Abstract
Nearly all extant land plants possess stomata, the epidermal structures that mediate gas exchange between the plant and the environment. The developmental pathways, cell division patterns, and molecules employed in the generation of these structures are simple examples of processes used in many developmental contexts. One specific module is a set of “master regulator” basic helix-loop-helix transcription factors that regulate individual consecutive steps in stomatal development. Here, we profile transcriptional changes in response to inducible expression of Arabidopsis (Arabidopsis thaliana) FAMA, a basic helix-loop-helix protein whose actions during the final stage in stomatal development regulate both cell division and cell fate. Genes identified by microarray and candidate approaches were then further analyzed to test specific hypothesis about the activity of FAMA, the shape of its regulatory network, and to create a new set of stomata-specific or stomata-enriched reporters.
Stomata are pores in the plant epidermis that serve as the main conduits for gas exchange between the plant and the atmosphere. Stomata are found in virtually all land plants, and their activity, when considered collectively, plays a pivotal role in global carbon and water cycles (Hetherington and Woodward, 2003). Individual plants use these pores to efficiently balance photosynthesis with water availability. They do so in the short term by regulating the aperture of existing stomata, but in the long term, they rely on developmental processes to alter the number of stomata on emerging leaf surfaces.
Each step of stomatal development, from initiation to terminal differentiation, is highly organized and regulated (for review, see Bergmann and Sack, 2007; Abrash and Bergmann, 2009; Nadeau, 2009; Peterson et al., 2010). In dicots like Arabidopsis (Arabidopsis thaliana), stomatal formation begins with asymmetric divisions of protodermal cells to generate dispersed meristemoids. Meristemoids may continue dividing asymmetrically several times before differentiating into a guard mother cell (GMC), the immediate precursor of the paired guard cells of the stomatal complex. Grasses, in contrast, create GMCs by asymmetric divisions in distinct cell files. The GMC then recruits subsidiary cells from the neighboring cell files before completing its division into a pair of guard cells (Hernandez et al., 1999; Liu et al., 2009). As seen in these two examples, diversity and flexibility are the hallmarks of the early stages in stomatal development, but later, these trajectories converge on a common final step wherein the GMC undergoes a single symmetric division, and the resultant daughters undergo significant shape and gene expression changes to become the two functional guard cells of the mature stoma (Fig. 1A). Not only is the transition from GMC to guard cells the most conserved developmental process in stomatal formation, but current data indicate that the underlying genetic control of the transition is also conserved (Liu et al., 2009).
Figure 1.
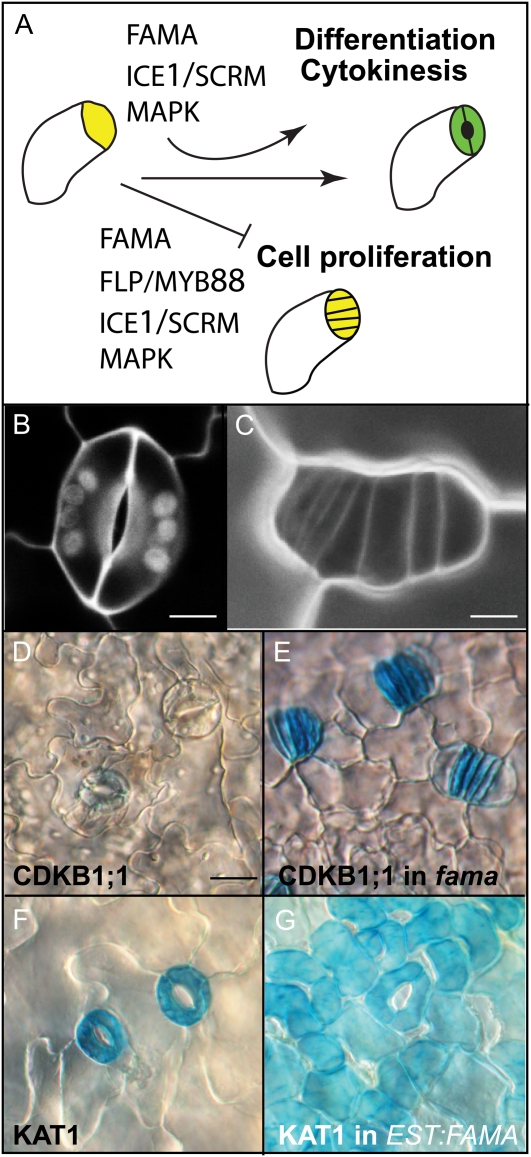
Overview of FAMA function in stomatal development. A, Diagram of the stage in stomatal development, the GMC (yellow)-to-guard cell (green) transition, regulated by FAMA and other noted genes. In the absence of FAMA, GMCs divide again and fail to differentiate. Arrows indicate promoting, and the T-bar repressing, influences. B, Confocal image of wild-type stoma consisting of two guard cells and a pore between them; white discs inside guard cells are chloroplasts. C, Confocal image of a fama-1 “tumor” consisting of GMCs. Cell outlines in B and C are visualized with propidium iodide. D and E, CDKB1;1pro:GUS expression in wild-type (D) and fama-1 (E) cotyledons at 10 d postgermination (dpg). CDKB1;1pro:GUS expression is down-regulated in mature stomata but still active (blue) in fama tumors. F and G, KAT1pro:GUS expression marks mature guard cells in wild-type cotyledons at 7 dpg (F) and ectopic guard cells produced by ESTpro:FAMA seedlings at 7 dpg (G). Bars = 6 μm in B and C and 20 μm in D. E to G are at the same scale as D. [See online article for color version of this figure.]
Genes regulating stomatal development are best known in Arabidopsis. Here, a combination of cell-cell signaling mediated through ligands and cell surface receptors is coordinated with (and sometimes directly impinges on) the activity of transcription factors (for review, see Rowe and Bergmann, 2010). Studies of the MYB transcription factors FOUR LIPS (FLP) and MYB88 (Lai et al., 2005; Xie et al., 2010) and the basic helix-loop-helix (bHLH) transcription factor FAMA (Ohashi-Ito and Bergmann, 2006) indicate that the transition from a division-competent state to one where a single symmetric division is followed immediately by a terminal differentiation program is highly regulated. FLP and MYB88 are primarily required for ensuring that division ceases (Lai et al., 2005; Xie et al., 2010), whereas FAMA is required for both division and differentiation. The GMC-to-guard cell stage is also regulated by another bHLH, INDUCER OF CBF EXPRESSION1/SCREAM (ICE1/SCRM; Kanaoka et al., 2008), by CYCLIN-DEPENDENT KINASE B1;1 (CDKB1;1; Boudolf et al., 2004) and CDKB1;2 (Xie et al., 2010), and by MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) cascade members YODA, MAPK kinase 7, and MAPK kinase 9 (Lampard et al., 2009).
FAMA has the characteristics of a genetic “master regulator” in that it is both necessary and sufficient for guard cell fate. In the leaf epidermis, FAMA expression is first observed in GMCs, peaks in newly divided guard cells, and disappears as guard cells mature (Ohashi-Ito and Bergmann, 2006). Arabidopsis fama null mutants make meristemoids and GMCs, but the GMCs cannot progress to become guard cells and instead continue rounds of symmetric division while maintaining GMC marker expression, eventually forming small epidermal “tumors” (Fig. 1, C and E; Ohashi-Ito and Bergmann, 2006). These fama plants arrest as small pale seedlings. Ectopic FAMA expression, on the other hand, is sufficient to confer at least partial guard cell identity on other cells, causing them to express guard cell-specific markers, develop pore structures, and change their morphology to approximate kidney-shaped guard cells (Ohashi-Ito and Bergmann, 2006). These ectopic FAMA-induced guard cells can form without first undergoing a symmetric precursor division (Fig. 1G).
FAMA, like the majority of the approximately 160 Arabidopsis bHLHs, contains His (H), Glu (E), and Arg (R) residues in the core bHLH region and, furthermore, requires them for normal function (Ohashi-Ito and Bergmann, 2006). These residues have been shown in other bHLHs to facilitate interaction with DNA at G-box (CACGTG) elements (Martínez-García et al., 2000; Toledo-Ortiz et al., 2003; Qian et al., 2007). The G-box cis-element, alone or in tandem, is found upstream of the start sites of genes controlling a variety of developmental and homeostatic processes (Menkens et al., 1995; Chakravarthy et al., 2003; Chandrasekharan et al., 2003). FAMA has an N-terminal region that serves as an activation domain in a heterologous system, and overexpression of a fusion of FAMA to an EAR-repression domain produces the fama phenotype in wild-type plants, consistent with a general role of FAMA as a transcriptional activator (Ohashi-Ito and Bergmann, 2006). bHLH transcription factors, however, often form and work in multiple complexes, each of which may have different targets and different regulatory consequences (activating/repressing) when bound to those targets. In plants, bHLH proteins are known to interact with a wide variety of other proteins such as R2-R3-type MYBs and phytochromes, and different bHLHs form homodimers or heterodimers (with one or more partners) or both (Payne et al., 2000; Toledo-Ortiz et al., 2003; Zhang et al., 2003; Kanaoka et al., 2008). In stomatal development, the MYBs FLP and MYB88 and two bHLHs, ICE1/SCRM and SCRM2, are coexpressed with FAMA in leaves. FAMA does not interact with these MYBs (Ohashi-Ito and Bergmann, 2006), but it can form complexes with ICE1/SCRM and SCRM2 in the yeast two-hybrid assay and in bimolecular fluorescence complementation assays in plants (Kanaoka et al., 2008).
Stomata are functionally important, the transition between division competence and terminal differentiation is a widely faced regulatory problem, and bHLHs are a large fraction of the plant transcription factor repertoire. Analysis of FAMA in the GMC-to-guard cell transition, therefore, has the potential to illuminate key questions of broad interest. Previous data have identified a few potential partners and targets of FAMA (Ohashi-Ito and Bergmann, 2006; Kanaoka et al., 2008), and high-throughput studies in Arabidopsis and Brassica have significantly advanced our knowledge of the mature guard cell transcriptome, translatome, and proteomes (Yang et al., 2008; Zhang et al., 2003; Mustroph et al., 2009). How guard cells are made, and how FAMA directs this transition, however, are not well defined. In this study, we identified FAMA targets and regulators of guard cell development by microarray-based transcript profiling of genes differentially modulated during a time course of FAMA induction. We further identified a subset of these genes as potential direct targets of FAMA through expression and chromatin immunoprecipitation (ChIP) analysis. Our results from these targets lead to specific hypotheses about the activity of FAMA and the shape of its regulatory network.
RESULTS
Generation of Transcriptional Profiles in Response to FAMA Induction
FAMA was previously shown to induce morphologies and gene expression patterns typical of guard cells in epidermal cells and, to a lesser extent, in mesophyll cells throughout the plant (Ohashi-Ito and Bergmann, 2006). Because of this ability to drive multiple cell types into guard cell identity, we used this previously described estrogen-inducible expression system in a time-course experiment to identify genes that were differentially regulated by FAMA and/or by the acquisition of guard cell identity (for details, see “Materials and Methods”). Two time points were analyzed, 4 h and 48 h postinduction; these time points were chosen to enrich for potential direct targets of FAMA (4 h) and to capture the set of genes expressed when guard cells differentiate (48 h). Five-day-old seedlings were used as the starting material for induction. No morphological changes were observed after induction of FAMA expression for 4 h, but examination of these plants several days after induction treatment revealed some ectopic stomata, suggesting that this short induction was sufficient to induce FAMA activity (data not shown). Subsequent monitoring of FAMA expression confirmed strong induction of expression after 4 h (Fig. 2, cluster I). Three biological replicates for each sample and control at each time point were collected. RNA was isolated from whole seedlings at these time points, processed, and labeled using standard protocols and hybridized to Affymetrix ATH1 arrays (see “Materials and Methods”).
Figure 2.
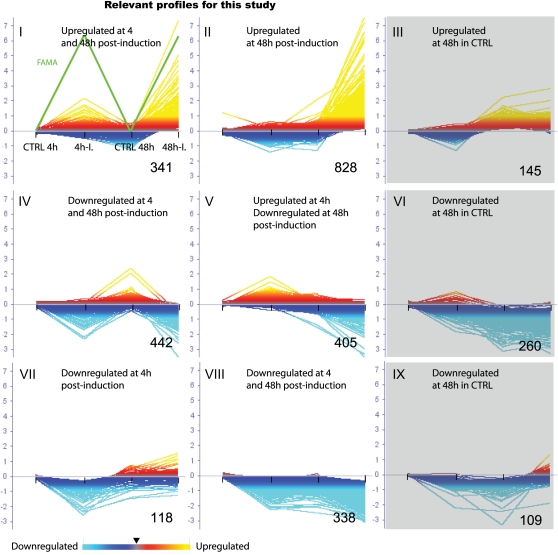
K-means clustering analysis of the 2,986 genes showing altered expression in response to FAMA. The four treatments (control 4 h, FAMA induction 4 h, control 48 h, FAMA induction 48 h) are represented from left to right, respectively, on the x axis of each cluster. On the y axis is the log2 of the relative gene expression level compared with control expression at 4 h (CTRL 4 h). In each cluster, a general description is given at the top and the number of genes is indicated in the bottom right corner. Three shaded clusters are of less interest in this study (rationale in text). Cluster I, Genes that were up-regulated at 4 and 48 h compared with controls at similar time points. FAMA expression levels are indicated by the green trace. Cluster II, Genes that were strongly up-regulated at 48 h but not significantly changed at 4 h relative to respective controls. Cluster III, Genes whose expression level is higher in the 48-h control than in the 4-h control (not of interest). Cluster IV, Genes that were strongly down-regulated at 4 and 48 h relative to respective controls. Cluster V, Genes that were up-regulated at 4 h but down-regulated at 48 h relative to respective controls. Cluster VI, Genes whose expression level is lower in the 48-h control than in the 4-h control (not of interest). Cluster VII, Genes that were down-regulated at 4 h but whose expression level was mainly unchanged or slightly increased at 48 h relative to respective controls. Cluster VIII, Genes that were strongly down-regulated at both 4 and 48 h relative to respective controls. Cluster IX, Genes whose expression level exhibited a mixed response, with down-regulation at 48 h as a common feature (not of interest). [See online article for color version of this figure.]
Analysis of Microarray Data
We identified genes modulated by FAMA expression in the microarray data using two-way ANOVA and Benjamini and Hochberg multiple testing correction with P < 0.001 and three different fold cutoff (FC) values (2, 1.5, and 1.2; Supplemental Fig. S1). These FCs yielded gene lists containing 942, 1,969, and 2,986 genes modulated 4 and/or 48 h after FAMA induction, respectively. After examining the resultant gene lists for “known” stomata regulators, we chose a FC value of 1.2 (Supplemental Table S1) for subsequent analyses. To identify dominant trends in the data, this filtered data set was clustered using a K-means clustering algorithm (matrix distance, Pearson correlation), leading to the identification of nine different clusters (Fig. 2).
After 4 h of FAMA induction, 357 genes were up-regulated (with FC = 1.2). A significant fraction (44%; n = 164) of these genes was also up-regulated at 48 h, but, interestingly, 25% (n = 89) were down-regulated at the 48-h time point (Fig. 2, clusters I, II, and V; Supplemental Fig. S1). Because of the highly restricted expression pattern of FAMA, this latter class is most consistent with the behavior of genes involved in promoting the GMC-to-guard cell developmental transition, while the genes that remain up-regulated might be predicted to also have a role in guard cell identity or function. We also predicted that the 4-h up classes would be enriched in direct FAMA targets; in support of this idea, we found a significant enrichment of G-boxes in the 2-kb upstream regions of these early up-regulated genes relative to 30,067 2-kb “promoter regions” that represent the whole genome (23% as calculated by the Athena data-mining tool; O’Connor et al., 2005). This was true when we considered the whole group (110/357, 30%; P < 10−6) or just the 4-h up and 48-h down class (27/89, 30%; P < 10−4). These early genes encode proteins with a variety of functions. Some expected classes based on FAMA’s phenotypes were transcription factors and cell cycle controllers, receptors, and signaling proteins (Table I; Supplemental Table S2). However, there were significant numbers of genes associated with cell wall modification and metabolic processes, and the most significantly enriched Gene Ontology (GO) categories corresponded to catalytic (GO:0003824) and glycosyltransferase (GO:0016758 and GO:0016757) activities (Supplemental Table S1).
Table I. Genes up-regulated after 4 h of FAMA induction organized by selected GO term classifications.
| AGI Code | Gene Name | Gene Description |
| Transcription factors | ||
| AT1G22490 | bHLH protein | |
| AT1G52890 | NAC transcription factor | |
| AT1G48150 | MADS box transcription factor | |
| AT1G62700 | NAC domain transcription factor | |
| AT1G71130 | Member of the ERF subfamily B-5 of ERF/AP2 transcription factor family | |
| AT1G25340 | MYB116 | Putative transcription factor (MYB116) |
| AT2G22540 | AGL22 | SHORT VEGETATIVE PHASE |
| AT2G33860 | ARF3 | ETTIN gene; encodes a protein with homology to DNA-binding proteins that bind to auxin response elements |
| AT2G40350 | Member of the DREB subfamily A-2 of ERF/AP2 transcription factor family | |
| AT2G47260 | WRKY23 | WRKY transcription factor |
| AT2G29060 | Scarecrow transcription factor family protein | |
| AT3G57600 | Member of the DREB subfamily A-2 of ERF/AP2 transcription factor family | |
| AT3G47500 | CDF3 | Dof-type zinc finger domain-containing protein |
| AT3G15540 | IAA19 | Primary auxin-responsive gene |
| AT3G24140 | FAMA | bHLH protein |
| AT4G11140 | CRF1 | Member of the ERF subfamily B-5 of ERF/AP2 transcription factor family |
| AT5G54470 | Zinc finger (B-box type) family protein | |
| AT5G26870 | AGL26 | AGAMOUS-LIKE 26 |
| AT5G65590 | Dof-type zinc finger domain-containing protein | |
| AT5G08130 | bHLH protein | |
| AT1G10610 | bHLH protein | |
| AT2G40340 | DREB subfamily A-2 of ERF/AP2 transcription factor | |
| AT2G20570 | GLK1 | Golden2-like1 transcription factor |
| AT3G54390 | Transcription factor | |
| AT2G40435 | bHLH like protein | |
| Other DNA- or RNA-binding proteins | ||
| AT2G45850 | DNA-binding family protein | |
| AT2G38610 | KH domain-containing protein | |
| AT3G15790 | MBD11 | Protein containing methyl-CpG-binding domain |
| AT1G20020 | LFNR2 | Ferredoxin:NADP(H) oxidoreductase |
| AT3G13700 | RNA-binding protein, putative | |
| AT1G08390 | Nucleic acid binding | |
| Receptors and signaling proteins | ||
| AT1G66100 | Thionin protein | |
| AT5G36910 | Thionin protein | |
| AT3G45780 | NPH1 | Blue light photoreceptor |
| AT1G03440 | Leu-rich repeat family protein | |
| AT1G07410 | RAB-A2B | RAB GTPASE HOMOLOG A2B |
| AT1G60630 | Leu-rich repeat family protein | |
| AT2G24130 | Leu-rich repeat transmembrane protein kinase | |
| AT3G25020 | RLP42 | Receptor-like protein 42 |
| AT4G22540 | OXYSTEROL-BINDING PROTEIN-RELATED PROTEIN 2A | |
| AT4G03010 | Leu-rich repeat family protein | |
| AT5G54510 | DFL1 | IAA-amido synthase |
| AT5G53890 | Leu-rich repeat receptor kinase (LRR-RK) | |
| AT5G57050 | ABI2 | Protein phosphatase 2C |
| AT1G26600 | CLE9 | Homologous to the Clavata3 gene |
| AT3G24982 | Leu-rich repeat family protein | |
| AT4G29240 | Leu-rich repeat family protein | |
| AT1G32320 | MKK10 | MAPK kinase |
| Kinases | ||
| AT1G21210 | Cell wall-associated Ser/Thr kinase | |
| AT2G04300 | Leu-rich repeat protein kinase | |
| AT3G18810 | Protein kinase | |
| AT4G26540 | Protein kinase | |
| AT4G24480 | Ser/Thr protein kinase | |
| AT5G57610 | Protein kinase | |
| AT5G53450 | ORG1 | OBP3-responsive gene 1 (ORG1) |
| AT5G45820 | CIPK20 | CBL-interacting Ser/Thr protein kinase |
| AT1G10760 | GWD | Encodes an α-glucan, water dikinase |
| Transporters | ||
| AT1G24400 | AATL2 | High-affinity transporter for neutral and acidic amino acids |
| AT2G29940 | PDR3 | PLEIOTROPIC DRUG RESISTANCE3 |
| AT2G28900 | OEP16-1 | AtOEP16; a 16-kD plastid outer membrane protein involved in plastid import of protochlorophyllide oxidoreductase A |
| AT2G38460 | IREG1 | IRON-REGULATED PROTEIN1 |
| AT2G32390 | GLR3.5 | Ionotropic Glu receptor ortholog |
| AT2G28260 | CNGC15 | Cyclic nucleotide-gated channel |
| AT3G53420 | PIP2;1 | Plasma membrane intrinsic protein subfamily PIP2 |
| AT3G16240 | TIP2;1 | Delta tonoplast intrinsic protein |
| AT3G47750 | ABCA4 | Member of ATH subfamily |
| AT3G47760 | Member of ATH subfamily | |
| AT3G55130 | WBC19 | Protein of the ABC transporter White-Brown Complex (WBC) |
| AT3G05030 | NHX2 | Sodium proton exchanger |
| AT4G05110 | ENT6 | Equilibrative nucleoside transporter |
| AT4G12030 | 2-Keto acid transporter | |
| AT4G17340 | TIP2.2 | TONOPLAST INTRINSIC PROTEIN 2;2 (TIP2;2) |
| AT4G10770 | Oligopeptide transporter | |
| AT5G58070 | Temperature-induced lipocalin TIL1 | |
| AT1G04690 | POTASSIUM CHANNEL β-SUBUNIT (KAB1) | |
| AT1G72150 | KV-BETA1 | Cell plate-associated protein |
| AT1G18220 | PUP9 | Purine transporter |
| AT4G05120 | ENT3 | Equilibrative nucleoside transporter AtENT3 |
| Cell organization and biogenesis/cell wall enzymes | ||
| AT1G68560 | XYL1 | Bifunctional α-l-arabinofuranosidase/β-d-xylosidase |
| AT1G70710 | Endo-1,4-β-glucanase | |
| AT2G36710 | Pectinesterase | |
| AT2G13290 | Glycosyl transferase | |
| AT3G49220 | Pectinesterase | |
| AT4G20050 | Polygalacturonase | |
| AT4G23920 | UGE2 | UDP-d-Glc 4-epimerase |
| AT5G09760 | Pectinesterase | |
| AT5G63800 | MUM2 | β-Galactosidase |
| AT5G19730 | Pectinesterase | |
| AT1G53840 | PME1 | Pectin methylesterase |
| AT1G05310 | Pectinesterase | |
| AT2G15470 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein | |
| AT5G20740 | Invertase/pectin methylesterase inhibitor | |
| AT5G65730 | Xyloglucan:xyloglucosyl transferase | |
| AT5G62360 | Invertase/pectin methylesterase inhibitor | |
| AT3G48580 | Xyloglucan:xyloglucosyl transferase | |
| AT3G16850 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein | |
| AT3G61490 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein | |
| AT3G17130 | Invertase/pectin methylesterase inhibitor | |
| AT2G15460 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein | |
| AT2G15450 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein | |
| AT1G10640 | Polygalacturonase | |
| AT1G48100 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) | |
At the 48-h time point, many more genes were up-regulated, and each to a much greater extent than at 4 h. For example, the fold change in expression in the top 20 genes from the 4-h sample ranged between 4.7× and 2.0×, whereas the top 20 from 48 h ranged between 188.7× and 19.6× (Fig. 2; Supplemental Table S1). Most (1,203, 88.0%) of these 48-h up genes were not significantly different from controls at 4 h (Fig. 2, clusters I, II, III, and VII). We reasoned that this later time point was capturing the trans-differentiation of cells throughout the plant into guard cells and, therefore, that these “late up-regulated” genes were likely to encode components present in maturing stomata. This hypothesis was confirmed by comparing these data with profiles derived from mature guard cell protoplasts (Yang et al., 2008). Seventy-five percent (1,025/1,367) of the genes up-regulated after 48 h were expressed in guard cells. This overlap was significantly different (P < 0.01) from the overlap between guard cells and a larger control sample (48%, 2,700/5,650) derived from root cells (Birnbaum et al., 2003). The intersection of the guard cell transcriptome with genes up-regulated at 4 h (55%, 197/357) or down-regulated at 4 h (53%, 601/1,126) or 48 h (58%, 840/1,453) was not significantly different from the control (root) population.
FAMA induction also triggered a significant down-regulation of a large number of genes at 4 h (n = 1,126; Fig. 2, clusters II, III, IV, VII, VIII, and IX) and 48 h (n = 1,453; Fig. 2, clusters IV, V, VI, and VIII). While this trend could be due to a direct role of FAMA as a transcriptional repressor on many targets, we noticed that many of the genes in this group have tissue-specific expression in nonstomatal lineage cells. For example, this group included root-specific MYB49 (At5g54230) and HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING1 (At1g13300) genes. Furthermore, 9 of 40 of the most highly down-regulated genes after 4 h were designated as root specific by microarray expression analysis on sorted cells (Birnbaum et al., 2003). The simplest explanation for these results is that as cells are being converted to guard cell identity, they lose their previous identities, and with them, their specific gene expression. Because of this potential developmental confound, we focused more heavily on the up-regulated genes in subsequent analyses.
Expression Patterns of Differentially Expressed Genes
Because FAMA is expressed in GMCs and young guard cells, we expected endogenous targets promoting stomatal differentiation to be expressed in these cells. Because ectopic expression of FAMA creates guard cells, we also predicted that our transcriptional profile, especially at 48 h, would uncover genes expressed in the mature guard cell. Both of these predictions were borne out by the appearance of previously known stomatal genes in our differentially regulated gene lists. Genes expressed in developing stomata include FAMA’s potential partner ICE1/SCRM (At3g26744; up-regulated 1.5-fold after 48 h). Genes expressed in, and required for, guard cell function were also identified. These include KAT1 (At5g46240; 3.5-fold up at 48 h), a guard cell-specific inward K+ channel, whose expression pattern in leaves is often used as a marker for mature guard cell identity (Nakamura et al., 1995); KAT2 (At4g18290; 3.6-fold up at 48 h), a redundantly acting homolog of KAT1 (Pilot et al., 2001); GORK (At5g37500; up-regulated 3.5-fold at 48 h), a guard cell-specific major outwardly rectifying K+ channel (Hosy et al., 2003); and abscisic acid signal transducers MPK12 (At2g46070; 8.5-fold up at 48 h) and MPK9 (At3g18040; 2.5-fold up at 48 h; Jammes et al., 2009).
Production of Reporters Indicated New Genes Expressed in GMCs and Guard Cells
Finding known stomata-expressed genes suggested that we would be able to use our data set to identify new stomatal lineage-enriched genes. As a first survey of expression patterns, we generated transcriptional reporters using approximately 2.5-kb 5′ regulatory sequences to drive the expression of GFP and/or GUS for 28 genes (Fig. 3). These genes were chosen based on timing and degree of modulation by FAMA expression, predicted gene function, and comparison with other stomatal expression data sets described in the previous section.
Figure 3.
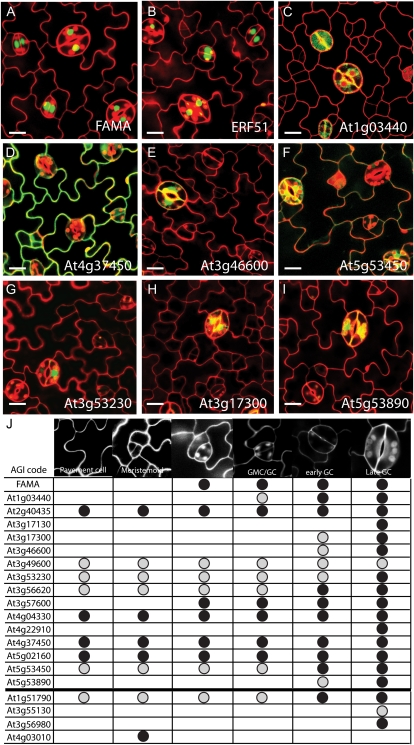
Reporter line analysis of candidate FAMA targets by confocal microscopy. A to I, Confocal images of the adaxial epidermis from the first leaf of Columbia seedlings. Reporter expression is in green, and cell outlines, as visualized by propidium iodide, are in red. Red discs in stomatal lineage cells are due to chloroplast autofluorescence. A to C, Expression of translational reporters at 9 d postgermination: FAMApro:FAMA-GFP (A), At3g57600pro:At3g57600-GFP (B), and At1g03440pro:At1g03440-YFP (C). D to I, Expression of transcriptional reporters at 10 d postgermination: At4g37450pro:GFP (D; note nonspecific expression throughout the epidermis), At3g46600pro:GFP (E), At5g53450pro:GFP (F), At3g53230pro:GFP (G), At3g17300pro:GFP (H), and At5g53890pro:GFP (I). Bars = 12 μm. J, Table highlighting the stomatal cell differentiation stage at which reporter expression was observed. Black circles indicate a relatively high level of expression, and gray circles show a low level assessed by comparing the intensity of the fluorescence in a given cell type with other cells and background levels; “high” and “low” designations are not indicative of a gene’s absolute expression level. Lines for which no signal was detected in the epidermis are not reported in this table. The thick horizontal line separates genes for which GFP reporters were made (above) from GUS reporters (below). [See online article for color version of this figure.]
Top priority genes were transcription factors, especially of the bHLH and HLH classes, given the fact that this family plays key roles in several stomatal cell fate decision points and bHLHs often participate in interconnected regulatory loops. Canonical bHLHs included bHLH039 (At3g56980; up 4.8-fold at 48 h), bHLH101 (At5g04150; up 4.6-fold at 48 h), and At5g08130 (up 1.4-fold at 4 h and 2.7-fold at 48 h). The most highly up-regulated gene at 4 h (At2g40435; 4.7-fold up at 4 h and 26.6-fold up at 48 h) resembles an HLH protein, and we also included this in the analysis. We made a reporter for the most highly up-regulated transcription factor at 48 h, ethylene response factor 51 (ERF51; At3g57600; Nakano et al., 2006), and also generated tools to follow the expression of cell cycle regulators, signal transduction elements, and cell wall-modifying proteins. We chose the remaining genes based solely on expression profiles generated in our experiment and in comparison with other high-throughput approaches targeted toward guard cells (specific gene names and Arabidopsis Genome Initiative codes are given in Fig. 3 and Supplemental Table S4).
Of the 28 reporters, we could detect expression of 21 in the leaf epidermis, and 19 among those were expressed in guard cells (Fig. 3). Eleven genes were specifically expressed in the stomatal lineage (meristemoids, GMCs, and/or guard cells). The majority of these genes (seven) were expressed only in guard cells, with expression peaking in mature guard cells. The expression pattern of ERF51 (At3g57600) and a leucine-rich repeat protein (LRR; At1g03440), however, more closely mirrored that of FAMA. Expression of these genes began in GMCs nearing their transition to guard cells and peaked in young guard cells, although reporter expression of these two genes persisted in maturing guard cells longer than the FAMA reporter (Fig. 3, A–C). Our reporter results indicate that induction of FAMA expression could enrich for genes expressed in the late stomatal lineage and potential FAMA target genes.
Another test of the connection between these genes and FAMA is monitoring their expression when FAMA is inactivated. We were concerned, however, that the poor viability of the fama mutant might confound results if we compared our overexpressors with these plants. We instead profiled plants expressing ESTpro:FAMA-EAR, an inducible dominant negative construct that recapitulates the fama mutant phenotype (Ohashi-Ito and Bergmann, 2006), grown under the same conditions as the 48-h samples. Many (but not all) of the genes we chose to follow more extensively were oppositely regulated in the ESTpro:FAMA and ESTpro:FAMA-EAR plants (Supplemental Table S3).
ChIP Reveals Potential Direct FAMA Targets
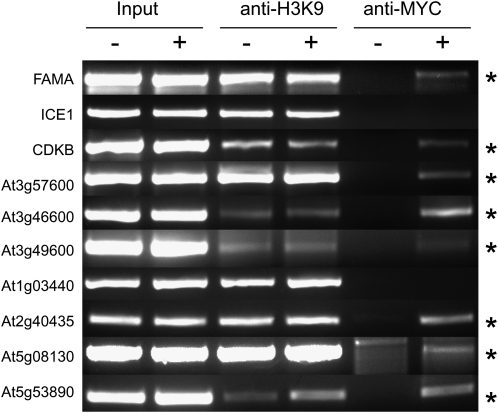
Differential expression following FAMA induction and expression in late stomatal lineage cells are characteristic of direct targets; however, to test this directly, we performed ChIP using FAMA and the 5′ regions of candidate targets. To specifically pull down FAMA/DNA complexes, we created a MYC-tagged version of the estrogen-inducible FAMA construct used for the transcriptional profile and demonstrated that it was functional in inducing the formation of single-celled stomata (Supplemental Fig. S2). FAMA overexpression was then induced by 10 μm β-estradiol treatment for 4 or 48 h, and the efficiency of induction was assayed by phenotypic analysis and a western blot against the MYC epitope (data not shown). For each of 10 promoter regions tested, induced and noninduced samples were immunoprecipitated with antibodies against MYC and histone H3 (as a positive control). The abundance of the PCR product obtained from the MYC immunoprecipitation with and without induction was then compared. ChIP was performed on regulatory regions of three candidate genes, CDKB1;1, FAMA, and ICE1/SCRM, and seven genes chosen from the microarray and reporter results, with an exon of the PIP2;1 gene (At3g53420) used as a negative control for immunoprecipitation specificity. As we expected direct targets to be in the 4-h induction, we focused primarily on this time point; however, we did also retest several of our candidates at 48 h to reflect the stage at which they were most highly expressed. For these experiments, we considered a result positive only if there was no PCR amplification of the target in the noninduced sample.
We initially used ChIP analysis to test whether FAMA could directly regulate CDKB1;1 expression. CDKB1;1 is a cyclin-dependent kinase that promotes the completion of stomatal lineage cell divisions (Boudolf et al., 2004). FAMA-promoted terminal cell differentiation leads to a steep down-regulation of CDKB1;1 expression level, and in fama mutants, CDKB1;1 reporter expression persists (Fig. 1, D and E; Ohashi-Ito and Bergmann, 2006). Here, we show that this effect on expression may be direct, as FAMA-MYC is associated with the approximately 400 bp 5′ regulatory region of CDKB1;1 (Fig. 4) but not with the negative controls.
Figure 4.
Identification of direct FAMA targets by ChIP. Semiquantitative PCR was performed on 5′ regulatory regions of 10 candidate FAMA target genes (specific primers are listed in Supplemental Table S2). The source of DNA is noted in the column headings: “Input” represents DNA isolated prior to the ChIP procedure as an amplification control; “anti-H3K9” represents DNA isolated after ChIP with anti-histone H3 K9 (general chromatin) and is a positive control for the ChIP protocol; and “anti-MYC” contains DNA isolated after ChIP with antibodies to recognize the epitope tag on ESTpro:FAMA-MYC. All samples were collected both with and without a 4-h estrogen induction. FAMA was called present at a given promoter region (asterisks to the right) only when amplification was always detected in input and H3K9 controls but only detected in the anti-MYC ChIP after Estpro:FAMA-MYC induction.
One of the most extensively studied transcriptional regulatory paradigms was established around the animal muscle bHLH, MyoD. MyoD modulates its own expression and that of its partner bHLHs (for review, see Berkes and Tapscott, 2005). We used ChIP analysis to test whether either of these regulatory features was also found in the FAMA and ICE1/SCRM module. Both the FAMA and ICE1/SCRM promoters contain G-box elements. We previously showed that FAMA was not absolutely required for its own expression (Ohashi-Ito and Bergmann, 2006); however, these data did not rule out other autoregulatory relationships such as negative and positive feedback to fine-tune expression. ChIP assays showed that FAMA was indeed associated with its own promoter (Fig. 4); however, we did not see an association with the 1,000-bp 5′ regulatory region of ICE1/SCRM (Fig. 4).
We next used ChIP to detect in vivo interactions of FAMA with promoter regions of new candidate targets. We continued to address the relationship among bHLH and HLH factors by testing At5g08130 and At2g40435, the latter of which, while highly up-regulated by FAMA, is fairly broadly expressed at low levels in the leaf epidermis (Fig. 3; expression values less than 50 in leaves [e-FP browser; Toufighi et al., 2005; our uninduced samples]). FAMA was associated with the 5′ regulatory regions of both genes (Fig. 4). Of considerable interest as direct targets were the two genes expressed in patterns mirroring FAMA, ERF51 and the LRR At1g03440. While we found enrichment of FAMA at the ERF51 promoter after 4 h of induction (Fig. 4), this was not true for At1g03440, although after 48 h we did detect some association (data not shown). Our final choices for ChIP were expressed late in guard cell development, the up-regulated LRR-receptor-like kinase At5g53890 and down-regulated SCRL17 (At3g46600). In both cases, FAMA was associated with the 5′ regulatory regions (Fig. 4), suggesting that it may not just set off an initial wave of gene expression but plays a continued role in guard cell development.
Functional Analysis of Differentially Expressed Genes
After identifying potential target genes based on expression pattern and FAMA-ChIP results, we sought to assign functions to these genes and to assess their contribution to a FAMA-led differentiation program. The fama phenotype of GMCs that are division competent but differentiation defective could arise from misregulation of many independent downstream programs, each of modest effect, or may be coordinated in a steep hierarchy with a few major regulators (also likely transcription factors), each controlling major subprograms. These two scenarios predict different phenotypic consequences of mutations in FAMA downstream genes.
Among the previously characterized genes we also identified in our study, a few qualitatively mirror the effects of misregulating FAMA. For example, scrm1-2 infrequently produces epidermal tumors that consist of redividing GMCs (Kanaoka et al., 2008), and elimination of CDKB1;1 and CDKB1;2 produces some single-celled stomata that resemble those formed by FAMA overexpression (Xie et al., 2010). Many of the genes highly up-regulated at 48 h when ectopic guard cells are present are likely to contribute more to guard cell function than to development. These genes include signaling pathway components MPK9 and MPK12 (Jammes et al., 2009) and ion channels KAT1 (Nakamura et al., 1995) and GORK (Hosy et al., 2003) that modulate stomatal pore aperture.
To test potential functions of the not previously characterized genes identified in the microarray experiment, we analyzed stomatal lineage phenotypes and overall growth in 99 T-DNA insertion lines (representing 82 genes) and 17 CMV35S promoter-driven overexpression lines corresponding to a subset of those genes of interest from the array analysis (Supplemental Tables S4 and S5). Individual seedlings were screened for cell division and cytokinesis abnormalities such as arrested GMCs, malformed guard cells, stomatal clusters, or altered stomatal index (number of stomata/total epidermal cells). Cotyledons and leaves of 20 5- to 10-d-old seedlings were analyzed for each line and genotypes confirmed as noted in Supplemental Table S5. Under the screening criteria we applied, deregulated expression of no single previously uncharacterized gene produced a reproducible stomatal development phenotype (data not shown).
One explanation for the lack of phenotypes is that these genes may share functions with close relatives; to address this possibility, we made higher order mutants with paralogs of the two genes expressed in GMCs, ERF51 and the LRR At1g03440. ERF51 is closely related to three other genes, ERF49 (At1g75490), ERF50 (At5g18450), and ERF52 (At2g40220). Based on microarray data, these homologs are found in guard cells but are also expressed more broadly (e-FP browser; Toufighi et al., 2005). The expression pattern of LRR At1g03440 mirrors FAMA, but we noticed that its closest paralog, At4g03010, was also up-regulated in our array (4.1-fold at 48 h) and is also stomatal lineage expressed (Fig. 3). Multiple mutant combinations of T-DNA insertion lines were constructed by crossing, and genotypes were confirmed by PCR; however, we did not observe any stomatal development phenotype in the double LRR mutant, nor did we find reproducible, highly penetrant stomatal phenotypes in quadruple mutant plants bearing T-DNA insertions in ERF49-52 (data not shown).
DISCUSSION
FAMA encodes a bHLH transcription factor that acts as a master controller of the GMC-to-guard cell transition, regulating both cell division and cell fate. In this role, FAMA shares properties with animal bHLHs that guide muscle and neural development, with its close paralogs MUTE and SPCH involved in stomatal development, and with members of other transcription factor families that regulate formative divisions (Levesque et al., 2006; Cui et al., 2007; Welch et al., 2007; Breuninger et al., 2008; Willemsen et al., 2008). In each of these situations, the same question arises: how do these transcription factors initiate the cascade of events that culminate in precise cell identities and division behaviors?
In the experiments reported here, we used inducible expression of FAMA to identify potential targets of this transcription factor and took advantage of FAMA’s ability to redirect many cells into guard cell identity to identify genes that are enriched in this specialized cell type. We substantiated these findings by demonstrating that the expression of 11 previously uncharacterized genes was highly enriched in the stomatal lineage. We further showed that FAMA can be associated with the 5′ regulatory regions of eight genes, consistent with these genes being its direct targets. Our microarray study effectively identified previously characterized stomata-expressed proteins, many of which lead to distinctive phenotypes when misregulated (Nakamura et al., 1995; Pilot et al., 2001; Kanaoka et al., 2008; Jammes et al., 2009). We were unable, however, to identify novel stomatal phenotypes associated with any single T-DNA insertion line or with multiple mutants of ERF51 and LRR relatives.
Recent large-scale studies of transcription factors involved in light signaling (Lee et al., 2007), brassinosteroid response (Sun et al., 2010), or root development (Levesque et al., 2006) indicate that the regulatory networks downstream of major regulators are likely to be broad. Our results add to this growing observation. As an example, in the case of the cell fate regulator SHORT ROOT (SHR), Levesque et al. (2006) defined eight potential direct targets of this GRAS domain transcription factor, including one known target, SCARECROW (SCR). Later studies of some of these factors pointed to important regulatory relationships between SHR and the new targets (Welch et al., 2007), but it is interesting that with the exception of SCR, loss-of-function mutations in these targets do not exhibit morphological phenotypes on their own (Strader et al., 2004; Levesque et al., 2006; Welch et al., 2007). This theme of lack of observable phenotypes is repeated in other microarray studies on developmental regulators (Nawy et al., 2005). Based on these previous studies with transcription factor families and our own data with the genes regulated by FAMA (and the ERF subfamily in particular), it appears that multiple overlapping functions of close relatives are the norm.
In our experiments, it is also possible that the lack of observable morphological phenotypes may be because the genes we identified affect guard cell functions such as stomatal opening and closing, ion transport, or alter cell wall composition. Given our primary interest in developmental regulators and the fact that functional redundancy has also been documented in these responses (e.g. channels KAT1/KAT2 [Pilot et al., 2001] and MPK12/MPK9 [Jammes et al., 2009]), we did not pursue extensive physiological analyses in this study. We also cannot rule out the possibility that there are genes whose altered expression would lead to stomatal development defects in our data set and we simply did not discover them because we sampled only the currently available T-DNA insertion resources.
Refinement of FAMA Activity Based on Its Relationship to Targets
As a member of the HER-containing group of bHLHs, FAMA is predicted to bind to a G-box sequence motif; these motifs are sufficiently short, however, that they occur by chance every 4 kb and thus are predicted to be present in the regulatory regions of many genes. We previously demonstrated that the HER motif was required for FAMA activity (Ohashi-Ito and Bergmann, 2006); however, our attempts to confirm in vitro FAMA binding to G-box sites (or to identify others by SELEX) by electromobility shift assays using purified FAMA protein have not been successful (K. Ohashi-Ito, unpublished data), consistent with the findings that FAMA poorly homodimerizes, but efficiently heterodimerizes, with several bHLH partners (Ohashi-Ito and Bergmann, 2006; Kanaoka et al., 2008). The best opportunities to find FAMA targets, therefore, were through identifying the genomic regions associated with FAMA by ChIP. In this study, we selected a small number of differentially regulated genes and found that FAMA indeed was associated with their 5′ regulatory regions. Eight of the 10 genes possess G-boxes (or the related E-boxes) within the FAMA-associated regions, but we could demonstrate an association with two (CDKB1;1 and At3g49600) that do not. We also found that FAMA could interact directly both with genes that were up-regulated and genes that were down-regulated by its induction, indicating that FAMA could serve as a repressor as well as an activator in vivo. Such dual behavior is not uncommon; for example, WUSCHEL is primarily a repressor, but in combination with a specific partner, it is part of an activating complex (Ikeda et al., 2009).
ChIP data revealed that FAMA is able to bind to its own promoter. FAMA also binds directly to the regulatory regions of some bHLH and HLH-like proteins it induces (though not of putative partner ICE1/SCRM). Many transcription factors are regulated by feedback loops in which they regulate their own expression; for example, other bHLHs (such as MyoD [Weintraub et al., 1994]) induce the expression of dimerization partners that, in addition to initiating cascades of downstream gene expression, later feedback regulate themselves. In the case of FAMA, it is particularly interesting that the most highly up-regulated early gene (At2g40435) is not a bHLH but resembles the antagonistic HLH proteins that dimerize with, and inactivate, bHLHs (Weintraub et al., 1994). Previously, we showed that a FAMApro:GUS reporter was still expressed in the GMC tumors of a fama mutant, indicating that FAMA is not absolutely necessary to promote its own expression (Ohashi-Ito and Bergmann, 2006); however, we have noticed that the normal window of FAMA expression is very tight and that even modest overexpression of FAMA can drive cells into differentiation without division, forming malfunctional one-celled stomata (Ohashi-Ito and Bergmann, 2006). Based on these observations, an attractive model is that FAMA ensures its temporally restricted activity by both inducing antagonists and binding to its own promoter as part of negative feedback loops. FAMA’s potential “active” partner ICE1/SCRM is also up-regulated in response to FAMA induction, although it is not a direct FAMA target. ICE1/SCRM normally is expressed earlier than FAMA and persists in many stomatal lineage cells (Kanaoka et al., 2008), and this higher expression level may be an indirect effect of the induction of guard cell identity by FAMA.
Implications for Combinatorial Control of the GMC-to-Guard Cell Transition
The transition from GMC to guard cell and the subsequent maturation of the guard cells into mature, highly responsive and functional valves is an important control point and is regulated by a number of different genes in addition to FAMA. The nature of the products encoded by these genes suggests the involvement of signaling cascades (YODA and downstream MAPKs; Lampard et al., 2009), cell cycle regulators CDKB1;1 and -1;2 (Boudolf et al., 2004; Xie et al., 2010), as well as other transcription factors of the bHLH (Ohashi-Ito and Bergmann, 2006; Kanaoka et al., 2008) and MYB classes (Lai et al., 2005; Xie et al., 2010). We have an ongoing interest in establishing functional relationships among these different elements. Some of these factors are likely to act upstream of FAMA. For example, signaling through MAPK cascades promotes guard cell formation and expression of a dominant negative version of YODA phenocopies the fama mutant (Lampard et al., 2009). We previously showed a direct connection between MAPK signaling and FAMA’s paralogue SPCH (Lampard et al., 2008), but the relationship between MAPKs and FAMA differs in two important ways. First, FAMA is not a target of MPK3 and MPK6 (Lampard et al., 2008), and second, phosphorylation of SPCH results in down-regulation of its activity, opposite what would be predicted for FAMA. As the experiments in this paper examine potential transcriptional control by FAMA, we monitored the expression of YODA, MKK7, and MKK9 to determine if they might participate in a feedback loop with FAMA, but their expression levels are not altered by FAMA induction (Supplemental Table S1).
Recent studies on FLP and MYB indicate that these proteins directly regulate the transcription of many cell cycle genes, including CDKB1;1, as they regulate the GMC-to-guard cell transition (Xie et al., 2010). Here, we have demonstrated that FAMA is also associated with the CDKB1;1 regulatory region, and a vast literature supports the existence of MYB/bHLH partnerships for plant transcriptional control (Goff et al., 1992; Payne et al., 2000; Zimmermann et al., 2004). Despite the common expression pattern and biological function, however, no physical interaction could be found between FLP/MYB88 and FAMA (Ohashi-Ito and Bergmann, 2006), genetic experiments pointed to these transcription factors acting in largely parallel pathways (Ohashi-Ito and Bergmann, 2006), and there is no evidence that either class regulates the expression of the other (Xie et al., 2010; Supplemental Table S1). Identifying the precise binding sites of FAMA relative to the MYBs on the well-defined CDKB1;1 promoter may point to a new way that MYBs and bHLHs can promote specific transcriptional responses in plants.
A number of trends are revealed by the analysis of the regulators downstream of FAMA. First, there are no genes whose misregulation accounts for a large part of the FAMA phenotype, suggesting that the network downstream of FAMA is quite broad. Consistent with this, our initial ChIP analysis showed that FAMA was associated with the promoters of genes representing other transcription factors, cell cycle regulators, and genes we might expect to be downstream effectors of the differentiation process, a suite of levels similar to that seen for SHR direct targets (Levesque et al., 2006; Welch et al., 2007). In Figure 5, we outline a working model of FAMA activity based on the experimental data presented in this paper.
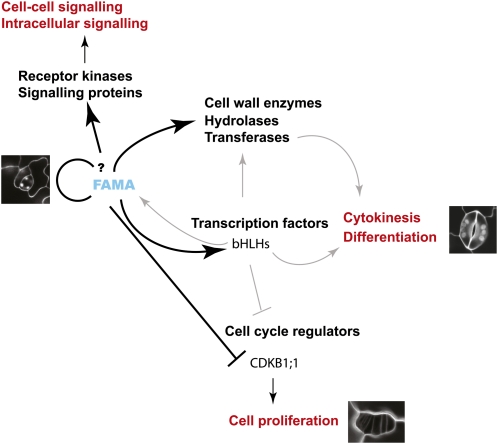
Figure 5.
Model for the FAMA-mediated control of stomatal guard cell differentiation. To form typical two-celled stomata, genes promoting guard cell identity must be up-regulated while cell division processes must be coordinately down-regulated during the transition from the GMCs. FAMA expression is strong in late GMCs and early guard cells. With this temporal and spatially restricted timing, FAMA may both directly activate differentiation genes while repressing cell cycle control genes such as CDKB1;1 to limit GC division. In this model, FAMA acts as both transcriptional activator and repressor. The question mark indicates uncertainties concerning the exact nature of the regulatory feedback loops detected in our assays. Black lines represent trends that are backed up by ChIP data and/or reporter line analysis, while gray lines represent plausible but yet-to-be-demonstrated regulatory steps. [See online article for color version of this figure.]
In the future, genome-scale approaches using ChIP-seq could significantly extend the network immediately downstream of FAMA. In addition, analysis of the promoter occupancy by FAMA’s potential partner ICE1/SCRM could demonstrate whether there is an obligate relationship between these two transcription factors or whether each also participates in its own activities. The latter interpretation is suggested by the fact that ICE1/SCRM is involved in roles outside of the stomatal lineage (Chinnusamy et al., 2003) and that the penetrance of the guard cell phenotype in ice1-2/scrm null mutant plants is quite low (only two to four defective GMCs per leaf; D.C. Bergmann, unpublished data). Monitoring the promoter occupancy by the new bHLH and HLH genes induced by FAMA relative to occupancy by FAMA itself would determine whether feed-forward and feedback regulation manifested by bHLH partner switching is a part of this system.
MATERIALS AND METHODS
Construction of Lines and Choice of Induction Times
We tested FAMA in several inducible expression systems available for use in Arabidopsis (Arabidopsis thaliana), including ethanol (Deveaux et al., 2003), estrogen (Zuo et al., 2000), and glucocorticoid (Lloyd et al., 1994; Craft et al., 2005). After preliminary trials of all three systems, we found that the estrogen system (Estpro:FAMA) consistently gave a strong, reliable induction of the FAMA transcript in germinating seedlings without inducing severe stress responses. Two time points were selected, 4 and 48 h, based on the rationale that such time points should help identify either direct targets of FAMA (4 h) and/or guard cell-expressed genes (48 h).
Microarray Analysis
RNA was obtained from seedlings treated as follows. Sterilized seeds of transgenic Estpro:FAMA in the Columbia ecotype were sown on half-strength Murashige and Skoog (MS) + 10 g L−1 Suc plates and then kept at 4°C for 3 d. Seedlings were grown on vertically oriented plates in a 22°C incubator under 24 h of light for 5 d. Approximately 100 mg of seedlings per sample was then transferred with forceps to plates containing MS + Suc + 10 μm β-estradiol (or new MS + Suc plates for controls). After 4 or 48 h on new plates, samples were collected (three independent replicates for each sample), frozen in liquid nitrogen, and stored at –80°C. From this point on, the three replicates from each time point and genotype were processed in parallel. RNA was isolated with the RNeasy Plant Mini Kit (Qiagen), and samples were processed and labeled with standard protocols as provided by the manufacturer (Enzo) and hybridized to Affymetrix ATH1 array chips using protocols as described by Bergmann et al. (2004). The array images were analyzed with the Affymetrix Microarray Suite 5.0 with the target intensity set to 500. The expression levels of the genes were analyzed using GeneSpring 4.2 software (Silicon Genetics). Genes that consistently increase or decrease in their expression levels relative to the expression level in the corresponding wild-type samples were identified using the Significance Analysis of Microarrays software with delta value greater than 10 and false discovery rate less than 0.01%. K-means clustering using Jexpress 9.1 (Molmine) was used to generate the clusters in Figure 2 with the following parameters: nine clusters built from a maximum of 200 iterations using a Pearson correlation as distance matrix. Data were prefiltered, as recommended by Orlando et al. (2009), to remove genes that were not expressed above the cutoff or that were expressed uniformly across all the measurements.
ChIP
Seedlings expressing the ESTpro:FAMA-MYC construct were used for the ChIP assays. Seedlings were grown in liquid culture (MS medium supplemented by 10 g L−1 Suc) on a rotating table (90 rpm) with a 16-h-light/8-h-dark regime at 21°C for 6 d. Seedlings were collected 4 or 48 h after induction by 10 μm β-estradiol. Noninduced ESTpro:FAMA-MYC plants were used as a negative control in the experiment. In both cases, FAMA-MYC protein levels were assessed by western blot. No signal using anti-MYC antibodies was ever detected for the noninduced control, whereas a band could be observed for the EST-induced seedlings. ChIP assays were carried out on 1 g of tissue according to the protocol of Gendrel et al. (2005) with only minor modifications. For immunoprecipitation, 150 μg of protein extract was incubated overnight with anti-MYC (Covance; 9E10 Monoclonal Antibody) or anti-histone H3 (dimethyl K9; Abcam; Ab1220) antibodies at a 1:100 dilution. At the end of the experiment, the final DNA extracts were diluted five times in water, and 1 μL from the diluted DNA was used as template in a PCR. A small aliquot of untreated sonicated chromatin was reverse cross-linked and used as the total input DNA for both induced and control samples. The PCR program was as follows: 5 min at 95°C, 35 cycles of 15 s at 95°C, 30 s at 55°C, and 30 s at 72°C, followed by a 5-min final elongation step at 72°C. PCR primers were designed to amplify a 200-bp sequence from the promoter region of the genes of interest.
T-DNA Screening
Ninety-nine T-DNA insertion mutant alleles of 82 different FAMA-inducible genes were obtained from the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu; Supplemental Table S5). Stomatal phenotypes were scored initially in cotyledons and first leaves of live 5- to 10-d-old plate-grown seedlings using bright-field microscopy. Confirmation of T-DNA insertions was performed by PCR using primers designed using the iSECT tools (salk.signal.edu) and standard genotyping protocols (Lukowitz et al., 2000). Double and higher order mutants were generated by crosses and PCR genotyping in subsequent generations. Supplemental Table S1 summarizes the different T-DNA lines screened in this study.
Reporter and Overexpression Constructs and Observations
Translational and transcriptional reporter lines were made by PCR amplifying a 2.5-kb promoter region using D-TOPO cloning-compatible primers from a genomic DNA extract (Supplemental Table S4). In most cases, a NotI restriction site was introduced on the reverse primer to facilitate subsequent cloning steps. PCR fragments were recombined into a pENTR vector using pENTR Directional TOPO Cloning Kit (Invitrogen) and further recombined into pMDC107 (GFP) or pMDC163 (GUS) destination vector (Curtis and Grossniklaus, 2003). For overexpression or fusion protein constructs, a cDNA of interest was obtained by PCR amplification from total cDNA (extracted from 7-d-old seedlings) or directly from Arabidopsis Biological Resource Center clones and transferred, if needed, to a Gateway-compatible entry clone. When necessary, the 2.5-kb promoter region of the cDNA of interest was inserted upstream of the initial ATG by utilizing a NotI site in the vector. Plant transformation vectors pMDC107, pMDC163, pMDC43, pMDC83, pMDC32, and p35GY (Kubo et al., 2005) were used as noted in Supplemental Table S2. Every construct was verified by DNA sequencing and introduced into Agrobacterium tumefaciens strain GV3101. Arabidopsis plants were then transformed by floral dipping (Clough and Bent, 1998). GUS staining and fluorescent protein observations were performed using standard protocols as described (Ohashi-Ito and Bergmann, 2006) and visualized on a Leica DM5000 microscope or a Leica SP5 confocal microscope. For fluorescent protein fusions, cell walls were visualized by bathing seedlings in 0.02 mg mL−1 propidium iodide (Molecular Probes P3566) for 1 to 5 min. Images were converted from Leica file format with NIH ImageJ and false colored and prepared for publication with Photoshop (Adobe) by increasing brightness and enhancing contrast. All digital enhancements were applied equally across all regions of a given image.
Microarray data sets are MIAME compliant and have been submitted to the Gene Expression Omnibus (GEO) database under the GEO Series accession number GSE21786 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21786). All named genes are associated with an Arabidopsis Genome Initiative code at first mention in the text.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. FAMA-modulated genes at alternate FC choices.
Supplemental Figure S2. Demonstration of Est:FAMA-MYC activity.
Supplemental Table S1. Microarray data filtered for cluster analysis with FC = 1.2.
Supplemental Table S2. Additional GO classes of genes up-regulated after 4 h of FAMA induction.
Supplemental Table S3. Selected genes significantly modulated by FAMA-EAR.
Supplemental Table S4. Reporter and 35S lines created in this study.
Supplemental Table S5. T-DNA lines analyzed in this study.
Supplemental Table S6. Primer sets used in the ChIP assay.
Supplementary Material
References
- Abrash EB, Bergmann DC. (2009) Asymmetric cell divisions: a view from plant development. Dev Cell 16: 783–796 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR. (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. (2007) Stomatal development. Annu Rev Plant Biol 58: 163–181 [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. (2005) MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16: 585–595 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Barrôco R, de Almeida Engler J, Verkest A, Beeckman T, Naudts M, Inzé D, De Veylder L. (2004) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Despres C, Martin GB. (2003) The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15: 3033–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC. (2003) Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J 33: 853–866 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I. (2005) New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J 41: 899–918 [DOI] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveaux Y, Peaucelle A, Roberts GR, Coen E, Simon R, Mizukami Y, Traas J, Murray JA, Doonan JH, Laufs P. (2003) The ethanol switch: a tool for tissue-specific gene induction during plant development. Plant J 36: 918–930 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6: 864–875 [DOI] [PubMed] [Google Scholar]
- Hernandez ML, Passas HJ, Smith LG. (1999) Clonal analysis of epidermal patterning during maize leaf development. Dev Biol 216: 646–658 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI. (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Hosy E, Vavasseur A, Mouline K, Dreyer I, Gaymard F, Porée F, Boucherez J, Lebaudy A, Bouchez D, Very AA, et al. (2003) The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc Natl Acad Sci USA 100: 5549–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Nadeau JA, Lucas J, Lee EK, Nakagawa T, Zhao L, Geisler M, Sack FD. (2005) The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17: 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Lukowitz W, Ellis BE, Bergmann DC. (2009) Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC. (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC. (2009) Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 266: 436–439 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR. (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci 20: 506–510 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA. (2009) Stomatal development: new signals and fate determinants. Curr Opin Plant Biol 12: 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura RL, McKendree WL, Jr, Hirsch RE, Sedbrook JC, Gaber RF, Sussman MR. (1995) Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol 109: 371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ. (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Brady SM, Koch JD, Dinneny JR, Benfey PN. (2009) Manipulating large-scale Arabidopsis microarray expression data: identifying dominant expression patterns and biological process enrichment. Methods Mol Biol 553: 57–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KM, Rychel AL, Torii KU. (2010) Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell 22: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot G, Lacombe B, Gaymard F, Cherel I, Boucherez J, Thibaud JB, Sentenac H. (2001) Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J Biol Chem 276: 3215–3221 [DOI] [PubMed] [Google Scholar]
- Qian W, Tan G, Liu H, He S, Gao Y, An C. (2007) Identification of a bHLH-type G-box binding factor and its regulation activity with G-box and box I elements of the PsCHS1 promoter. Plant Cell Rep 26: 85–93 [DOI] [PubMed] [Google Scholar]
- Rowe MH, Bergmann DC. (2010) Complex signals for simple cells: the expanding ranks of signals and receptors guiding stomatal development. Curr Opin Plant Biol 13: 548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM. (2004) Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA 101: 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Weintraub H, Genetta T, Kadesch T. (1994) Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev 8: 2203–2211 [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Bauch M, Bennett T, Campilho A, Wolkenfelt H, Xu J, Haseloff J, Scheres B. (2008) The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev Cell 15: 913–922 [DOI] [PubMed] [Google Scholar]
- Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JA, Sack FD, Grotewold E. (2010) Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22: 2306–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) Technical advance. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.