Abstract
A multidimensional image navigation and display software was designed for display and interpretation of large sets of multidimensional and multimodality images such as combined PET-CT studies. The software is developed in Objective-C on a Macintosh platform under the MacOS X operating system using the GNUstep development environment. It also benefits from the extremely fast and optimized 3D graphic capabilities of the OpenGL graphic standard widely used for computer games optimized for taking advantage of any hardware graphic accelerator boards available. In the design of the software special attention was given to adapt the user interface to the specific and complex tasks of navigating through large sets of image data. An interactive jog-wheel device widely used in the video and movie industry was implemented to allow users to navigate in the different dimensions of an image set much faster than with a traditional mouse or on-screen cursors and sliders. The program can easily be adapted for very specific tasks that require a limited number of functions, by adding and removing tools from the program’s toolbar and avoiding an overwhelming number of unnecessary tools and functions. The processing and image rendering tools of the software are based on the open-source libraries ITK and VTK. This ensures that all new developments in image processing that could emerge from other academic institutions using these libraries can be directly ported to the OsiriX program. OsiriX is provided free of charge under the GNU open-source licensing agreement at http://homepage.mac.com/rossetantoine/osirix.
Keywords: DICOM viewer, 3D, image fusion, dynamic series, open-source software
THE RAPID EVOLUTION of digital imaging techniques and the increasing number of multidimensional and multimodality studies constitute a challenge for PACS workstations and image display programs. With the improvement of spatial resolution of multi-detectors CT scanners and the emergence of multimodality exams like PET-CT1 or Cardiac-CT,2 traditional two-dimensional image viewers and image display programs are becoming unsuitable for interpretation of these large sets of images. For most tomographic imaging techniques such as magnetic resonance imaging (MRI) and CT, the traditional 2D acquisition technique of cross-sectional slices is evolving into 3D volume acquisitions with isotropic voxel sizes resulting in very large data sets. The conventional way of reviewing these images slice-by-slice is too cumbersome for interpreting the 800 to 1,000 slices that can be acquired with multi-detector CT scanners. These large sets of images require additional image processing and reformatting to make them suitable for efficient and rapid image navigation and image interpretation. In most cases this can only be achieved on high-end dedicated 3D workstations that can provide thick-slab maximum intensity projections (MIP), orthogonal and oblique multiplanar reformatting (MPR), and 3D volume and surface rendering.3 These recent changes in the acquisition modalities require radiologists to use expensive dedicated 3D processing workstations to properly interpret these exams.4 Furthermore, access to these visualization and rendering tools is usually limited to high-end users in a radiology department, preventing referring physicians, surgeons, and other care providers to benefit from the extraordinary value of the multidimensional imaging techniques for decision making and patient management In most cases static snapshots and preselected movie clips must be exported from the 3D processing workstations to be distributed to other users outside of a radiology department.
The goal of our project is to develop a completely new software platform that will allow users to efficiently and conveniently navigate through large sets of multidimentional data without the need for high-end expensive hardware or software. We also elected to develop our system on new open-source software libraries, thus by allowing the system to be easily adaptable to a variety of hardware platforms. We also benefit from our experience several years ago of having developed one of the first free DICOM viewers, OSIRIS, running on Macintosh and Windows personal computers.5,6 The success and wide adoption of the OSIRIS software has encouraged us to extend our effort into a completely new program more suitable for navigating through large multidimensional data sets. The name given to the new software, OsiriX, marks the transition to a completely new platform with the added “X” indicating the migration to the new Macintosh operating system version 10, also called MacOS X. The X also indicates the compatibility with underlying Unix platform and the adoption of the open-source paradigm.
In our design we also wanted to take advantage of the significant progress in performance and flexibility of 3D animation on personal computers mostly driven by the computer graphics and game industry. Most video games today are developed on OpenGL14 graphic libraries that benefit from hardware acceleration and processing capabilities of today’s ultra-fast video cards. Also, OpenGL, because it is an industry standard, adapts automatically to any hardware configuration and take advantage of any hardware accelerator that is provided for video display of 2D and 3D data.
In the scientific community, new open-source libraries have emerged for the visualization of multidimensional data. The Visualization Toolkit or VTK7 is a well-recognized and widely adopted software library that runs on multiple platforms and has been used for numerous scientific and medical applications so far. The recent adjunction of the Insight Segmentation and Registration Toolkit or ITK,8 mostly funded by the US National Library of Medicine as part of the Visible Human Project,9 adds a wealth of additional rendering and image processing tools for medical applications. These new open-source software toolkits offer powerful functions to do complex images manipulations, and great performance for real-time 3D image visualization.
With the introduction of more advanced image processing and visualization functions, imaging software applications tend to become more complex and usually include numerous processing functions that are not necessarily useful for all users. Depending on the type of images being evaluated and the interpretation task to be performed, most users would only use a fraction of the tools or features of a typical image processing and 3D visualization program. The purpose of our project was to explore new techniques that would allow easy customization of the program for different users and applications. Each user should be able to customize the software to include only the functions and tools needed for their specific purposes.
METHODS, TECHNIQUES, AND RESULTS
The Open-Source Paradigm
The OsiriX software program is developed as a stand-alone application for the MacOS X operating system. It includes an image database that is updated automatically when new images are downloaded to a specific directory (Fig. 1). Images can either be pushed from the PACS using a DICOM “store” function or they can be “pulled” by a DICOM query-retrieve function of the program. Image files can also be manually copied from off-line media or from other network sources. The OsiriX software was developed based on an open architecture built on existing open-source components as described in Figure 2. The main components are these:
“GNUstep / Cocoa”10,11: an object-oriented and cross-platform framework for the development of the graphical user interface. This framework allows the user to quickly design and develop complex graphic user interfaces. It is distributed in open-source format under the name of GNUstep. Cocoa is the name given by Apple Computer in the MacOS X environment. It is developed in Objective-C language.12 Objective-C is an object-oriented language that has the benefits of C++ but without its complexity. Objective-C language also provides powerful memory management: it uses a retain/release scheme. It allows the user to automatically retain or release memory blocks. This prevents any undesired memory hugs and memory overflows (memory leaks) of the type that often occur in the development of graphic programs that manipulate large sets of image data. The open-source and cross-platform compiler “GNU Compiler Collection” (GCC)13 is used to compile this Objective-C framework.
“OpenGL”14: an industry standard graphic library for 3D image visualization functions. OpenGL was originally developed by SGI (Silicon Graphics) under the name of IrixGL and was only available to high-end dedicated Unix workstations that came with extremely high price tags. With the evolution of the 3D games market, OpenGL has been adapted by graphic boards manufacturers for the personal computing market, dominated by two main manufacturers: NVIDIA15 and ATI.16 All their graphic video boards are OpenGL compatible.
allows the user to take advantage of hardware acceleration by 3D graphic cards when available. It is the only truly cross-platform 3D library available that is designed for hardware acceleration. It is currently available for Linux, IRIX, Windows, and MacOS X; there is even an OpenGL version for the Palm operating system. OpenGL has been developed for 3D rendering, but it also performs extremely well in 2D rendering functions. On most operating systems 2D images are displayed faster through OpenGL than by using the standard graphic functions of the native operating system. By adopting OpenGL, large sets of over 1000 CT slices can be displayed in a few seconds.
“The Visualization Toolkit” (VTK)7: an object-oriented open-source and cross-platform library for 3D image processing and visualization that is widely adopted in the scientific community. This toolkit offers a large number of functions for the manipulation and display of 3D data sets. It is built over OpenGL to ensure hardware accelerated rendering on different platforms. VTK offers a complete set of functions for the manipulation and display of 3D data sets: multiplanar reconstruction (MPR), maximum intensity projection (MIP), volume rendering with transparency, and surface rendering with 2D or 3D texturing. VTK also offers a complete set of function to display and modify 3D vectorial graphics: it allows the display and modification of region-of-interest as well as other measures in 3D data sets. Finally, it fully supports all the blending functions of OpenGL to easily create 3D image fusion between two different data sets. VTK is actually the only true cross-platform and hardware accelerated 3D rendering library for personal computers. Under open-source licensing, VTK is used and developed by a large number of users, allowing its fast and robust evolution.
The “Insight Segmentation and Registration Toolkit” (ITK)8 is an extended set of libraries for specific medical image processing. It is an extension of the VTK library and is based on the same framework. It was developed to solve some of today’s problems in medical imaging such as image segmentation and multimodality image registration. It provides a full set of 2D and 3D processing algorithms adapted for these tasks. The different library components allow developers to easily add image fusion and organs segmentation in image processing software programs. This library is, like VTK, under active open-source licensing and benefits from updates and improvements provided by a large number of users.
“Papyrus toolkit”17 for DICOM files management (Fig. 1): This public domain library developed at the University of Geneva offers all the necessary functions to read and write DICOM files including all meta-data. This library is distributed with the entire ANSI C18 source code, easily adaptable to any platform. It provides a complete set of functions to manage the complex and cumbersome DICOM image file format. It allows for the extraction of image data but also for all numeric and textual meta-data associated to a DICOM file. The Papyrus toolkit is a convenient extension to VTK and ITK libraries.
“DICOM Offis”19 for DICOM network functions: This cross-platform library supports the DICOM communication protocols that make it possible to query, send, retrieve, and receive DICOM images within a PACS network. The complete C++ source code is also available. This library provides a complete set of functions that greatly facilitate the management and implementation of the extremely complex and convoluted DICOM network protocol.
“Altivec” of PowerPC20 is a low-level assembly function for accelerating digital signal processing (DSP) functions. Because our goal is to provide the maximum level of acceptability and adoption by the users, we have emphasized the need for high performance and real time interaction. To achieve the best performance, we adopted Altivec as the only platform-specific and non-open-source technology that is available only for PowerPC platforms. We used this assembly language feature in very limited portions of our program to accelerate key features such as convolutions, image projections, and complex mathematical data conversions. We also wrote these functions in standard ANSI C18 in order to compile the project on non-PowerPC platforms. Altivec is a “Single Instruction, Multiple Data” (SIMD) unit of PowerPC processors. A SIMD system packs multiple data elements into a single register and performs the same calculation in all of them at the same time. It offers great performance when manipulating large sets of data such as 3D volume data. One disadvantage is the complexity of software development restricted to 128 bit registers for all operations. However, the advantage is the acceleration in performance of some functions by a factor of 10 or 20.
“Quicktime”21 for exchange of multimedia image file formats. Many commercial DICOM viewers or 3D workstations allow the user’s to see and manipulate images, but they lack a convenient way to export the images to standard multimedia file formats. This limits the ability to communicate the resulting images to other users and applications. To support the maximum possible number of existing image and graphic file formats in OsiriX, the cross-platform Quicktime library was used. It allows the export of any image data to all standard multimedia image formats such as TIFF, Photoshop, JPEG, JPEG2000, and BMP22 and any image sequence to movies file formats such as AVI, MPEG, and MPEG4. Furthermore we also used the QuickTime library to enable importing of non-DICOM images into OsiriX, thereby allowing users to benefit from the display and processing tools provided in OsiriX.
An important and challenging aspect of the development of this project was to integrate all of these technologies. OpenGL, VTK, ITK, DICOM Offis, Papyrus, and Quicktime are C/C++ cross-platform toolkits. These components were not designed to be fully compatible, and some inconsistencies and incompatibilities between header files or compiler flags made the integration somewhat difficult and laborious. Once past the steep learning curve in assessing the structure of the various components, the major advantage of the integrated product is the wealth of functions and features that are invaluable for any software developer of medical imaging applications.
Figure 1.
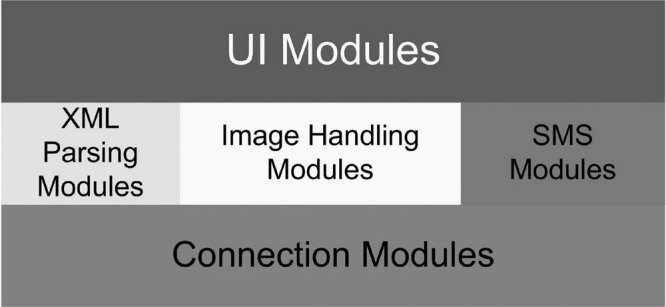
Main window of the OsiriX program providing a listing of available images in the database (upper left corner) and sets of thumbnail images of the different series (right panel), as well as a preview window of any selected sets of images (lower left corner).
Figure 2.
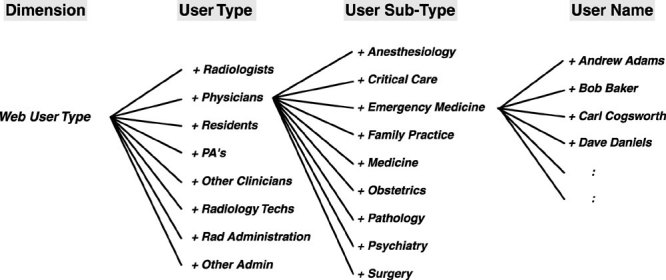
General architecture of the OsiriX program showing some of the open-source components and libraries that were used.
The most important aspect of the project is that all the components function together within a simple and user-friendly graphic user interface. To achieve the best possible user interface design, we chose to develop the OsiriX program on a Macintosh platform to benefit from its well-known user interface features and convenience and ease of use. Additionally, the latest development environment provided by Apple greatly facilitates the rapid development of interactive graphic applications. The first version of OsiriX was developed in less than 6 months including complex 2D and 3D functions, DICOM files, and network management and multimedia file formats management. One of the key features of the latest Macintosh graphic user interface is the ability to interactively add or delete some of the software features by simply dragging icons from a retractable palette (Fig. 3) to and from a window toolbar. Each icon can represent a simple program function or a complex processing tool. This feature alone offers a major advantage over other computer platforms, allowing easy customization of the program adaptable to different users’ needs.
Figure 3.
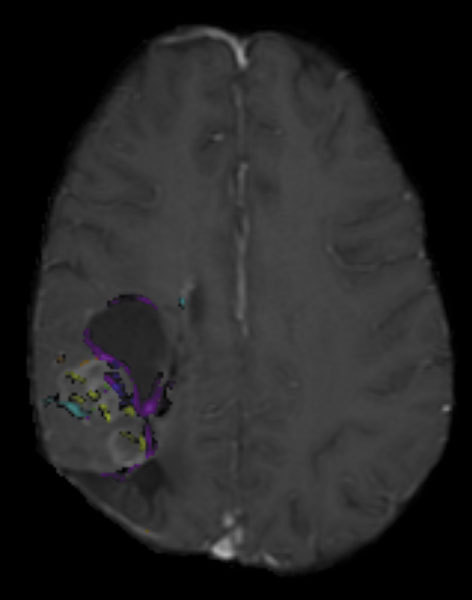
Example of user interface customization allowing the user to drag and drop tools and functions represented by a list of icons that can be added and removed from the tool bar.
From Parallel Processing to Grid Computing
The OsiriX program offers all the basic image manipulation functions of zoom, pan, intensity adjustment, and filtering with real-time performance. Additional functions such as multiplanar projection, convolution filters, variable slice thickness adjustments, volume rendering, minimum and maximum intensity projections, and surface rendering are also accessible in quasi-real-time, depending on the hardware used, as well as on the number of slices to reconstruct. For example, on a dual-GS 2Â GHz PowerMac computer, OsiriX can to render two images per second of a 400 CT slices set in volume-rendering mode. All these basic functions being handled by the OpenGL library essentially rely on the processing capabilities of the video hardware and require very minimal processing from the central processor unit (CPU). In essence the most basic image manipulations are processed in parallel by the video graphic processor unit (GPU), and the performance therefore does not depend on the computer processor unit (CPU).
The program enables the rapid review of very large sets of images and does not rely on pre-loading of the images from the disk to memory before review. As soon as an images serie is selected, images will automatically be displayed on the screen in a cine loop using a “streaming” technique, enabling direct display of the images at a very rapid pace. For example, on a G4 1 GHz mobile PowerBook computer, OsiriX is able to completely load a 355 CT images series in less than 10 seconds (loading rate: 35 images per second) while displaying the first image on the screen in less than 1 second. This streaming technique allows for an unlimited number of image series to be displayed simultaneously in separate windows. The software architecture was tested on very large image series consisting of thousands of images without any limitations. The OsiriX program was developed and mainly tested on the following standard off-the-shelf computers: PowerPC G4 laptop from Apple Computer, running at 1.25 GHz with 768 MB of RAM, with a built-in original 64 MB graphic board from ATI (ATI Mobility Radeon). Even on this standard laptop, the basic image manipulations were extremely fast and performed in real time without noticeable delays in response. For performance comparisons, we also implemented the software on a PowerPC dual G5 processors desktop computer from Apple Computer, running at 2 GHz with 1 GB of RAM and with a 128 MB graphic board from ATI (ATI Radeon 9600 Pro): performance was significantly better on the dual processor unit, with sometimes up to a factor of 6–10 faster performance for computer intensive tasks such as MIP and volume rendering, mostly due to the faster graphic board and dual processor architecture. The other basic functions of image manipulation and display such as intensity and contrast, zoom and pan, and navigation through large image sets were not noticeably different, as they are already fast in real time on the standard laptop computer. The program was also tested on lower end computers such as older generation laptops based on 800 MHz G4 PowerPC and 512 Mb of RAM, as well as a desktop 1.2 GHz G4 with 1 Gb of memory. No perceivable difference in performance could be identified except for moderately-slower 3D rendering and maximum intensity projection functions. The OsiriX software is designed to be multithreading compliant, taking advantage of parallel processing capabilities of multiprocessors when available. In particular, all graphics manipulations and rendering functions are tilled in separate threads and distributed to each available processor when possible. The support of multiprocessor architecture is seamless, performing particularly well on the new generation of dual processor G5 computers: the HyperTransport technology developed by Apple23 greatly accelerates memory transfers from RAM to processors (up to 12.8 gigabytes/second). The RAM to processor transfer is particularly critical for all 3D medical imaging rendering techniques, and significant improvement in performance can be perceived on multiprocessor platforms.
Another potential improvement in performance could be expected for very computational-intensive image processing and rendering techniques by using Grid Computing technology federating several networked computers. Grid Computing technology would allow multiple computers to be networked to perform complex processing tasks such as high-resolution volume rendering and surface rendering of very large data sets.
Navigating the Fifth Dimension
The development of this new DICOM viewer was motivated by the necessity to a have a tool more suitable for multidimensional and multimodality imaging studies such as PET-CT and ultra-fast multidetector Cardiac-CT.24 Essentially these new imaging techniques provide image data that are beyond the traditional 3D anatomical data. The combined PET-CT adds a new dimension to the data that represent the metabolic activity of the tracer, and blending this functional parameter represents a fourth dimension that the users need in order to navigate. The ultrafast multidetector CT scanners can add a temporal dimension to the data by sequentially acquiring images over time to display temporal behavior such as a beating heart or transit of a tracer or contrast agent across a vascular tree or an organ. The newer generation of PET-CT scanners can now provide data sets that represent all five dimensions for cardiac PET-CT studies. We therefore designed our software to allow users to interactively navigate in the five dimensions.
Although the software does not yet support any realignment technique for images obtained from different modalities, it does provide a very simple and very intuitive way to generate fused images by blending two image sets that are prealigned. For image fusion, the user needs to select and adjust a color scale for one of the image sets that will be overlaid over another image set. Both sets must be open at the same time on the screen, and the fusion can be initiated simply by dragging and dropping the title bar of the window containing the overlay image (PET for example) over the window of the basic images (CT for example) as shown in Figure 4. The program will automatically generate a set of fused images by color blending the two image sets. This new set is still fully synchronized with the original images, and any changes in contrast or intensity in either of the two sets will be reflected in the fused image set, allowing the user to adjust the best rendering setting of the fused image. The fused image set can also be manipulated like any other image data set, and image processing functions such as MIP rendering or MPR can be applied to the four-dimensional data (Fig 5).
Figure 4.
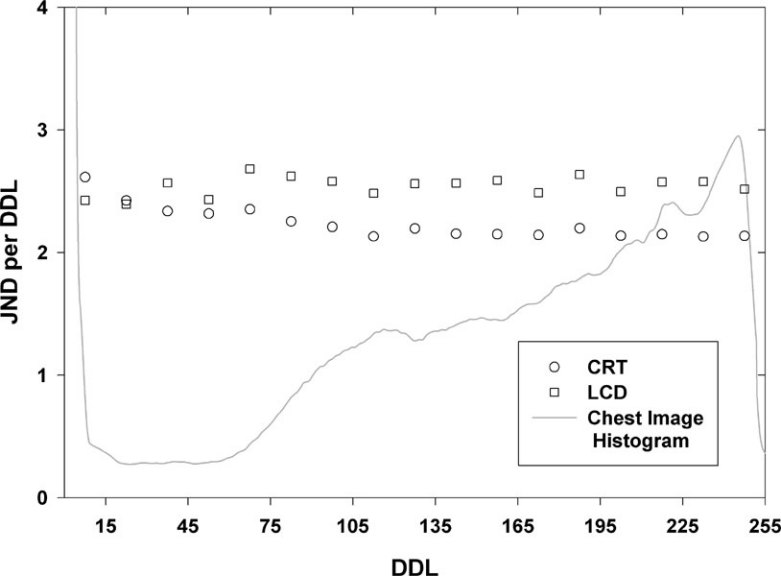
Diagram showing the simple process used for image fusion. (1) The overlay image (PET image) is selected and (2) a color scale is applied. (3) The image fusion is initiated by drag-and-drop of the PET window title bar over the CT window resulting in (4) a fused image set obtained by color blending of the two original sets. The image fusion is instantaneous and the user can continue to navigate through the whole set of fused images and apply any of the processing tools such as intensity and contrast adjustment, slice thickness adjustment, MIP and MPR.
Figure 5.
Example of maximum intensity projection rendering of 4-dimensional data obtained by fusion of PET and CT image sets.
To facilitate and improve the navigation and image manipulation functions in five-dimension exams, we explored innovative solutions using advanced joystick and multidimensional navigation devices that can be used in conjunction with the standard mouse and keyboard functions. We elected to use a special multipurpose video editing jog-wheel device (Fig 6) that is widely used by professional video editors. This allows rapid navigation in multiple data sets and multiple dimensions with a single hand. The addition of this low-cost pointing device increases the ability of the user to navigate rapidly and to switch among different functions in real time.
Figure 6.
Integration of a video-editing jog wheel device allowing the users to rapidly navigate through multidimensional data sets. The upper buttons above the jog wheel are used to select the data dimension of the navigation function that the jog wheel applies to. The user can therefore rapidly change from 3D navigation to slice thickness adjustment to contrast and intensity adjustment without having to use on screen cursors or sliders.
By using this jog-wheel device we were able to develop an interactive graphic user interface that allows navigation in these five dimensions in all rendering modes: the classic 2D viewer, but also in MPR or volume rendering modes (Fig 7). The user is able at any time to move in any of the five dimensions: for example it is possible to modify the dynamic and the fusion parameters while navigating in a 3D MPR view.
Figure 7.
Example of three dimensional multiplanar reformatting (MPR) showing the real-time navigation panel on the right allowing the user to easily navigate and position the selected reformatted slice shown in the main window. The example here shows 4-dimensional data obtained by fusion of PET and CT data.
DISCUSSION AND CONCLUSIONS
Radiology imaging modalities are evolving from conventional sets of 2D tomographic slices to 3D volumetric acquisition extending to a fourth or fifth dimension with temporal and functional data that can be acquired with ultrafast CT and MR scanners with combined PET/CT scanners. To allow radiologists and clinicians to conveniently and efficiently interpret these large exam sets, traditional image viewers have to be re-designed and tailored to a new paradigm of multidimensional image navigation visualization, and manipulation. By combining the performance of new hardware components and the wealth of existing open-source image processing and manipulation tools, it is now possible to develop a new generation of high-performance multidimensional image viewers for off-the-shelf personal computers. These advanced navigation and visualization tools were traditionally only accessible on expensive dedicated 3D workstations, thereby restricting their use to specialist radiologists. With easier access to these multidimensional navigation and visualization tools in standard personal computers, they should soon become complementary tools for routine interpretation of complex diagnostic studies.
One of the key features of the OsiriX software is its flexible user interface allowing users to customize the program by adding and removing tools and functions from the tool bar and menus of the program. This also allows the creation of “customized” versions of the program for specific groups of users. Users who are not computer experts can adopt simpler versions of the software containing only a limited number of essential tools. Specialized versions of the program could be easily customized for specific medical specialties or for specific clinical applications without the need for any additional programming or software development.
The multithreading ability of the software that allows OsiriX to take full advantage of multiprocessor computers ensures a scalability of performance that can be expected from future generation computers. The same software architecture will also allow a migration toward the emerging technology of Grid Computing. With Grid Computing technology25, 26 it is possible to significantly enhance image processing performance by using clusters of computers. The grid technology is already used for large 3D virtual reality software applications to greatly accelerate the rendering time. In the computer animation movie industry, including PIXAR27 and other industry leaders, Grid Computing technology has taken a prominent role. Most of the rendering is performed based on clusters of computers. Grid Computing is also used extensively in large multicentric bioinformatic projects for genome analysis.28, 29 We anticipate that Grid Computing technology could significantly improve the performance of 3D medical imaging rendering techniques. The VTK library already supports Grid Computing rendering through the open-source and cross-platform standard Message Passing Interface (MPI).30 Apple Computer recently announced a new technology, X-Grid,31 that will facilitate the management and configuration of large clusters of computers. With the X-Grid technology it will be easy to connect all computers of a radiology department to share complex processing and rendering tasks during processor idle time. In large academic radiology departments it is common to have a relatively large number of computers that are only partially used and remain idle for significant amounts of time. The Grid Computing technology makes it possible to take advantage of idle computer time for performing computational intensive tasks needed for 3D rendering applications across the network. By adopting the X-Grid technology, OsiriX, which is already multithread compliant, could benefit from a significant improvement in performance if used in a network of multiple computers.
OsiriX is distributed freely as open-source software under the GNU licensing scheme at the following Web site: http://homepage.mac.com/rossetantoine/osirix.
References
- 1.Voge WV, Oyen WJ, Barentsz JO, et al. PET/CT: panacea, redundancy, or something in between? J Nucl Med. 2004;45(Suppl 1):15S–24S. [PubMed] [Google Scholar]
- 2.Flohr T, Ohnesorge B, Bruder H, et al. Image reconstruction and performance evaluation for ECG-gated spiral scanning with a 16-slice CT system. Med Phys. 2003;30:2650–2662. doi: 10.1118/1.1593637. [DOI] [PubMed] [Google Scholar]
- 3.Salgado R, Mulkens T, Bellinck P, et al. Volume rendering in clinical practice, a pictorial review. JBR-BTR. 2003;86(4):215–220. [PubMed] [Google Scholar]
- 4.Kirchgeorg MA, Prokop M. Increasing spiral CT benefits with postprocessing applications. Review. Eur J Radiol. 1998;28:39–54. doi: 10.1016/S0720-048X(98)00011-4. [DOI] [PubMed] [Google Scholar]
- 5.Ligier Y, Funk M, Ratib O, et al. The OSIRIS user interface for manipulating medical images. In: Springer-Verlag, et al., editors. Picture archiving and communication system (PACS) in medicine. Evian: NATO ASI Series. Berlin, Heidelberg: Springer-Verlag; 1991. pp. 395–399. [Google Scholar]
- 6.Ratib O, Ligier Y, Mascarini C, et al. Multimedia image and data navigation workstation. RadioGraphics. 1997;17:515–521. doi: 10.1148/radiographics.17.2.9084087. [DOI] [PubMed] [Google Scholar]
- 7.The Visualization Toolkit (VTK): http://public.kitware.com/VTK/, Accessed February 20, 2004
- 8.The Insight Segmentation and Registration Toolkit (ITK): http://itk.org/, Accessed February 20, 2004
- 9.Ackerman MJ, Yoo TS. The Visible Human Data Sets (VHD) and Insight Toolkit (ITK): Experiments in Open Source Software. Proc AMIA Symp. 2003;.:773. [PMC free article] [PubMed] [Google Scholar]
- 10.GNUstep Framework: http://www.gnustep.org/, Accessed February 20, 2004
- 11.Cocoa Framework: http://developer.apple.com/, Accessed February 20, 2004
- 12.Objective-C language; http://theory.uwinnipeg.ca/gnu/libobjects/objective-c_toc.html, Accessed February 20, 2004
- 13.GNU GCC Compiler. http://gcc.gnu.org, Accessed February 20, 2004
- 14.OpenGL: http://opengl.org, Accessed February 20, 2004
- 15.NVIDIA, Inc., Company: http://www.nvidia.com, Accessed February 20, 2004
- 16.ATI, Inc., Company; http://www.ati.com, Accessed February 20, 2004
- 17.Papyrus Toolkit, Digital Imaging Unit, Geneva University Hospital: http://www.expasy.ch/UIN/html1/projects/papyrus/papyrus.html, Accessed January 10, 2004
- 18.ANSI C: American National Standard for Information. http://www.lysator.liu.se/c/rat/title.html, Accessed January 10, 2004
- 19.DICOM Offis Toolkit: http://dicom.offis.de, Accessed January 10, 2004
- 20.Altivec / PowerPC: http://www.ibm.com/powerpc, Accessed January 10, 2004
- 21.Quicktime. Apple Computer: http://www.quicktime.com, Accessed February 20, 2004
- 22.Wiggins RH, Davidson C, Harnsberger R. Image File Formats: Past, Present, and Future. Radiographics. 2001;21:789–798. doi: 10.1148/radiographics.21.3.g01ma25789. [DOI] [PubMed] [Google Scholar]
- 23.HyperTransport: http://www.hypertransport.org/, Accessed February 20, 2004
- 24.Saito K, Saito M, Komatu S, et al. Real-time four-dimensional imaging of the heart with multi-detector row CT. Radiographics. 2003;23:E8–8. [PubMed] [Google Scholar]
- 25.Grid Computing: http://www.gridcomputing.com/, Accessed February 20, 2004
- 26.Avery P. Data grids: a new computational infrastructure for data-intensive science. Philos Transact Ser A Math Phys Eng Sci. 2002;15;360:1191–1209. doi: 10.1098/rsta.2002.0988. [DOI] [PubMed] [Google Scholar]
- 27.Pixar Company: http://www.pixar.com, Accessed February 20, 2004
- 28.Rowe A, Kalaitzopoulos D, Osmond M, et al. The discovery net system for high throughput bioinformatics. Bioinformatics. 2003;19(Suppl 1):1225–1231. doi: 10.1093/bioinformatics/btg1031. [DOI] [PubMed] [Google Scholar]
- 29.Cummings L, Riley L, Black L, et al. Genomic BLAST: custom-defined virtual databases for complete and unfinished genomes. FEMS Microbiol Lett. 2002;216:133–138. doi: 10.1016/S0378-1097(02)00955-2. [DOI] [PubMed] [Google Scholar]
- 30.Message Passing Interface: http://www.lam-mpi.org/, Accessed February 20, 2004
- 31.X-Grid: http://www.apple.com/acg/xgrid, Accessed February 20, 2004


