Abstract
Malaria caused by Plasmodium falciparum results in approximately 1 million annual deaths worldwide, with young children and pregnant mothers at highest risk. Disease severity might be related to parasite virulence factors, but expression profiling studies of parasites to test this hypothesis have been hindered by extensive sequence variation in putative virulence genes and a preponderance of host RNA in clinical samples. We report here the application of RNA sequencing to clinical isolates of P. falciparum, using not-so-random (NSR) primers to successfully exclude human ribosomal RNA and globin transcripts and enrich for parasite transcripts. Using NSR-seq, we confirmed earlier microarray studies showing upregulation of a distinct subset of genes in parasites infecting pregnant women, including that encoding the well-established pregnancy malaria vaccine candidate var2csa. We also describe a subset of parasite transcripts that distinguished parasites infecting children from those infecting pregnant women and confirmed this observation using quantitative real-time PCR and mass spectrometry proteomic analyses. Based on their putative functional properties, we propose that these proteins could have a role in childhood malaria pathogenesis. Our study provides proof of principle that NSR-seq represents an approach that can be used to study clinical isolates of parasites causing severe malaria syndromes as well other blood-borne pathogens and blood-related diseases.
Introduction
Malaria is endemic in over 100 countries, and 3.3 billion people are at risk of contracting this disease (1). While only a small percentage of clinical cases are fatal, cumulatively they amount to approximately 1 million deaths per year, so that malaria remains one of the leading worldwide causes of mortality. Severe malaria, caused mainly by Plasmodium falciparum, is most frequent in young African children who have not yet developed naturally acquired immunity but is also a concern for nonimmune travelers in malaria-endemic areas (2). In addition, adult women living in malarious areas who are otherwise clinically immune become susceptible to a severe form of the disease during pregnancy (3).
Severe malaria might be caused by parasite forms that are more virulent than those that cause mild disease (4). In the case of pregnancy malaria, severe symptoms result from the accumulation of P. falciparum–infected red blood cells (pRBC) in the placenta, mediated by binding to host receptor chondroitin sulfate A (CSA) (5). The ability of pRBC to bind to CSA correlates with the export of VAR2CSA/PFL0030c protein to the host cell surface (reviewed in ref. 3). VAR2CSA protein is encoded by 1 of the approximately 60 P. falciparum erythrocyte membrane protein 1 (PfEMP1) or var genes present in the parasite genome (6). Other PfEMP1 proteins displayed on the surface of pRBC have been shown to interact with a range of host receptors (such as CD36, ICAM1, VCAM1, PECAM1/CD21, heparan sulfate, etc.), resulting in adhesion to the vascular endothelia (7). Sequestration allows P. falciparum to avoid splenic clearance and has been associated with severe syndromes, such as cerebral malaria (7, 8).
PfEMP1 proteins belong to a larger group of variant surface antigens (vsa) that also includes the rifin, stevor, PfMC-2TM, and surfin gene families. Although the functions of these other vsa have not been well defined, they are exported to the host cell surface, and it has been suggested that, like PfEMP1s, they could have a role in protective immunity (9). Hundreds of non-vsa parasite proteins have been shown or predicted to be exported beyond the parasite’s vacuolar membrane and into the host cell (10, 11). Although little is known about the function of the majority of these exported proteins, some have been implicated in the process of translocation of other parasite proteins, in the creation of parasite-derived structures in the cytoplasm of pRBC, or in the remodeling of the pRBC surface. This extensive remodeling of the host cell has been proposed to be essential for the parasite’s virulence (12).
The identification of genes or proteins related to severe disease could guide intervention strategies, such as vaccines designed to prevent severe malaria and death. For this reason, earlier studies sought to associate the expression of particular parasite genes or proteins with disease severity by comparing the transcriptome and proteome of parasites obtained from different types of malaria episodes. Two recent DNA microarray studies failed to identify parasite genes whose expression was related to disease severity (13, 14). In contrast, microarray and proteomic studies of pregnancy malaria have identified sets of genes and proteins, including VAR2CSA, that are more abundant in parasites isolated from pregnant women than in those from children (15, 16) or that are more abundant in placental parasites than in a laboratory-adapted reference strain (17).
In this study, we characterize the transcriptional profile of in vitro cultures of P. falciparum parasites by next-generation sequencing (NGS). Due to its digital nature, NGS is not subject to saturation or cross-hybridization and thus displays a much broader dynamic range than traditional microarrays (18). NGS does not necessarily require a priori knowledge of the underlying genomic sequence and allows the identification of new genes and the refinement of gene models. Therefore, NGS is ideally suited to investigate gene expression in field isolates of P. falciparum, including that of variable surface antigens that are not fully captured by array designs that are necessarily based on previously determined sequences.
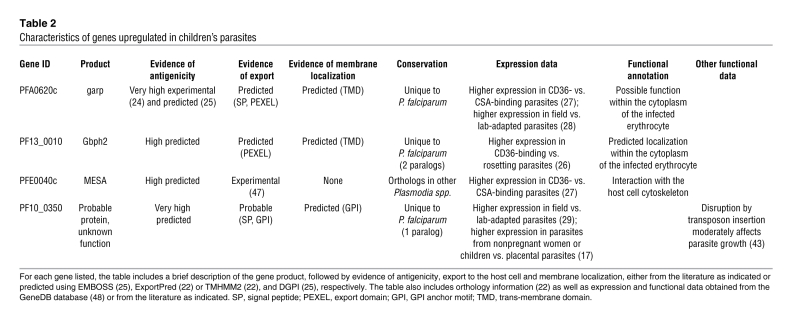
The predominant human-derived RNA species in P. falciparum–infected erythrocytes corresponds to globin mRNA (19) and rRNA, which remains a major contaminant even after polyA+ purification (20). To avoid amplification of these very abundant RNAs, which would decrease the complexity of the resulting libraries, we used the not-so-random sequencing (NSR-seq) method (21) to construct NGS libraries from laboratory-adapted strains of P. falciparum cultured in human erythrocytes. We show that this technique excludes the majority of human globin and rRNA and enables the identification of parasite genes whose transcription levels vary among field isolates. In addition, NSR-seq detects most genes previously discovered by microarray analysis as upregulated in parasites infecting pregnant women, including the pregnancy malaria vaccine candidate var2csa (15), demonstrating that this technique is suitable for the identification of differentially regulated parasite genes. More importantly, NSR-seq identifies 4 genes that are expressed preferentially by children’s parasites (glutamic acid-rich protein [garp/PFA0620c]; mature-parasite-infected erythrocyte surface antigen [MESA/PfEMP2/PFE0040c]; glycophorin-binding protein 2 [Gbph2/PF13_0010]; and probable protein PF10_0350]), which might have roles of particular importance in the pathogenesis of childhood malaria. All 4 of these genes encode proteins that are known or predicted to be exported beyond the parasitophorous vacuole and into the host cell (22). Exported proteins have been implicated in malaria virulence due to their ability to modify the host cell, including alterations in membrane rigidity; in parasite binding to receptors in the vascular endothelia, the placenta, and other erythrocytes; and in evasion of immune system clearance (reviewed in ref. 23). Garp has been experimentally demonstrated to be very antigenic (24), while the other 3 proteins have high predicted antigenicity (25). Three of the genes upregulated in children’s parasites (garp, Gbph2, and probable protein PF10_0350) are unique to P. falciparum (22), which causes the most severe form of human malaria, suggesting that these proteins could have a role in virulence that is absent in other parasite species. Finally, the expression of these 4 genes has been reported to vary in parasites selected for their ability to bind particular host receptors (26, 27) and in field parasites when compared with laboratory strains (28, 29). Future experiments will attempt to validate these proteins as biomarkers of disease severity and as targets for intervention strategies designed to reduce the severity of childhood malaria. The results described herein lay the foundation for NSR-seq analysis of fresh field isolates of P. falciparum in order to identify other genes whose expression might associate with malaria severity.
Results
NSR-seq of in vitro–cultured P. falciparum reference strain 3D7.
In anticipation of studies with clinical P. falciparum isolates, we modified the recently described NSR-seq protocol (21) and profiled the transcriptome of P. falciparum parasites grown in vitro in human erythrocytes. To deplete highly abundant but uninformative host transcripts, we excluded random hexamer sequences with perfect matches to human 18S and 28S rRNA genes (2,781 hexamers) as well as those that matched human α and β globin mRNA (an additional 384 hexamers). The remaining 931 hexamers (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI43457DS1) were synthesized in the forward and reverse directions 3′ of different universal tail sequences to preserve the strand specificity of the sequence tags generated during library construction.
This NSR oligonucleotide set was used as described (21) to generate double-stranded cDNA from polyA+ purified RNA obtained from an unsynchronized in vitro culture of P. falciparum reference strain 3D7 (Supplemental Table 2). The resulting NSR-seq library was submitted to ultra-high-throughput sequencing using a single Illumina (Solexa) GA2 flow cell lane and generated 36-mer sequence tags or “reads” that were aligned simultaneously to P. falciparum and Homo sapiens sequences using the BowTie algorithm and custom settings that allow, on average, 2 mismatches per read (30). The bioinformatic flowchart used for read assignment is diagrammed in Supplemental Figure 1: after trimming 4 nucleotides off the 5′ end (consisting of 2 barcode nucleotides, a random nucleotide, and the first position of the NSR hexamer, which has a higher error rate than the rest of the read), reads that matched an index containing rRNA and globin sequences were removed from further analyses or prefiltered. The remaining reads were first aligned to the parasite and host genomes, and those that did not yield matches were aligned to theoretical transcriptomes of both species in order to capture reads that span splicing sites (see Methods for a detailed explanation). Reads that were not captured in any of these steps were classified as unaligned.
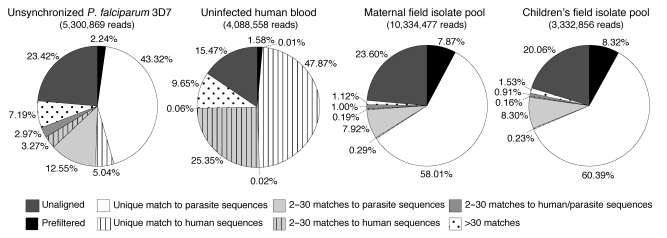
The P. falciparum reference strain 3D7 sample yielded 5.3 million reads, the vast majority of which (97.8%) did not match rRNA or globin genes (Figure 1 and Supplemental Table 3). Over 2.5 million reads (48%) could be assigned unambiguously to a unique location in the parasite or the human genomes and transcriptomes. About 26% of the reads matched multiple parasite and/or human sequences; those that produced 2–30 matches (18.8%) were assigned proportionally to all the corresponding genomic coordinates. Data analysis was performed either exclusively with uniquely aligned reads (“unique reads”) or with reads with 1–30 matches (referred to as “assigned reads”). Assigned reads had an average of 1 mismatch per read: 38.5% of assigned reads aligned to genome sequences with no mismatches, and an additional 27.4% aligned with 1 mismatch (Supplemental Table 3). Finally, reads with more than 30 alignments (7.2%) as well as reads that did not match either parasite or host sequences (23.4%) were excluded from further analyses. The majority of these unaligned reads corresponded to low quality sequences with ambiguous nucleotide calls, while approximately 37% of them matched the sequence of oligonucleotides used during library construction and approximately 6% matched non-3D7 P. falciparum sequences (Supplemental Table 4). Less than 0.01% of the unaligned reads matched possible contaminants such as E. coli, Mycoplasma spp., and homopolymeric sequences.
Figure 1. Read allocation for the 4 samples analyzed by NSR-seq in this study.
The numbers below the sample names correspond to the total number of reads obtained for each sample. The pie charts indicate the percentage of total reads that were unaligned (dark gray) and prefiltered (black) as well as those that had a unique match to parasite (white) or human sequences (white with vertical hatching), 2–30 matches to parasite (light gray) or human sequences (light gray with vertical hatching), 2–30 matches to human and parasite sequences (medium gray), or more than 30 matches (dotted). For reads with 30 or fewer matches, the lack of shading denotes reads that match parasite sequences exclusively, vertical hatching indicates reads that match human sequences exclusively, and stippling indicates reads that match both parasite and human sequences.
Overall, the data correlate extremely well with predicted gene models for P. falciparum. Over 99% of the reads assigned to the parasite mapped to annotated genes, and only 0.65% mapped to intergenic regions (Supplemental Table 5). The majority of these latter reads extend into the 3′ UTRs of predicted genes and are in agreement with EST evidence, while a few are indicative of genes that are not in the current genome annotation or support modification of gene models (Supplemental Figure 2, D–F). The remaining intergenic reads corresponded to singletons and were most likely not indicative of novel genes but resulted from positional biases (see Discussion). The vast majority (96.6%) of reads that align to the parasite genome corresponded to the coding strand of predicted genes, and less than 3.4% mapped to the noncoding strand, confirming that the NSR-seq approach maintains strand-specificity information (Supplemental Table 5). Other important characteristics of this data set, including characteristic gene coverage and detection levels, distribution of reads between coding and noncoding regions, strand specificity, and examples of reads that fall outside of coding regions, are included in Supplemental Figure 2.
NSR-seq analysis of uninfected human blood.
To estimate the percentage of human sequence reads from the infected blood samples that may be assigned erroneously to parasite genes, we generated and sequenced a NSR-seq transcript library from a sample of polyA+ RNA purified from whole human blood of 5 healthy adult volunteers (Supplemental Table 2). We obtained over 4 million reads, 48% of which could be assigned unambiguously to a single genomic location, while 35% mapped to multiple coordinates and 15% yielded no alignments (Figure 1 and Supplemental Table 3). Almost 3 million reads yielded between 1 and 30 matches and were therefore assigned to coordinates in the parasite and/or human genomes.
Almost 99.9% of the assigned reads matched human sequences exclusively; 0.08% aligned to both human and parasite sequences, and 0.04% aligned exclusively to parasite sequences (Supplemental Table 6). This number is very similar to our estimation of error rate, which revealed that 0.02% of human reads would be assigned to parasite sequences. In comparison, of the 3.5 million assigned reads from the P. falciparum–infected erythrocyte sample, 12.4% matched human sequences exclusively, 83.2% matched parasite sequences exclusively, and 4.4% matched both parasite and human sequences. Of the predicted human genes, over 48% received at least 1 read, 32% received at least 10 reads, and approximately 10% received at least 100 reads, whereas less than 7% of the predicted genes in the parasite’s genome received at least 1 read, and the number of genes that received 10 or more reads was negligible (Supplemental Table 5). Taken together, these analyses indicate that the erroneous assignment of human reads to parasite sequences and vice versa occurs at a very low frequency.
Comparison of NSR-seq with other RNA sequencing approaches.
To evaluate the ability of the NSR-seq method to reduce the proportion of human globin and 18S/28S rRNA transcripts from pRBC samples, we compared our data with 2 recently published data sets that also reported RNA sequencing (RNA-seq) of P. falciparum parasites cultured in vitro (20, 31). The first study used random nonamers to generate a transcript library from polyA+ RNA (20), while the second study compared different depletion strategies aimed at diminishing the proportion of abundant transcripts from total RNA (31). For each of these studies, we downloaded the reads obtained from the experimental condition that most closely resembled ours (respectively, the P. falciparum Not-Marched Control [NMC] sample, ref. 20, and the combined Affinity Depletion/Exonuclease [ADE] sample, ref. 31), and we analyzed them in our bioinformatic pipeline.
The NSR approach generated the highest proportion of aligned reads (74.34%), compared with 52.99% aligned reads for the ADE sample and 17.76% aligned reads for the NMC sample (Supplemental Table 7). Our analysis confirmed that, as originally reported, over 65% of the reads in the NMC sample could not be aligned to either parasite or human sequences, while the ADE approach yielded only 10.24% unaligned reads, compared with 23.42% unaligned reads in our NSR approach. Next, we counted all reads that matched human or parasite 18S/28S rRNA sequences, independently of whether they were unambiguously assigned to particular chromosomal coordinates or whether they mapped exclusively to either species, since the human and parasite genomes contain multiple copies of the basic rRNA unit, and the sequences of the rRNA genes are highly similar across both species. Although the NMC library was generated from polyA+-purified RNA, over 15% of the reads corresponded to 18S or 28S rRNA genes, whereas only 2.25% of the reads in the NSR library matched 18S or 28S rRNA genes. In spite of the combined affinity/exonuclease depletion approach, the ADE sample contained the largest proportion of reads that matched 18S or 28S rRNA genes (36.75%). This value is higher than the one reported by Otto et al. (31), who only counted reads that aligned to parasite rRNA. Finally, we compared the number of reads that matched human globin in all 3 data sets. Whereas 0.25% of the reads in the NMC sample mapped to globin mRNA, only 0.01% and 0.02% of the reads in the NSR sample and the ADE sample, respectively, corresponded to globin (Supplemental Table 7). These analyses suggest that the NSR-seq protocol efficiently minimizes the number of reads that correspond to globin RNA and rRNA, resulting in the highest proportion of aligned reads.
Next, we investigated whether the use of a reduced set of hexamer sequences influenced our ability to amplify parasite-encoded mRNAs by calculating their ability to bind to all predicted protein-coding P. falciparum genes. Over 98% of the protein-coding genes predicted in the parasite’s genome contained more than 20 hexamer-binding sites (HBSs) per 100 bp (Supplemental Figure 3B and Supplemental Table 8), suggesting that the NSR-seq oligonucleotide set is sufficiently complex to amplify the vast majority of P. falciparum genes. We also considered excluding an additional 521 hexamers that have perfect matches to P. falciparum 18S and 28S. However, we determined that the resulting set of 410 hexamers available for RNA amplification is not sufficiently complex to yield adequate coverage, since, for this further reduced set, none of the parasite genes contained more than 20 binding sites per 100 bp, and over 99% of the genes have fewer than 10 HBSs per 100 bp (Supplemental Figure 3B and Supplemental Table 8).
We also compared relative gene abundance in the NSR, NMC, and ADE data sets, as measured by the normalized number of reads received by each protein-coding gene (Supplemental Figure 3, C and D). The Spearman correlation coefficients between the NSR and NMC data sets and the NSR and ADE data sets, respectively, are 0.63 and 0.66, while the correlation between the NMC and ADE data sets is 0.73. The higher correlation observed between the NMC and ADE data sets stems from the fact that they were obtained from parasites collected 44 and 48 hours after invasion of erythrocytes, respectively (ref. 20 and Llinás et al., personal communication), while the NSR sample corresponds to an unsynchronized culture with a higher proportion of early intraerythrocytic stages. To further illustrate this point, we analyzed the transcriptional profiles of all 3 samples using 2 in-house algorithms (described in Supplemental Methods) that estimated the stage composition of a mixed parasite culture. The first one (Supplemental Figure 3D) estimated the cell cycle progression of parasites measured in hours after invasion of the erythrocyte and is based on published transcriptional expression profiles of in vitro–cultured erythrocytic stages of P. falciparum (32). The second one (Supplemental Figure 3E) estimated the contribution of each cell cycle stage to the observed gene expression pattern and is based on a different microarray study that includes nonerythrocytic stages (33). These analyses confirmed that the NMC and ADE samples are enriched for late intraerythrocytic cycle (IEC) stages, while the NSR sample corresponds to a mixed population containing a higher proportion of early IEC stages.
Finally, we estimated gene coverage by comparing the number of bases in exons that received reads in all 3 data sets (Supplemental Table 9). While the NSR data set had the largest number of aligned reads, the ADE approach resulted in more exonic bases receiving reads (e.g., 52.1% of bases in exons received at least 1 read for ADE, vs. 13.5% for NSR and 12.6% for NMC; 16.7% received at least 5 reads for ADE, vs. 5% for NSR and 2% for NMC). This is reflected in the less uniform coverage yielded by NSR-seq, although this approach maintains strand specificity, in contrast to the 2 other standard RNA-seq approaches that have been applied to P. falciparum (Supplemental Figure 4A). Importantly, the NSR-seq read alignment pattern is highly reproducible and independent of expression level (ref. 21 and Supplemental Figure 4B), thus allowing comparison of expression of the same transcript between different samples.
These results demonstrate that (a) the NSR-seq protocol greatly reduces the proportion of the total available sequence space consumed by uninformative human rRNA and globin mRNA reads, thereby maximizing the proportion of reads that correspond to protein-coding parasite genes of interest; (b) NSR-seq easily differentiates parasite and human transcripts; and (c) the use of NSR oligonucleotides for construction of the transcript library does not limit our ability measure the expression of the vast majority of parasite genes, although coverage along individual genes is not as uniform as with standard RNA-seq protocols. Taken together, these observations suggest that NSR-seq constitutes an effective tool for comparing the expression of P. falciparum genes between samples in the presence of human transcripts.
NSR-seq of maternal and children’s parasites recently adapted to culture.
Based on these results, we next applied NSR-seq to identify differentially regulated genes among clinical isolates. We chose to compare the transcriptomes of parasites that had been isolated from the peripheral blood of pregnant women and children and recently adapted to short-term in vitro culture. Previous microarray analyses of comparable samples identified a small subset of genes upregulated in maternal parasites (15) and provided a basis for comparison with NSR-seq analyses. In order to obtain sufficient quantities of polyA+ RNA for library construction, we pooled RNA purified from in vitro culture-adapted field isolates recently collected from 6 pregnant women and 8 children, respectively (Supplemental Table 2). Thin blood smears revealed that 56% of the pRBC in the pooled maternal parasite cultures and 59% of those in the pooled children’s parasite cultures contained ring-stage parasites; that 41% and 37% of the pRBC, respectively, contained trophozoites; and that only 3% of the pRBC in either pool contained schizonts. In addition to this microscopic evaluation, comparison of the gene expression profiles to available microarray data (32, 33) from in vitro–cultured P. falciparum strains using the algorithms described above suggested that these 2 samples had almost identical contributions from different IEC stages of the parasite’s life cycle (Supplemental Figure 5, A and B).
The maternal parasite sample yielded 10.3 million reads, while the sample corresponding to children’s parasites yielded 3.3 million reads (Figure 1 and Supplemental Table 3). Over 86% of these reads had no or only 1 mismatch to human or parasite sequences. Between 58%–60% of the reads aligned unambiguously to 1 location, 10%–11% of the reads aligned to multiple locations, and 20%–22% of the reads did not align to any location. The lower number of reads assigned to multiple locations as compared with that of the unsynchronized P. falciparum reference strain 3D7 sample correlates with the very low amount of human material in the field isolate samples (less than 1%; Supplemental Table 6). Approximately 8% of the reads aligned to 18S or 28S rRNA, and less than 0.01% of the reads aligned to globin.
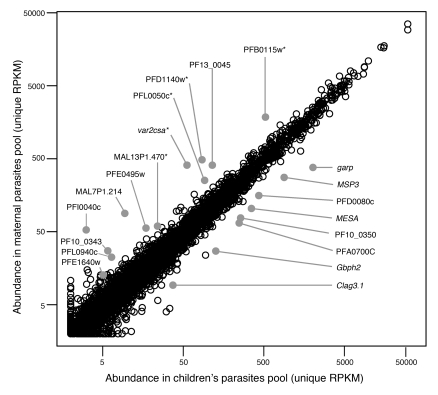
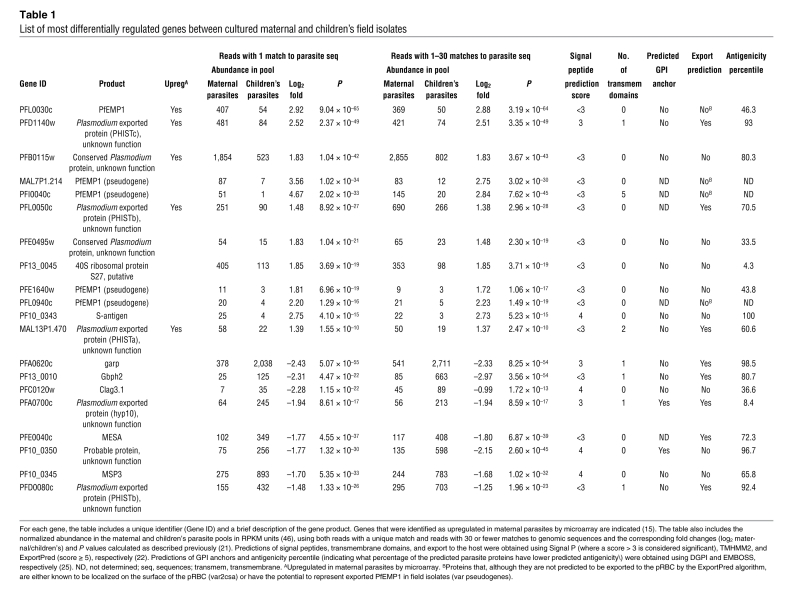
Based on the number of aligned parasite reads, most genes were not differentially regulated between maternal and children’s isolates (Figure 2, Supplemental Figure 5C, and Supplemental Table 10). A small subset of genes departs from the line of equivalence, indicating different levels of transcript in the sample pools. To identify the most differentially regulated genes, we selected 20 genes with P values of less than 1 × 10–9 and at least 2.5-fold difference in expression (i.e., log fold > 1.32), including 12 genes that are upregulated in the maternal samples pool and 8 genes that are upregulated in the children’s samples pool (Figure 2 and Table 1). The 12 genes upregulated in maternal parasites include var2csa, which encodes the best characterized pregnancy malaria vsa (34, 35) and 4 other members of the var gene family (MAL7P1.214, PFI0040c, PFE1640w, and PFL0940c) that are pseudogenes in reference strain 3D7. Other upregulated genes in the maternal parasites include hypothetical protein PFB0115w and 3 members of the variant gene family of exported pHIST proteins (PFD1140w, PFL0050c, and MAL13P1.470). Together with var2csa, these 4 genes were previously determined to be upregulated in parasites freshly obtained from the peripheral blood of pregnant women versus children using microarray analyses (15). The remaining 3 genes encode S-antigen (PF10_0343), whose sequence is known to vary widely between isolates (36), putative 40S ribosomal protein S27 (PF13_0045), and a protein of unknown function (PFE0495w).
Figure 2. Comparison of gene expression for P. falciparum isolates collected from 8 infected children and 6 pregnant women, shortly after adaptation to in vitro culture.
A scatterplot comparing the abundance of all parasite protein-coding genes by NSR-seq in maternal and children’s parasites is shown. The number of unique reads that aligned to the genomic sequence of each gene was normalized by the total number of reads and the length of each gene, as described previously (46). Genes selected as differentially regulated in the maternal or children’s parasites and listed in Table 1 are indicated by filled gray circles. Asterisks denote genes whose increased expression has been previously associated with pregnancy malaria parasites by microarray assays (15). RPKM, reads per kilobase of exon model per million mapped reads.
Table 1 .
List of most differentially regulated genes between cultured maternal and children’s field isolates
The 8 genes that were most significantly upregulated in children’s parasites include garp; Gbph2; MESA; PF10_0350, which encodes a probable protein of unknown function; cytoadherence-linked asexual gene 3.1 (Clag3.1/PFC0120w); merozoite surface protein 3 (MSP3/PF10_0345); and PFA0700c and PFD0080c, which encode 2 exported proteins of unknown function that belong to the hyp10 and pHISTb protein familes, respectively. Reanalysis of our published microarray study that compared parasite gene expression from peripheral blood samples obtained from malaria-infected pregnant women and children (15) showed that only garp and probable protein PF10_0350 are significantly upregulated in children’s parasites (Supplemental Table 13), suggesting that NSR-seq constitutes a more sensitive approach for the identification of differentially regulated genes in these samples.
Confirmation of NSR-seq gene expression data on fresh isolates.
Next, we used quantitative real-time PCR (qRT-PCR) on fresh parasite samples to confirm the 6 most highly upregulated genes of children’s parasites identified by NSR-seq. These samples were from 6 pregnant women and 12 children and did not include any of the samples used to prepare pools for NSR-seq studies (Supplemental Table 2). We measured var2csa as a positive control known to be upregulated in field isolates of maternal parasites (15) and seryl-tRNA synthetase (PF07_0073) as a housekeeping gene to normalize results (34). The list of genes and controls selected for qRT-PCR validation, as well as the sequence of the oligonucleotides used for their amplification, appears in Supplemental Table 11.
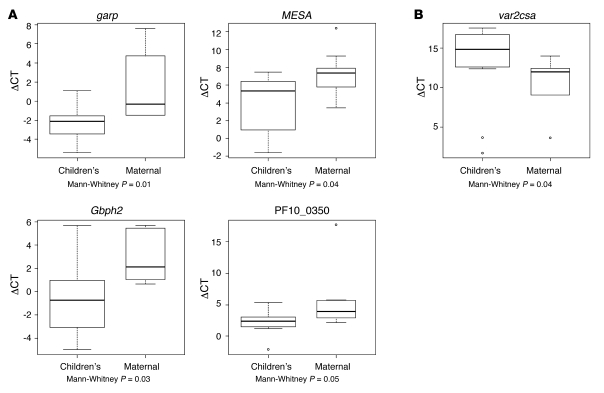
Four of the genes that associate with children’s parasites in the NSR-seq data (garp, MESA, Gbph2, and probable protein PF10_0350) showed significant upregulation (Mann-Whitney, P ≤ 0.05) in children’s parasites by qRT-PCR (Figure 3A). Two other genes (Clag3.1 and PFA0070c; Supplemental Figure 6) were not upregulated in children’s field isolates compared with maternal field isolates (see Discussion). As expected, var2csa is expressed at significantly higher levels by maternal field isolates (Mann-Whitney, P = 0.04; Figure 3B).
Figure 3. qRT-PCR analysis of differential expression of genes upregulated in maternal and children’s pools by NSR-seq validated in fresh field isolates.
(A) Genes upregulated in children’s isolates (garp, MESA, Gbph2, and probable protein PF10_0350). (B) Example of a gene upregulated in maternal parasites (var2csa). A ΔCt value was obtained for each sample by subtracting the Ct value for each gene from that of housekeeping gene seryl-tRNA synthetase included in the same plate. The box plots display the median ΔCt (horizontal bars) and interquartile range (top and bottom of the box) for 6 maternal and 12 children’s samples. The small dots indicate outlier values. The Mann-Whitney test was applied to estimate whether the difference between groups is statistically significant, and the resulting P values are displayed under each plot.
We also examined the expression of the proteins encoded by garp, MESA, Gbph2, and probable protein PF10_0350 in parasites isolated from malaria-infected children and pregnant women by mass spectrometry (Supplemental Table 12). Peptides corresponding to garp were detected by LC-MS/MS in a significantly higher proportion of parasite samples collected from children as compared with those collected from pregnant women (respectively, 14 out of 34 samples vs. 1 out of 18 samples; c2, P = 0.007). Similarly, peptides corresponding to MESA were detected in 85% (29 out of 34 samples) collected from children but only in 50% (9 out of 18 samples) collected from pregnant women (c2, P = 0.006). No peptides corresponding to Gbph2 or probable protein PF10_0350 were detected in parasites isolated from either children or pregnant women, and therefore differential expression could not be assessed. The mass spectrometry results demonstrate the predictive power of NSR-seq in the identification of genes whose expression associates with particular parasite forms.
Discussion
Malaria disease might be influenced by the ability of parasites to express specific subsets of proteins, as has been demonstrated for the parasites causing pregnancy malaria (3). Therefore, the identification of genes or proteins whose increased expression is indicative of disease severity could guide intervention strategies aimed at lessening the symptoms of malaria in nonimmune and partially immune individuals. Microarray-based transcriptional analyses have been productive in the characterization of proteins that associate with pregnancy malaria parasites (15) but to date have failed to identify parasite genes related to malaria disease in children (13, 14).
In this study, we successfully adapted the recently developed NSR-seq method (21) to profile P. falciparum gene expression in human blood samples. To evaluate the ability of NSR-seq to identify differentially regulated genes, we compared the transcriptomes of parasites collected from pregnant women versus children, soon after being adapted to in vitro culture. Comparable isolates have been previously analyzed by microarray assays by our group, yielding a subset of genes whose increased expression associates with maternal parasites (15). The robustness of the NSR-seq technique is demonstrated by its ability to identify the majority of these maternal parasite genes. The list of the genes that showed the highest upregulation in maternal parasites includes var2csa (7.6 fold, P = 9.04 × 10–65), which encodes the best-characterized pregnancy malaria vsa (17, 34, 35), as well as 4 other PfEMP1 family members that are pseudogenes in the reference strain 3D7. Further analysis of the sequence of PFI0040c and MAL7P1.214 suggests that reads assigned to these pseudogenes likely correspond to exon 2 of var2csa. In contrast, the same analysis confirmed the increased expression of PFL0940c and PFE1640w in clinical isolates from pregnant women, suggesting that they are bona fide genes. Indeed, PFE1640w, which has been previously implicated in pregnancy malaria, is not a pseudogene in other P. falciparum strains (37), while PFL0940c expression has been detected in several P. falciparum strains (22). Four other genes previously identified by microarray studies as enriched in parasites isolated from the peripheral blood of pregnant women (15) were amongst the most upregulated genes in maternal parasites by NSR-seq (Figure 2 and Table 1). They correspond to hypothetical protein PFB0115w and 3 members of the variant gene family of exported pHIST proteins (PFD1140w, PFL0050c, and MAL13P1.470). The other 2 genes identified by microarrays as upregulated in maternal parasites (i.e., pHIST protein PFI1785w and conserved membrane protein MAL13P1.320; ref. 15) were not significantly different between the 2 samples by NSR-seq analysis (Supplemental Table 10).
Notably, NSR-seq allowed us to describe for the first time a set of 4 genes that are enriched in children’s parasites (Figure 3A and Table 2). PFA0620c encodes garp, a protein of unknown function that has very high antigenicity (24) and is predicted to be exported to the cytoplasm of the host cell (22). Reanalysis of our existing microarray data set (15) indicates that this gene is expressed at higher levels in fresh parasites isolated from children as compared with those from pregnant women (log2 fold = –1.23, P = 1.84 × 10–8; Supplemental Table 13). Moreover, mass spectrometry analyses revealed that peptides from the corresponding protein were detected in a significantly (P = 0.007) higher proportion of parasites isolated from children versus pregnant women (Supplemental Table 12). Interestingly, orthologs of this gene are absent from all other Plasmodium species (22), suggesting that its role is only required in P. falciparum, which is the only human malaria parasite capable of sequestering and causing severe symptoms. The expression of garp has also been reported to be higher in parasite isolates that bind host receptor CD36 when compared with parasites that bind CSA (27). The majority of parasites isolated from children bind CD36, while placental parasites always bind CSA (5). Finally, garp is expressed at higher levels in field parasites than in P. falciparum reference strains that have been adapted to long-term in vitro culture, suggesting that the function of this protein is not required outside of the human host (28). These data taken together suggest that garp protein carries out a function in the pRBC that is most important in childhood malaria and might be related to the ability of these parasites to bind to specific host receptors. Since garp protein is not predicted to possess a transmembrane domain or a glycosylphosphatidylinositol (GPI) anchor (although it is predicted to have a signal peptide) (22), further experiments are required to address its localization in the host cell as well as to elucidate the mechanism by which it might be involved in differential adhesion to receptors and pathogenesis.
Table 2 .
Characteristics of genes upregulated in children’s parasites
The second validated gene whose expression is increased in children’s parasites corresponds to MESA. MESA encodes an exported protein that is predicted to have moderately high antigenicity (25) and is known to localize to the internal side of the erythrocyte surface, at which it interacts with the host cell membrane cytoskeleton protein band 4.1 (38, 39). The prevention of this interaction results in the cytoplasmic accumulation of MESA and leads to parasite death (39). The MESA gene was not found to be differentially expressed among children’s and maternal samples in our reanalysis of existing microarray data (Supplemental Table 13). However, MESA peptides were detected using LC-MS/MS at a significantly higher level in parasites collected from children versus pregnant women (Supplemental Table 13). MESA gene abundance was much higher in CD36-binding versus CSA-binding parasites and was inversely related to that of var2csa (27). Although MESA protein levels reflected this difference, the subcellular localization of this protein was not altered in the parasites with different binding profiles. MESA might participate in the pathogenesis of childhood malaria by modifying parasite adhesion via its interaction with components of the host cytoskeleton that lie beneath the surface of the pRBC.
Another gene whose increased expression characterizes children’s parasites is Gbph2 (40). The protein encoded by this gene belongs to the P. falciparum–specific family of glycophorin-binding proteins, together with glycophorin-binding protein 130 precursor (GBP130/PF10_0159) and glycophorin-binding protein homologue (Gbph/PF14_0010) (22). All 3 family members are characterized by the presence of multiple tandem repeats of a 50–amino acid motif in the carboxy-terminal region, which, in GBP130, has been shown to be responsible for the interaction with host glycophorin during invasion of the host cell by merozoites (41, 42). Gbph2 is predicted to be exported beyond the confines of the parasitophorous vacuole (22) and to be among the top 20% of parasite proteins sorted by their predicted antigenicity (25), but its function (as well as that of Gbph) remains uncharacterized. Our previous microarray data (15) did not detect a statistically significant difference in the expression of Gbph2 gene in maternal and children’s parasites (Supplemental Table 13). Peptides corresponding to this protein were not detected in our proteomic studies, so we cannot assess whether Gbph2 protein is more abundant in children’s parasites. Gbph2 expression has been reported to be more than 10-fold higher in parasites selected for CD36 binding when compared with those selected for their ability to undergo rosetting (binding of pRBC to uninfected erythrocytes) (26).
Finally, we also determined that probable protein PF10_0350 is enriched in children’s parasites. This gene encodes a protein of uncharacterized function that is predicted to have a signal peptide, a GPI anchor, and high antigenicity (22, 25). We did not detect peptides corresponding to this protein using mass spectrometry, but reanalysis of our existing microarray data sets (15) indicated that probable protein PF10_0350 expression levels are significantly higher in parasites isolated from children’s versus those isolated from pregnant mothers (log2 fold = –3.15, P = 4.66 × 10–8; Supplemental Table 13). Probable protein PF10_0350 was previously reported to be downregulated in placental parasites when compared with parasites from nonpregnant women and children (17) and has been reported to be expressed at higher levels in field isolates than in laboratory-adapted strains (29), suggesting that its function might be important for binding of pRBC to nonplacental receptors in the human host. As with garp and Gbph2, this protein has no orthologs in other Plasmodium species (although it does have a paralog in P. falciparum, predicted exported protein of unknown function MAL7P1.171) (22). Little else is known about this gene, although it was recently reported that its mutation by transposon-based insertion resulted in a moderate attenuated growth of the corresponding parasite line, suggesting that its function is important for parasite survival (43). Further experiments, including the experimental verification of the predicted export of probable protein PF10_0350 to the host cell and analysis of its subcellular localization, are needed to better understand the potential role of this protein in childhood malaria pathogenesis.
As in all cDNA library-based approaches, a major concern when constructing RNA-seq libraries is the overrepresentation of certain transcripts, which can severely affect the coverage of genes of interest. Our approach to expression profiling is designed to exclude the majority of human globin and rRNA, which constitute the predominant RNA species in the sample, allowing the analysis of parasite gene expression in the presence of human cells. We first validated the technique on reference strain 3D7: more than two-thirds of the 5 million reads obtained could be assigned to the parasite or human genomes and almost 84% matched parasite sequences exclusively. Furthermore, we demonstrate that the assignment of parasite reads to human sequences and vice versa occurs at a very low frequency. In comparison with earlier studies using random oligonucleotides to analyze the gene expression of P. falciparum reference strain 3D7 by RNA-Seq (20, 31), the use of NSR oligonucleotides resulted in the largest proportion of aligned reads and the lowest percentages of reads matching globin and rRNA, while allowing the detection of the vast majority of P. falciparum genes (Supplemental Figure 3 and Supplemental Tables 7 and 8). These results indicate that NSR-seq is an effective tool for profiling parasite gene expression in the presence of human transcripts that should be applicable to the profiling of fresh parasite samples.
In comparison with the recently published ADE approach (31), NSR-seq yields sparser coverage across the length of parasite genes (Supplemental Figure 4A). This is due to the fact that NSR-seq library construction protocol requires the presence of short nucleotide tails upstream of the hexamer sequence for PCR-mediated amplification, which biases the binding of the NSR oligonucleotides and results in uneven coverage (21). While extensive coverage is required for other applications, such as gene annotation, uniform coverage along the length of a gene is not essential for the purpose of comparing expression of parasite genes between samples. Our data demonstrate that while the abundance of reads in each location along the length of the gene was different between samples, the pattern of read distribution along the length of a gene was extremely similar (Supplemental Figure 4B). In addition, by using NSR-seq, we were able to identify genes previously reported to be enriched in maternal samples by microarray analysis as well as a set of genes that characterize children’s parasites. NSR-seq preserves strand directionality and is technically simple, requiring a standard polyA+ purification step, followed by library generation using a subset of NSR oligonucleotides. Importantly, while the ADE approach yielded high-quality results in laboratory-adapted samples (31), the use of Terminator enzyme on RNA obtained from freshly collected field isolates of varying quality would result in the removal of partially degraded transcripts with unprotected 5′ ends from the sample. Future studies might compare our NSR-seq protocol with the affinity depletion/Terminator protocol. Preliminary results (Vignali et al., unpublished observations) suggest that NSR-seq is useful for the identification of differentially regulated genes in parasite samples extracted immediately after collection from malaria-infected patients.
In conclusion, NSR-seq represents a robust approach for whole-genome expression profiling of P. falciparum parasites that overcomes the limitations of earlier methods. In studies of the 3D7 reference P. falciparum clone and of recently adapted field isolates, NSR-seq yielded highly specific mRNA transcriptional information, as evidenced by overwhelming alignment of sequences to annotated genes in the correct orientation and limited assignment to intergenic, rRNA, and human globin sequences. Our sequencing and analysis pipeline correctly identified genes that are known to be preferentially expressed by pregnancy parasites, including a variant antigen gene, and revealed for genes expressed preferentially by children’s parasites. Notably, the genes that distinguish children’s parasites encode proteins of high antigenicity with putative export sequences that would direct them to the host-cell interface, and their variable transcription profiles in strains that differ in their binding properties suggest they could be involved in adhesion and sequestration. Further experiments will be required to better understand the potential roles of these proteins in malaria pathogenesis and immunity. Proteins that associate with severe disease could be targets for vaccines that prevent severe malaria or death. The new NSR-seq technology will be valuable for understanding whether differences in parasite gene expression may explain, in part, the differences in children’s outcomes, similar to the distinct transcriptome of P. falciparum parasites that cause placental malaria and disease during pregnancy.
Methods
Sample origin.
Parasite samples were donated by Tanzanian women (aged 16–45 years) and/or their children presenting at the Muheza Designated District Hospital or the Morogoro Regional Hospital in Tanzania. Sample donors were among those enrolled in ongoing cohort studies known locally as the Mother-Offspring Malaria Studies (MOMS). Women provided signed informed consent for themselves and/or their offspring before joining the study. Ethical approval of human care and use of samples for these experiments was obtained from the Western Institutional Review Board in Olympia, Washington, USA, and the National Medical Research Coordinating Committee in Dar es Salaam, Tanzania.
Fresh human blood used in these studies was provided by employee volunteers from Seattle BioMed after they provided written informed consent, under a protocol known as “SBRI Blood Donor Program,” approved by the Western Institutional Review Board.
Parasite culture and RNA isolation.
P. falciparum reference strain 3D7 as well as field isolates recently collected from malaria-infected women and children were cultured in vitro in erythrocytes isolated from whole blood as previously described (44). Red blood cells were collected by centrifugation, and the pellet was mixed with 5 volumes of TRIzol (Invitrogen) and stored at –80°C until RNA purification. The same procedure was applied to blood collected from uninfected adult volunteers.
Samples were thawed at room temperature and total RNA was isolated using the RNeasy Micro Kit (QIAGEN) according to the manufacturer’s instructions. Total RNA from uninfected samples as well as samples obtained from the peripheral blood of children and pregnant women infected with P. falciparum were pooled as described in Supplemental Table 2 and then subjected to polyA+ RNA purification using the Poly ATract Kit (Promega). RNA concentration and quality was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc.) and a 2100 Bioanalyzer (Agilent Technologies Inc.), respectively.
Oligonucleotides.
NSR primers were designed to deplete human cytoplasmic 18S and 28S rRNA and human globin mRNA sequences during cDNA library construction. We computationally identified 931 distinct hexamers without perfect complementarity to any of the following sequences: human 18S rRNA (U13369.1, nt 3657–5527), 28S rRNA (U13369.1, nt 7935–12969), α globin mRNA (NM_000558.3, NM_000517.4), and β globin mRNA (NM_000518.4), as obtained from NCBI. Each NSR hexamer was synthesized individually in the antisense and sense orientation with universal 5′ tail sequences to facilitate first-strand (5′-TCCGATCTCTN-[NSR reverse complement]-3′) and second-strand (5′-TCCGATCTGAN-[NSR]-3′) synthesis, respectively. Desalted NSR primers were adjusted to 100 μM with water before pooling. Terminal sites for sequencing by synthesis were added during PCR using distinct forward (5′- AATGATACGGCGACCACCGACACTCTTTCCCTACACGACGCTCTTCCGATCTCT-3′) and reverse (5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTGA-3′) primers. All oligonucleotides were synthesized by Operon Biotechnologies Inc.
Library generation and sequencing.
Transcriptome libraries were constructed with NSR primers, as previously described, with minor modifications (21). First-strand synthesis was performed on 150 ng polyA+-purified mRNA using 10 μM NSR primers and 200 units of Superscript III Reverse Transcriptase (Invitrogen) at 45°C for 90 minutes. The RNA template was subsequently degraded by RNase H (Invitrogen), and cDNA was purified using the QIAquick PCR Purification Kit (QIAGEN). All other steps, including second-strand synthesis and PCR amplification, were carried out as described previously. Directional, single-end sequencing of PCR products was performed using the Illumina GAII platform to generate 36-nt reads.
Bioinformatic analyses.
Reads were assigned to human and parasite sequences using the R software package “DuffyRNASeq,” which is build around the Bowtie algorithm (30) and can be downloaded from http://duffyrnaseq.sourceforge.net/. The method is described in detail in the Supplemental Methods and diagrammed in Supplemental Figure 1. Algorithms used to estimate cell cycle stage composition were based on published transcriptional expression profiles of stages of P. falciparum (32, 33) and are described in the Supplemental Methods.
Analysis of existing microarray data sets.
Intensity values for all probes were extracted separately from the Cy3 and Cy5 channels for slides corresponding to all the mother/child pairs included in reference 15, using the R LIMMA package and applying the normalized exponential background correction option. After RMA quantile normalization, all probes for each gene were compared for all children samples versus all maternal samples using a 2-sample t test to derive mean intensities, fold changes, and P values.
qRT-PCR verification of candidates.
Total RNA extracted from field isolates was amplified with the Ambion MessageAmp II Kit, and qRT-PCR was performed as described previously (15). The oligonucleotides used for amplification are listed in Supplemental Table 11 and were designed using NCBI’s Primer BLAST software or obtained from the literature (34). Relative quantification of RNA expression was calculated by the comparative Ct method of subtracting the Ct of the gene of interest from that of P. falciparum seryl-tRNA synthetase (PF07_0073) as a housekeeping gene control after normalizing the amount of cDNA in each qPCR reaction, as described previously (45). An RNA sample prepared from a laboratory-adapted P. falciparum isolate was included in all plates as a calibrator. For samples in which expression of a particular gene was undetectable, the Ct value was manually set to 40.
Mass spectrometry analyses.
Parasite samples collected from malaria-infected women and children were enriched for membrane fractions and processed by tandem mass spectrometry as previously described (16).
Statistics.
For each protein of interest, a contingency table was generated indicating whether peptides were detected or not in each parasite sample obtained from children or pregnant women. Pearson’s χ2 test was used to determine significance.
Supplementary Material
Acknowledgments
Theonest Mutabingwa led the team of MOMS Project clinicians and nurses who collected the clinical samples used in this study. We thank Fenella Raymond for suggesting the collaboration that led to this work; Christopher Mendez, Kathryn Williamson, and Emily Amos for culturing parasites; and Jason Wendler, Cate Speake, and Andrew Oleinikov for helpful discussions. This work was funded in part by a grant from the Foundation for the NIH through the Grand Challenges in Global Health initiative, the Bill and Melinda Gates Foundation (grant 29202), and the National Institute of Allergy and Infectious Diseases (grant R01AI052059 to P.E. Duffy).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(3):1119–1129. doi:10.1172/JCI43457.
Christopher D. Armour, Matthew C. Biery, and Christopher K. Raymond’s present address is: NuGEN Technologies Inc., Research and Development, San Carlos, California, USA.
John C. Castle’s present address is: Institute for Translational Oncology and Immunology, Mainz, Germany.
Heather Bouzek’s present address is: Department of Microbiology, University of Washington, Seattle, Washington, USA.
Tomas Babak’s present address is: Merck Research Labs, Boston, Massachusetts, USA.
References
- 1. World Health Organization. World Malaria Report 2008. Geneva, Switzerland: World Health Organization: 2008. [Google Scholar]
- 2. Doolan DL, Dobano C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. 2009;22(1):13–36. doi: 10.1128/CMR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith JD, Deitsch KW. Pregnancy-associated malaria and the prospects for syndrome-specific antimalaria vaccines. J Exp Med. 2004;200(9):1093–1097. doi: 10.1084/jem.20041974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKenzie FE, Smith DL, O’Meara WP, Riley EM. Strain theory of malaria: the first 50 years. Adv Parasitol. 2008;66:1–46. doi: 10.1016/S0065-308X(08)00201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. . Science. 1996;272(5267):1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 6. Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. . Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415(6872):673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 8. Mackintosh CL, Beeson JG, Marsh K. Clinical features and pathogenesis of severe malaria. Trends Parasitol. 2004;20(12):597–603. doi: 10.1016/j.pt.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 9. Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. . Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 10. Hiller NL, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306(5703):1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 11. Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306(5703):1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 12. Haase S, de Koning-Ward TF. New insights into protein export in malaria parasites. Cell Microbiol. 2010;12(5):580–587. doi: 10.1111/j.1462-5822.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 13. Daily JP, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. . Nature. 2007;450(7172):1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 14. Lemieux JE, et al. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. . Proc Natl Acad Sci U S A. 2009;106(18):7559–7564. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Francis SE, et al. Six genes are preferentially transcribed by the circulating and sequestered forms of Plasmodium falciparum parasites that infect pregnant women. . Infect Immun. 2007;75(10):4838–4850. doi: 10.1128/IAI.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fried M, Hixson KK, Anderson L, Ogata Y, Mutabingwa TK, Duffy PE. The distinct proteome of placental malaria parasites. Mol Biochem Parasitol. 2007;155(1):57–65. doi: 10.1016/j.molbiopara.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 17. Tuikue Ndam NG, et al. High level of var2csa transcription by Plasmodium falciparum isolated from the placenta. . J Infect Dis. 2005;192(2):331–335. doi: 10.1086/430933. [DOI] [PubMed] [Google Scholar]
- 18. Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5(7):585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 19. Goh SH, et al. The human reticulocyte transcriptome. Physiol Genomics. 2007;30(2):172–178. doi: 10.1152/physiolgenomics.00247.2006. [DOI] [PubMed] [Google Scholar]
- 20. Sorber K, et al. The long march: a sample preparation technique that enhances contig length and coverage by high-throughput short-read sequencing. PLoS One. 2008;3(10):e3495. doi: 10.1371/journal.pone.0003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Armour CD, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6(9):647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- 22. PlasmoDB. Plasmodium Genomics Resource Web site. http://www.plasmodb.org/plasmo/ . Updated July 15, 2010. Accessed September 15, 2010.
- 23. Cooke BM, Mohandas N, Coppel RL. Malaria and the red blood cell membrane. Semin Hematol. 2004;41(2):173–188. doi: 10.1053/j.seminhematol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 24. Triglia T, Stahl HD, Crewther PE, Silva A, Anders RF, Kemp DJ. Structure of a Plasmodium falciparum gene that encodes a glutamic acid-rich protein (GARP). . Mol Biochem Parasitol. 1988;31(2):199–201. doi: 10.1016/0166-6851(88)90170-3. [DOI] [PubMed] [Google Scholar]
- 25. Agüero F, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov. 2008;7(11):900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mok BW, et al. Comparative transcriptomal analysis of isogenic Plasmodium falciparum clones of distinct antigenic and adhesive phenotypes. . Mol Biochem Parasitol. 2007;151(2):184–192. doi: 10.1016/j.molbiopara.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 27. Ralph SA, et al. Transcriptome analysis of antigenic variation in Plasmodium falciparum--var silencing is not dependent on antisense RNA. . Genome Biol. 2005;6(11):R93. doi: 10.1186/gb-2005-6-11-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daily JP, et al. In vivo transcriptome of Plasmodium falciparum reveals overexpression of transcripts that encode surface proteins. . J Infect Dis. 2005;191(7):1196–1203. doi: 10.1086/428289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mackinnon MJ, et al. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. . PLoS Pathog. 2009;5(10):e1000644. doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Otto TD, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. . Mol Microbiol. 2010;76(1):12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. . PLoS Biol. 2003;1(1):E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young JA, et al. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. . Mol Biochem Parasitol. 2005;143(1):67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 34. Salanti A, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. . Mol Microbiol. 2003;49(1):179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 35. Viebig NK, et al. Disruption of var2csa gene impairs placental malaria associated adhesion phenotype. PLoS One. 2007;2(9):e910. doi: 10.1371/journal.pone.0000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saint RB, et al. Changes in repeat number, sequence, and reading frame in S-antigen genes of Plasmodium falciparum. . Mol Cell Biol. 1987;7(8):2968–2973. doi: 10.1128/mcb.7.8.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rowe JA, Kyes SA. The role of Plasmodium falciparum var genes in malaria in pregnancy. . Mol Microbiol. 2004;53(4):1011–1019. doi: 10.1111/j.1365-2958.2004.04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Black CG, Proellocks NI, Kats LM, Cooke BM, Mohandas N, Coppel RL. In vivo studies support the role of trafficking and cytoskeletal-binding motifs in the interaction of MESA with the membrane skeleton of Plasmodium falciparum-infected red blood cells. . Mol Biochem Parasitol. 2008;160(2):143–147. doi: 10.1016/j.molbiopara.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Magowan C, Coppel RL, Lau AO, Moronne MM, Tchernia G, Mohandas N. Role of the Plasmodium falciparum mature-parasite-infected erythrocyte surface antigen (MESA/PfEMP-2) in malarial infection of erythrocytes. . Blood. 1995;86(8):3196–3204. [PubMed] [Google Scholar]
- 40. Rudolph B, Nolte D, Knapp B. Isolation of a third member of the Plasmodium falciparum glycophorin-binding protein gene family. . Mol Biochem Parasitol. 1994;68(1):173–176. doi: 10.1016/0166-6851(94)00170-7. [DOI] [PubMed] [Google Scholar]
- 41. Bianco AE, et al. Putative glycophorin-binding protein is secreted from schizonts of Plasmodium falciparum. . Mol Biochem Parasitol. 1987;23(1):91–102. doi: 10.1016/0166-6851(87)90191-5. [DOI] [PubMed] [Google Scholar]
- 42. Ravetch JV, Kochan J, Perkins M. Isolation of the gene for a glycophorin-binding protein implicated in erythrocyte invasion by a malaria parasite. Science. 1985;227(4694):1593–1597. doi: 10.1126/science.3883491. [DOI] [PubMed] [Google Scholar]
- 43. Balu B, et al. piggyBac is an effective tool for functional analysis of the Plasmodium falciparum genome. . BMC Microbiol. 2009;9:83. doi: 10.1186/1471-2180-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193(4254):673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 45. Pfaffl MC Relative quantification. In: Dorak MT, eds.Real-time PCR . New York, New York, USA: Taylor and Francis; 2006:63–82. [Google Scholar]
- 46. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 47. Coppel RL, Lustigman S, Murray L, Anders RF. MESA is a Plasmodium falciparum phosphoprotein associated with the erythrocyte membrane skeleton. . Mol Biochem Parasitol. 1988;31(3):223–231. doi: 10.1016/0166-6851(88)90152-1. [DOI] [PubMed] [Google Scholar]
- 48. GeneDB. http://www.genedb.org/Homepage/ Pfalciparum. Updated September 1, 2010. Accessed September 15, 2010 . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.