2009; Schlereth et al., 2010). WEREWOLF (WER) encodes a negative regulator of the root hair cell fate (Lee and Schiefelbein, 1999). The gene was down-regulated upon Pi deficiency, consistent with the formation of extra root hairs both in normal and ectopic positions. wer mutants show an exaggerated response to Pi deficiency and form hairs in almost all epidermal cells (Müller and Schmidt, 2004). The increase in root hair number in the wer mutant under Pi-deficient conditions is due to an increase of root hairs in ectopic position, which is consistent with the assumption that decreased WER protein is the cause of an increased number of epidermal cells entering the root hair cell fate upon Pi deficiency. Coexpression analysis suggests that TMO7 is directly connected with the Myb-type transcription factor CAPRICE, a positive regulator of root hair cell fate that is expressed in nonhair cells and moves to hair cells to compete with WER for binding to a Myb-bHLH/bHLH-WD40 complex (Ishida et al., 2008; Schiefelbein et al., 2009; Fig. 9). WER is found in the same cluster, pointing to a possible role of TMO7 in cell fate decisions.
Figure 9.
Coexpression gene network around At1g74500. The gene network was determined with ATTED-II version 5.5 (http://atted.jp/). [See online article for color version of this figure.]
One of the genes identified in this study, CELLULOSE SYNTHASE-LIKE5 (CSLB5), encodes a gene similar to cellulose synthase. cslb5 mutants showed shorter root hairs under Pi-deficient conditions (Fig. 8). Interestingly, a gene with high similarity to CSLB5, CELLULOSE SYNTHASE5, was shown to be regulated by the homeobox transcription factor GLABRA2 (Tominaga-Wada et al., 2009). GLABRA2 is a negative regulator of root hair cell fate expressed predominantly in nonhair cells and regulated by WER. A likely scenario connects some well-known players in cell specification with the network of genes that induce the root hair phenotype that is typical of Pi-deficient plants.
Activation of the AP2 domain-containing transcription factor ETHYLENE RESPONSE DNA-BINDING FACTOR3 (EDF3) by ethylene was shown to inhibit cell elongation (Alonso et al., 2003). Consistent with such a role under Pi-deficient conditions, EDF3 was up-regulated upon Pi starvation. SKU5 SIMILAR15 (SKS15) belongs to a 19-gene family named SKS. The founding member of this family, SKU5, was suggested to participate in cell wall expansion in root cells, is located on the plasma membrane and the cell wall, and is mainly expressed in expanding tissue (Sedbrook et al., 2002). SKS15 is closely connected with two extension genes, a pectinesterase and EXPANSIN14 (http://atted.jp/), and was found to be strongly down-regulated upon Pi starvation (Table IV). It is thus tempting to speculate that SKS14 has a similar function under Pi-deficient conditions.
CONCLUSION
In summary, we present here a systems approach to distinguish unrelated genes from central players in Pi deficiency signaling, based on organ-specific coexpression relationships of Pi-responsive genes identified from microarray analysis. With this approach, we provide a proof of principle to infer complete scenarios that give hints to mechanisms involved in the signaling of Pi deficiency and into the initiation of processes that acclimate plants to Pi pools with low availability. We show that genes can be attributed to these processes with a higher degree of certainty than simply by their change in expression level and demonstrate further that some of the genes are robustly expressed within 1 h after the change in the Pi regime. The approach applied in this study will help to direct reverse genetic screenings and integrate genes that have been identified by genetic screenings into their positions in the puzzle of signaling or metabolic pathways.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) was grown hydroponically in a growth chamber at a constant relative humidity of 75% under a 10/14-h light/dark cycle (Osram E40/ES Plantastar; 300 μmol m−2 s−1) at 21°C (day) or 18°C (night). All mutant and wild-type seeds were obtained from the Arabidopsis Biological Resource Center (Ohio State University; http://www.arabidopsis.org/). Homozygous mutants were identified by PCR from genomic DNA. Two to four seeds were sown in 1.5-mL Eppendorf tubes that were filled with 1 mL of 0.25% agarose. The tip of the tube was cut off with a hot scalpel, and the tubes were placed in black plastic containers in contact with the nutrient solution that was constantly aerated. Each container contained approximately 45 Eppendorf tubes. At 20 d following sowing, the agarose plug was removed using a gentle stream of water. The nutrient solution was replaced weekly and at 24 h prior to beginning the experiment. The nutrient solution was composed of KNO3 (3 mm), MgSO4 (0.5 mm), CaCl2 (1.5 mm), K2SO4 (1.5 mm), NaH2PO4 (1.5 mm), H3BO3 (25 μm), MnSO4 (1 μm), ZnSO4 (0.5 μm), (NH4)6Mo7O24 (0.05 μm) CuSO4 (0.3 μm), and Fe-EDTA (40 μm) with pH adjusted to 6.0 with KOH (Schmidt, 1994). At 30 d after sowing, the plants were washed briefly in a 0.1 mm EDTA solution and transferred into either Pi-free or Pi-replete (controls) nutrient solutions. Following transfer, the roots were harvested after 0, 1, 6, or 24 h into RNALater (Applied Biosystems). The experiment was initiated 1 h into the light cycle. For each time point analyzed, approximately 20 plants were harvested. The total processing time was approximately 15 s. Control plants were treated similarly but were grown in the presence of Pi. Short-term Pi-starvation experiments were conducted with 14-d-old plants by exposing them to liquid Pi-free medium for 10, 30, or 60 min.
Real-Time qRT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and DNase treated with the TURBO DNA-Free Kit (Ambion) following the manufacturers’ instructions. cDNA was synthesized using 4 μL of DNA-free-RNA with oligo(dT)20 primer and the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) following the manufacturer’s instructions. The cDNA was then used as a template in a 20-μL reaction using Power SYBR Green PCR Master Mix (Applied Biosystems), and subsequent experiments were carried out on an ABI StepOne Real-Time PCR System (Applied Biosystems). Three independent technical replicates were performed for each sample. The comparative cycle threshold method was used to determine the relative amount of gene expression, with the expression of tubulin used as an internal control. The gene-specific primers for qRT-PCR are listed in Supplemental Table S4.
Microarray Experiments and Data Analysis
The Affymetrix gene chip Arabidopsis ATH1 Genome Array was used for microarray analysis. Total RNA from roots of control and Pi-deficient plants was isolated with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Nucleic acid quantity was evaluated by using a NanoDrop ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies). All RNA samples were quality assessed by using the Agilent Bioanalyzer 2100 (Agilent). Complementary RNA synthesis was performed by use of the Gene Chip One-Cycle Target Labeling Kit (Affymetrix). Gene chips were hybridized with 15 μg of fragmented complementary RNA. Hybridization, washing, staining, and scanning procedures were performed as described in the Affymetrix technical manual.
Raw signal data from the mircoarray experiments were imported directly into GeneSpring (version 7.0; Agilent). The software was used to normalize data per chip to the 50th percentile and per gene to the control samples. The data were then filtered using the following: (1) by removing genes that were flagged as absent in one of the replicates of the control plants for genes down-regulated in low-Pi conditions and in low-Pi plants for genes that are up-regulated by low Pi; and (2) by expression level to remove those genes that were deemed to be unchanging between values 0.5 and 2.0 (greater than 2-fold difference). To match the probe sets to genes, the file affy_ATH1_array_elements-2009-7-29.txt (ftp://ftp.arabidopsis.org/home/tair/Microarrays/Affymetrix/) was used.
Preprocessing and Statistical Analysis
Preprocessing and statistical analysis of data were performed using the Bioconductor packages AffylmGUI and Limma (Wettenhall et al., 2006; http://www.bioconductor.org/). Arrays were normalized using the Robust Probe-level Linear model provided in the AffylmGUI package using the default parameters. Differentially expressed genes were calculated using the “eBayes()” function in Limma. Correction for false discovery was performed by the method of Benjamini and Hochberg (1995). Statistical values including P values were determined as described by Smyth (2004).
Coexpression-Based Gene Clustering Using the MACCU Toolbox
Gene clustering was performed based on the root-specific expression correlations of Pi-responsive genes. We downloaded and robust multiarray averaged the microarray data of 2,671 ATH1 arrays from the NASCarray database (http://affymetrix.arabidopsis.info/) and normalized them by the RMA method using the RMA function of the Affy package of the Bioconductor software. Among the 2,671 arrays, 300 root-related arrays were manually identified (Supplemental Table S5), and a corresponding portion from the normalized data of the 2,671 chips was extracted. Based on the root-related portion of 300 arrays and the normalized data of the whole data set, we computed two clustering results of Pi-responsive genes with Pearson correlation thresholds of 0.8 and 0.7, respectively. Finally, root-specific networks were obtained by subtracting relationships between genes inferred from the latter data set from the cluster derived from the root-related data.
Root Hair Length and Density Measurements
Root hair length was determined by using a light microscope equipped with a scale in the ocular. One hundred root hairs from five roots were measured for each genotype and growth type. Root hair density was analyzed by counting the root hairs of 10 roots per time point and treatment in serial 2-mm sections starting from the root tip. Statistically significant deviations from the wild type were determined by Student’s t test (P > 0.001).
All microarray data from this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE25171).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Coexpression network based on 300 root-related microarray data sets.
Supplemental Figure S2. Coexpression network after removing non-root-specific edges.
Supplemental Table S1. Gene chip data from roots grown under Pi-sufficient and Pi-deficient conditions.
Supplemental Table S2. GO enrichments at various cutoff values for RM1.
Supplemental Table S3. Root hair length of T-DNA insertion mutants.
Supplemental Table S4. Primers used for qRT-PCR.
Supplemental Table S5. List of the root-related microarray experiments used for gene clustering.
Supplementary Material
Acknowledgments
We thank Tze Yu Huang and Kalen Lin for skillful technical assistance, Cole Lu for editing the manuscript, and the Bioinformatics Core Facility at the Institute of Plant and Microbial Biology for proficient support. Affymetrix gene chip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy/), supported by Academia Sinica.
References
- Alonso A, Rahmouni S, Williams S, van Stipdonk M, Jaroszewski L, Godzik A, Abraham RT, Schoenberger SP, Mustelin T. (2003) Tyrosine phosphorylation of VHR phosphatase by ZAP-70. Nat Immunol 4: 44–48 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19: 529–538 [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statis Soc B 57: 289–300 [Google Scholar]
- Berken A, Thomas C, Wittinghofer A. (2005) A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Zhigilei A, Gilroy S. (1997) Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta 203: 495–505 [DOI] [PubMed] [Google Scholar]
- Borner GH, Lilley KS, Stevens TJ, Dupree P. (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK. (2005) Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol 138: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. (2002) Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur J Biochem 269: 6278–6286 [DOI] [PubMed] [Google Scholar]
- Breen G, Grierson C. (2007) Predicting functional modules in root hairs that drive tip growth. The Arabidopsis Information Resource. http://www.arabidopsis.org/servlets/TairObject?type=publication&id=501722073 (June 20–23, 2007)
- Buckhout TJ, Yang TJ, Schmidt W. (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero B, Nakagawa Y, Jiang XY, Miura KJ, Li F, Raghothama KG, Bressan RA, Hasegawa PM, Pardo JM. (2009) The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol Plant 2: 535–552 [DOI] [PubMed] [Google Scholar]
- Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, Reiter WD, Ringli C. (2006) The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-l-rhamnose synthase. Plant Cell 18: 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Li S, Lord EM, Yang Z. (2006) Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Jin Y, Wussler C, Blancaflor EB, Motes CM, Versaw WK. (2008) Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol 177: 889–898 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY. (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS. (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T, Takamiya K, Ohta H. (2006) Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J 47: 238–248 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Li WF, Perry PJ, Prafulla NN, Schmidt W. (2010) Ubiquitin-specific protease 14 (UBP14) is involved in root responses to phosphate deficiency in Arabidopsis. Mol Plant 3: 212–223 [DOI] [PubMed] [Google Scholar]
- Ma Z, Walk TC, Marcus A, Lynch JP. (2001) Morphological synergism in root hair length, density, initiation and geometry for phosphorus acquisition in Arabidopsis thaliana: a modeling approach. Plant Soil 236: 221–235 [Google Scholar]
- Majewska-Sawka A, Nothnagel EA. (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K. (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Müller M, Schmidt W. (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Picault N, Arrigoni R, Besin E, Palmieri F, Hodges M. (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem J 410: 621–629 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Bucher M. (2002) Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book 1: 1–35, doi/10.1199/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. (2005) GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6: 167–180 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J, Kwak SH, Wieckowski Y, Barron C, Bruex A. (2009) The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J Exp Bot 60: 1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schmidt W. (1994) Root-mediated ferric reduction: responses to iron deficiency, exogenously induced changes in hormonal balance and inhibition of protein synthesis. J Exp Bot 45: 725–731 [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR. (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Shi H, Kim YS, Guo Y, Stevenson B, Zhu JK. (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Cho MH, Kim TL, Yoo J, Kim JI, Han YJ, Song PS, Jeon JS, Bhoo SH, Hahn TR. (2010) A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development. J Biol Chem 285: 32151–32159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. (2009) The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J 60: 564–574 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J. (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. (2006) AffylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22: 897–899 [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. (2009) Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 150: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fu Y. (2007) ROP/RAC GTPase signaling. Curr Opin Plant Biol 10: 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.