Abstract
Aim
Our aim is to understand the molecular mechanisms of the selective nonsteroidal anti-inflammatory drugs (NSAID), cyclooxygenase-2 (COX-2) inhibitors', higher “priority” to reduce synthesis of the vascular protector, prostacyclin (PGI2), compared to that of nonselective NSAIDs.
Main methods
COX-1 or COX-2 was co-expressed with PGI2 synthase (PGIS) in COS-7 cells. The Km and initial velocity (½t Vmax) of the coupling reaction between COX-1 and COX-2 to PGIS were established. The experiment was further confirmed by a kinetics study using hybrid enzymes linking COX-1 or COX-2 to PGIS. Finally, COX-1 or COX-2 and PGIS were respectively fused to red (RFP) and cyanic (CFP) fluorescence proteins, and co-expressed in cells. The distances between COXs and PGIS were compared by FRET.
Key findings
The Km for converting arachidonic acid (AA) to PGI2 by COX-2 coupled to PGIS is ~2.0 µM; however, it was 3-fold more (~6.0 µM) for COX-1 coupled to PGIS. The Km and ½t Vmax for COX-2 linked to PGIS were ~2.0 µM and 20 s, respectively, which were 2–5 folds faster than that of COX-1 linked to PGIS. The FRET study found that the distance between COX-2-RFP and PGIS–CFP is shorter than that between COX-1-RFP and PGIS–CFP.
Significance
The study provided strong evidence suggesting that the low Km, faster ½t Vmax, and closer distance are the basis for COX-2 dominance over COX-1 (coupled to PGIS) in PGI2 synthesis, and further demonstrated the mechanisms of selective COX-2 inhibitors with higher potential to reduce synthesis of the vascular protector, PGI2.
Keywords: Cyclooxygenase-1 (COX-1), COX-2, COX-2 inhibitor, Arachidonic acid (AA), Prostacyclin or prostaglandin I2 (PGI2), PGI2 synthase (PGIS) and FRET
Introduction
Arachidonic acid (AA) can be metabolized into biologically active prostanoids including prostacyclin (PGI2) and thromboxane A2 (TXA2) through the cyclooxygenase (COX)-1 (COX-1) and -2 (COX-2) pathways (Majerus 1983; Pace-Asciak and Smith 1983; Samuelsson et al. 1978; Smith 1986; Funk 2001; Ruan 2004; Ruan and Dogné 2006; Dogné et al. 2006). The synthesized TXA2 mediates platelet aggregation and vascular constriction, thus promoting myocardial infarction and stroke (Needleman et al. 1986; Granstrom et al. 1982; Patrono et al. 1990). Conversely, PGI2 counters these effects and is considered a major vascular protective mediator. In the cells, the detailed steps for the biosynthesis of these two mediators involve catalyzing AA into an intermediate, prostaglandin G2 (PGG2), and PGH2 by COX-1 or COX-2 and further isomerizing PGH2 to TXA2 or PGI2 by downstream individual TXA2 and PGI2 synthases, respectively (Majerus 1983; Pace-Asciak and Smith 1983; Samuelsson et al. 1978; Smith 1986; Funk 2001; Ruan 2004; Ruan and Dogné 2006; Dogné et al. 2006). PGI2 synthesized through the COX-1 and COX-2 pathways is important for protection against blood clots and vasoconstriction in many organs including the heart, kidneys, and brain. Reduced PGI2 production is associated with the use of nonsteroidal anti-inflammatory drugs (NSAIDs), and has a potential to cause organ ischemia. This particularly occurs during the use of selective COX-2 inhibitors (Vane 2002). The absence of COX-2 in platelets could increase the ratio of TXA2/PGI2 with the use of COX-2 inhibitors. This increased risk for cardiovascular events in clinical trials has been proposed (Martinez-Gonzalez and Badimon 2007). However, the molecular mechanisms behind the COX-2 inhibitors' greater reduction in PGI2 production than that of nonselective NSAIDs, which inhibit both COX-1 and COX-2, have not been settled yet. The majority of explanations have come from clinical trials and speculations. For example, Vane hypothesized a model, in which the specific inhibition of COX-2 activity (using a COX-2 inhibitor) would result in more AA that would be available for conversion into TXA2 (through COX-1 coupling to thromboxane A2 synthase (TXAS)), which promotes heart disease (Vane 2002). This implies that COX-2 has priority over COX-1 in terms of acting upon AA. Finding answers behind this “priority” is a key step toward uncovering the molecular mechanisms of COX-2 inhibition, a risk to heart disease and further validating Vane's model. In this study, we are focusing on the characterization of COX enzyme kinetics and the topological coordination for the coupling between either COX-1 or COX-2 with PGI2 synthase (PGIS) in the biosynthesis of PGI2 in live cells. The study has demonstrated that COX-2's coupling to PGIS, and in turn the conversion of AA into PGI2, has a lower Km value and a faster initial reaction velocity in comparison to that of COX-1. The topological distance of PGIS is closer to COX-2 than that of COX-1 in live cells. These two factors could be used to further speculate that in the presence of COX-1, COX-2, and PGIS, COX-2 could be preferred over COX-1 in coupling to PGIS, and thus dominate PGI2 biosynthesis. This could be the mechanism of specific COX-2 inhibitors having stronger capability to reduce PGI2 production and promote heart disease in clinical trials. Our study supports Vane's model, in which selective COX-2 inhibitors have more potential risks than that of nonselective NSAIDs in regard to promoting heart disease.
Experimental procedures
Materials
COS-7 cell line was purchased from ATCC (Manassas, VA). Medium for culturing the cell lines was purchased from Invitrogen. [14C]-Arachidonic acid (AA) was purchased from Amersham Biosciences. Other reagents were from Sigma.
Molecular cloning
For the wild type enzymes, the cDNAs of human COX-1, COX-2 and PGIS were cloned by a polymerase chain reaction (PCR) approach and then subcloned into a pcDNA3.1 vector with a CMV promoter (Deng et al. 2002, 2003; Ruan et al. 2005). The cloning of the hybrid enzymes, COX-1-10aa-PGIS and COX-2-10aa-PGIS were performed by the approaches described previously (Ruan et al. 2006, 2008a,b). Briefly, the cDNA of human COX-1 or COX-2 was linked to a 10 amino acid transmembrane (TM) linker connected to human PGIS, and then subcloned into pcDNA3.1 vector (Ruan et al. 2006, 2008a,b). For preparation of the enzymes tagged with fluorescent proteins, the cDNA of C-terminus of COX-1 or COX-2 was linked to the N-terminus of RFP, and the C-terminus of PGIS was linked to the N-terminal CFP, as was performed by PCR approach to form the COX-1- or COX-2-RFP and PGIS–CFP hybrid enzymes. The COX-1- or COX-2-RFP was further linked to the PGIS–CFP by PCR and ligation approaches.
Co-expression of the recombinant proteins in COS-7 Cells
The recombinant synthases were expressed in COS-7 cells as described (Deng et al. 2003; Ruan et al. 2005). Briefly, the cells were cultured in a 100-mm cell culture dish with high glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum at 37 °C in a humidified 5% CO2 incubator. The cells were then transfected with a purified cDNA of the recombinant protein by the Lipofectamine 2000 method (Ruan et al. 2006) following the manufacturer's instructions (Invitrogen). Approximately 48 h after transfection, the cells were used for fluorescence microscopy, enzyme assays, and Western blot analysis. For the co-transfection, a 3:2 ratio of the cDNAs of COX-2 or COX-1 and PGIS was used. The ratio used was based on the optimized co-expression of the two enzymes in the cells.
Electrophoresis and Western blot
The cells were sonicated in PBS and then centrifuged at 17,000 ×g for 30 min at 4 °C. The cell pellets containing microsomes were solubilized using a sample buffer containing 1% SDS, and separated by 7–10% (w/v) SDS-PAGE under denaturing conditions and then transferred to a nitrocellulose membrane. Bands recognized by specific primary antibodies against human COX-1, COX-2 and PGIS were visualized with horseradish peroxidase-conjugated secondary antibody and chromogenic peroxidase substrates (Ruan et al. 2006). The intensities of the bands in the immunoblot were used to normalize the enzyme activities as well.
Enzyme activity determination for COX coupled to PGIS using the high-performance liquid chromatography (HPLC)-scintillation analysis method (Ruan et al. 2006)
To determine the activity of the enzyme that converted AA into PGI2, the cells were incubated with different concentrations of [14C]-AA (0.3–60 µM) in a total reaction volume of 100 µL. After a 0.5–5 min incubation, the reaction was terminated by adding 200 µL of the solvent containing 0.1% acetic acid and 35% acetonitrile (solvent A). After centrifugation (17,000 ×g for 5 min), the supernatant was injected into a reverse phase C18 column (Varian Microsorb-MV 100-5, 4.5 × 250 mm) using the solvent A with a gradient from 35 to 100% of acetonitrile for 45 min at a flow-rate of 1.0 mL/min. The [14C]-labeled AA metabolites, including [14C]-6-keto-PGF1α (degraded PGI2) were monitored directly by a flow scintillation analyzer (Packard 150TR). The peak and the relative amount of [14C]-6-keto-PGF1α converted from [14C]-AA were confirmed and calibrated by 6-keto-PGF1α standards using the enzyme immunoassay described (Ruan et al. 2006).
Fluorescence microscopy and immunofluorescence staining
The cultured cells expressing COX-RFP and PGIS–CFP were directly observed by fluorescence microscopy using corresponding wavelengths of excitation and emission for the red and cyanic colors. For immunofluorescence microscopy, the transfected cells grown on a cover glass were washed with PBS and then incubated with 3.7% formaldehyde for 10 min. The cells were then blocked for 20 min before being incubated with 1% saponin for 20 min. This was followed by the addition of the primary antibody (10 µg/mL of affinity-purified anti- human COX-1, COX-2 or PGIS antibody) in the presence of 0.25% saponin and 10% goat serum for 1 h. After washing with PBS, the cells were incubated with the secondary antibodies labeled with FITC- or Rhodamine, and then viewed under an Olympus epifluorescence microscope (Ruan et al., 2006, 2008b).
FRET experiments
The fluorescence images of the live cultured cells expressing the FP-fused protein(s) were captured by the Olympus epifluorescence microscope using three sets of filters: for CFP (Ex440, m480), for RFP (Ex525, m580) and for FRET (Ex440, m580).
Results
Co-expression of COX-1 or COX-2 with PGIS in COS-7 cells
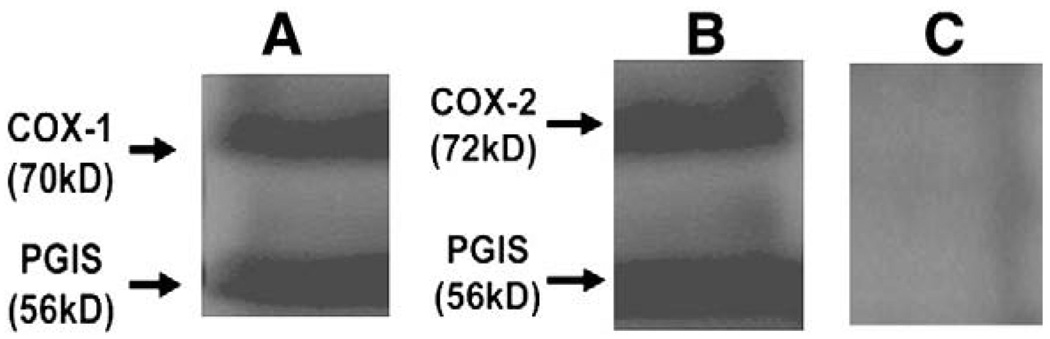
The previously cloned cDNAs of human COX-1, COX-2 and PGIS were subcloned into the mammalian expression vector, pcDNA3.1. COS-7 cells were used to co-express COX-1 or COX-2 with PGIS by gene transferring approach (Deng et al. 2002, 2003; Ruan et al. 2005). After 48-h of the transfection, the cells co-expressing the COX-2 or COX-1 with PGIS (Ruan et al. 2006) were confirmed by Western blot analysis using the antibodies against human COX-1, COX-2 and PGIS (Fig. 1). The correct molecular mass for the COX-1 (MW: 70 kDa) or COX-2 (MW: 72 kDa) co-expressed with PGIS (MW: 56 kDa) in the cells were shown (Fig. 1). The protein amounts loaded in the Western blot analysis were similar, and the differences shown in the Western blot are very small.
Fig. 1.
Western blot analysis of the overexpressed recombinant proteins in COS-7 cells. Approximately 48 h after cDNA transfection, the COS-7 cells were harvested for Western blot analysis. A total of 20 µg of the proteins of the cells co-transfected with the cDNAs of COX-1 and PGIS (A), and COX-2 and PGIS (B), or untransfected cells (C) was subjected to 10% SDS-PAGE and then transferred onto a nitrocellulose membrane, which were probed with specific rabbit anti-COX-1, -COX-2 and -PGIS antibodies and then stained with horseradish peroxidase-labeled goat-anti rabbit antibody. The numbers on the left side represent the molecular weight of the enzymes, which are indicated with arrows.
Comparison of the enzyme kinetics between COX-1 and COX-2 coupled to PGIS in biosynthesis of PGI2 in the cells
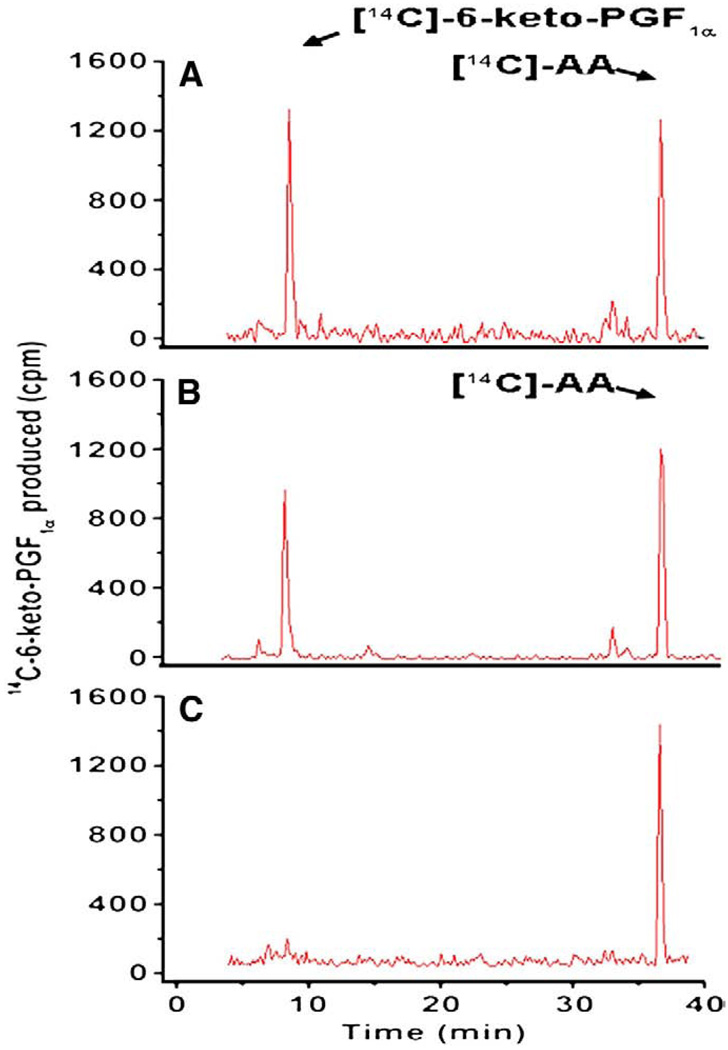
The catalytic activities for converting AA into PGI2 by the cells co-expressing COX-1 with PGIS, and COX-2 with PGIS were compared by enzyme coupling assays using [14C]-AA as an initial substrate. Both transfected cells converted [14C]-AA into [14C]-PGI2 (degraded into [14C]-6-keto-PGF1α) and had high efficiency with very little side products (Fig. 2). This demonstrated that the co-expressed COX-1 with PGIS, and COX-2 with PGIS were active, had no significant difference in general functions, and were suitable for comparison. Using the assay system, the Km value for converting AA into PGI2 by COX-2 coupled to PGIS was approximately 2.2 µM. In contrast, the Km value was approximately 6 µM for that of COX-1 coupled to PGIS (Table 1). It becomes clear that COX-2 coupled to PGIS has a “priority” to use the AA because its Km value is significantly lower than that of the COX-1 in the cell environment. The Km and Vmax determination and curves have been reported previously (Ruan et al. 2006, 2008a,b).
Fig. 2.
Determination of the triple catalytic reactions of COX coupled to PGIS in biosynthesis of PGI2. The COS-7 cells (~0.1×106) transfected with the recombinant cDNA(s) of COX-1 and PGIS (A), or COX-2 and PGIS (B) were washed three times, suspended in 0.01 M phosphate buffer, pH 7.4, containing 0.15% NaCl (PBS) and then incubated with [14C]-AA (10 µM) in a total volume of 30 µL. After 5 min, the reaction was terminated by the addition of 50 µL of 0.1% acetic acid containing 35% acetonitrile (buffer A), and centrifuged at 17,000 ×g for 5 min. The supernatant was separated by HPLC on a C18 column (4.5 × 250 mm) using buffer A with a gradient of 35–100% acetonitrile. The [14C]-AA metabolites were determined by a liquid scintillation analyzer built in the HPLC system. The retention times of [14C]-6-keto-PGF1α and [14C]-AA were calibrated by standards under the same conditions. The amounts of produced 6-keto-PGF1α represented the amounts of the produced PGI2. The produced [14C]-6-keto-PGF1α was quantitated based on the equation: 100 cpm is equal to approximately 0.5 pg of [14C]-6-keto-PGF1α. The untransfected COS-7 cells were used as a negative control (C).
Table 1.
Comparison of the kinetics for COX-1 or COX-2 coupled to PGIS in PGI2 biosynthesis as determined by enzyme coupling assays.
| Enzymes Co-expressed in COS-7 Cells |
Km (µM, converting A to PGI2) |
½ time of Vmax (s) |
Reference |
|---|---|---|---|
| COX-1 and PGIS | 6.0 | 120 | Ruan et al. 2008a |
| COX-2 and PGIS | 2.2 | <60 | Ruan et al. 2006 |
Comparison of the time course for the catalytic conversion of AA to PGI2 between COX-2 and COX-1 coupled to PGIS
Another factor determining the “priority” of COX-2 over COX-1 coupled to PGIS (using AA as substrate) is the initial reaction velocity of the catalytic reactions. A time course study has shown that half of the time for Vmax (1/2 t Vmax) for COX-2 coupled to PGIS (in producing PGI2) is less than 60 s. However, the time needed was doubled, (approximately 120 s) for that of COX-1 coupled to PGIS in order to produce the same amount of PGI2 (Table 1). This indicated that COX-2 is preferred over COX-1, therefore having the “priority” to use the available AA for PGI2 biosynthesis in cells.
Testing the COX-2 “priority” (coupled to PGIS) in converting AA into PGI2 using the newly engineered COX-1-10aa-PGIS and OX-2-10aa-PGIS hybrid enzymes
To further validate this COX-2 “priority” dominating the conversion of AA into PGI2, the previously engineered hybrid enzymes of COX-2 linked to PGIS (COX-2-10aa-PGIS) and COX-1 linked to PGIS (COX-1-10aa-PGIS) were used to mimic their coupling reactions. Because the co-expression of COX and PGIS in the COS-7 cells may not always be co-expressed with ideal levels at 1:1 ratio, the hybrid enzymes were constructed with one molecule of COX-1 or COX-2 to one molecule of PGIS. Thus, the hybrid enzymes are ideal models for kinetic studies of the COX-1 or COX-2 coupled to PGIS with 1:1 ratio in biosynthesis of PGI2. The COS-7 cells transfected with the single cDNA of COX-1-10aa-PGIS, or COX-2-10aa-PGIS were assayed for converting [14C]-AA to [14C]-PGI2. The Km values and ½t Vmax for the cells expressing COX-2-10aa-PGIS were approximately 2 µM, and 20 s, respectively, which were 2- and 5-fold faster than that of COX-1-10aa-PGIS, respectively (Table 2). This study has further validated that the COX-2 coupled to PGIS has a lower Km and a faster initial velocity in comparison with that of COX-1 coupled to PGIS. This could be the basis for the negative effects of the selective COX-2 inhibitors having side-effects such as reducing PGI2 production more so than that of nonselective NSAIDs, which have less potency to inactivate COX-2 coupled to PGIS.
Table 2.
Comparison of the kinetics for COX-1 or COX-2 linked with PGIS in COS-7 cells as determined by enzyme coupling assays.
| Enzyme reaction | Km (Converting AA to PGI2) |
½ Vmax (s) | Reference |
|---|---|---|---|
| COX-1-10aa-PGIS | 5 µM | 120 | Ruan et al. 2008a |
| COX-2-10aa-PGIS | 2 µM | 20 | Ruan et al. 2006 |
Comparison of the topological arrangement between COX-1 with PGIS and COX-2 with PGIS in the ER membrane
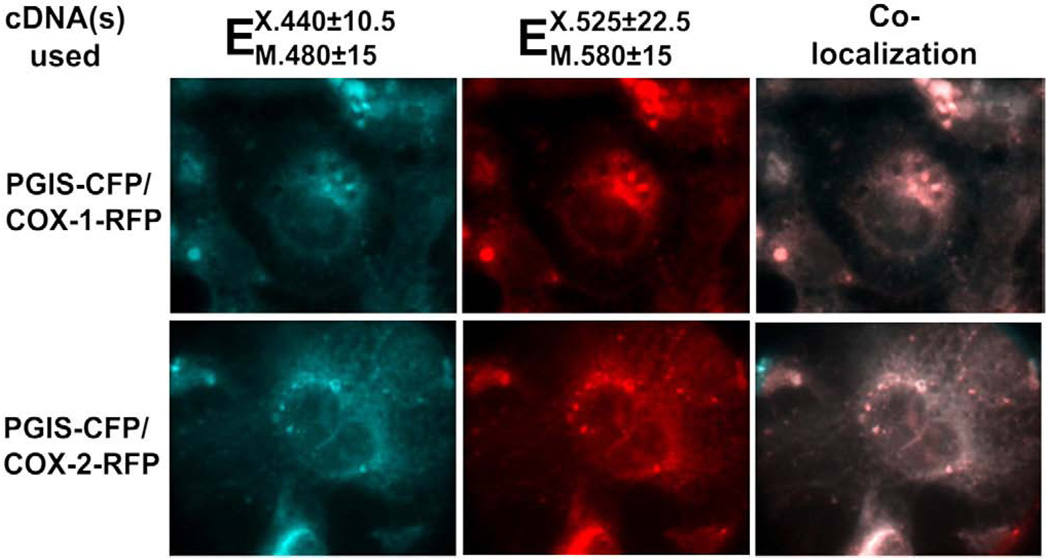
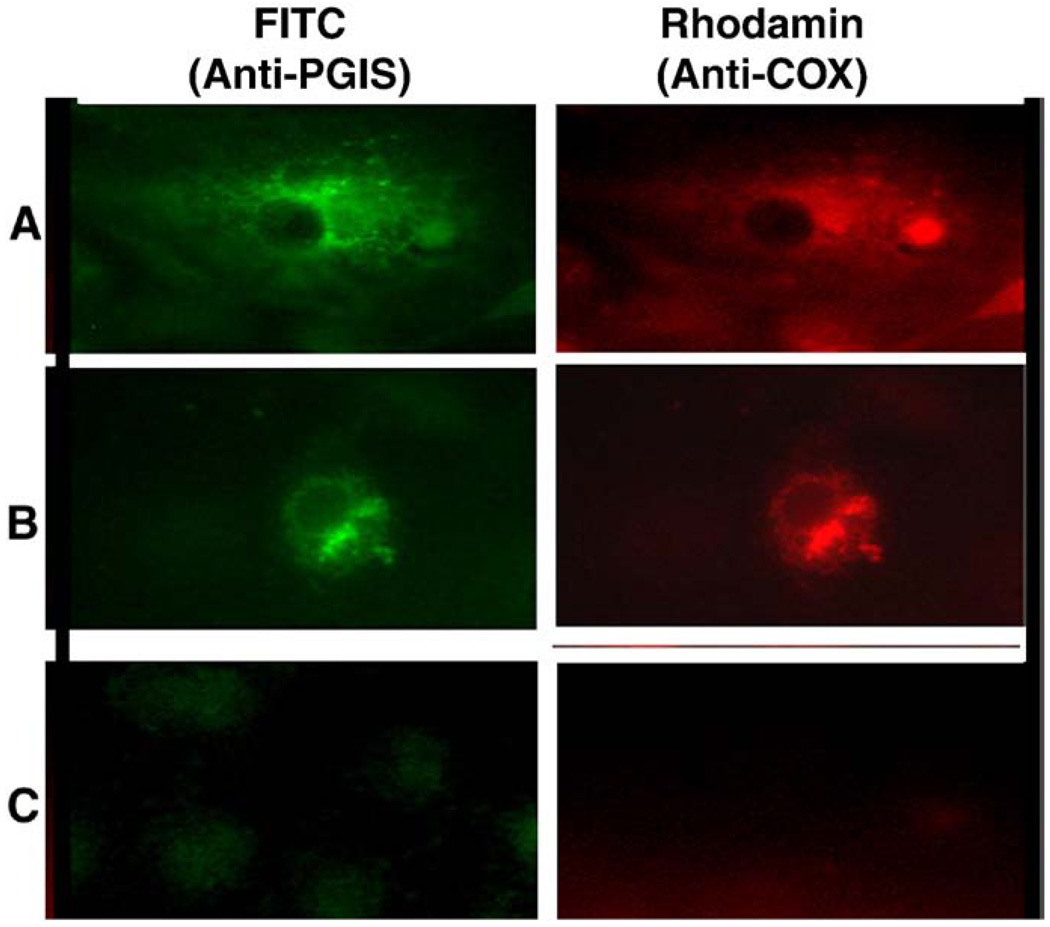
In order to further reveal the mechanisms behind the favored coupling of COX-2 with PGIS (compared to that of COX-1 with PGIS), COX-1 and COX-2 were fused to red fluorescence protein (RFP) and formed the fusion proteins, COX-1-RFP and COX-2-RFP, respectively. On the other hand, PGIS was fused to cyanic fluorescence protein (CFP) and formed the fusion protein PGIS–CFP. High resolution images were used to analyze the co-expression of the COX-2-RFP or COX-1-RFP with PGIS–CFP in the COS-7 cells by determining the subcellular co-localization of COX-1-RFP or COX-2-RFP with PGIS–CFP in the live cells. The cyanic color and red color represent the subcellular localization of COX-1 with PGIS, and COX-2 with PGIS in the cells (Fig. 3). Both, COX-1-RFP with PGIS–CFP, and COX-2-RFP with PGIS–CFP were mainly co-localized on the ER membrane and some nuclear membranes (Fig. 3, left and middle panes). By superimposing the red and cyanic colors together, the COX-1-RFP or COX-2-RFP was completely overlapped with PGIS–CFP as identified (Fig. 3, right). However, the COX-2-RFP with PGIS–CFP (Fig. 3, bottom panels) gave more nuclear localization than that of the COX-1-RFP with PGIS–CFP (Fig. 3 top panels). In order to confirm that the fusion enzymes of COX-1- (or COX-2)-RFP and PGIS–CFP have a similar topological arrangement to that of the wild type COXs and PGIS, the co-expressed wild type COX-1 with PGIS, and COX-2 with PGIS in COS-7 cells were double stained by immunofluorescence microscopy using Rhodamine red for COXs and FITC green for PGIS (Fig. 4). The overlapped red and green colors on the ER and nuclear membranes were very similar to that of the co-localized COX-RFP and PGIS–CFP (Fig. 4).
Fig. 3.
Fluorescence micrographs of COS-7 cell co-expressing PGIS–CFP (cyanic, left) and COX-1-RFP or COX-2-RFP (red, middle). The cells were transfected with cDNAs of PGIS–CFP and COX-1-RFP or COX-2-RFP using Lipofectamine2000 as described in the Experimental procedures. Subcellular localizations of COX-1 or -2-RFP and PGIS–CFP in the ER and nuclear membranes in the live cells were shown by fluorescence microscopy. The co-localization of the PGIS and COXs were identified by superimposition of the cyanic and red colors together (light pink, right).
Fig. 4.
Immunofluorescence micrographs of COS-7 cells co-expressing wild type COX-2 with PGIS (A) and COX-1 with PGIS (B). Non-transfected COS-7 cells are shown in (C). Immunostaining was performed using the first antibodies against PGIS or COXs and second antibodies labeled with FITC for PGIS (green) and with Rhodamine for COXs (red) for the cells treated with SLO (Ruan et al. 2006).
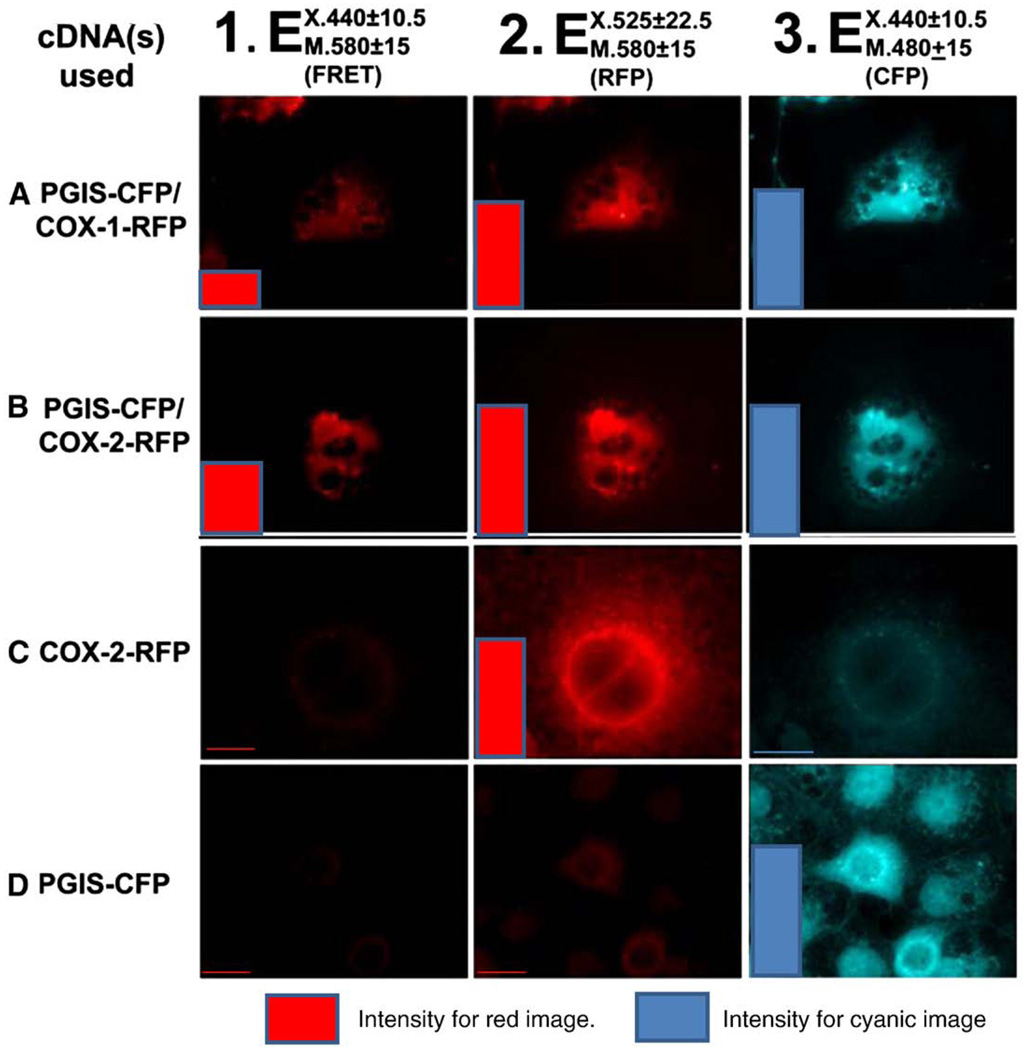
Comparison of the physical distances between COX-1 with PGIS, and COX-2 with PGIS
The distance between the COXs and PGIS on the ER membrane was measured by fluorescence resonance energy transfer (FRET) microscopy (Fig. 5). The similar intensities of the co-localized COX-RFP and PGIS–CFP in live cells were obtained by using the corresponding excitation and emission for COX-RFP (Fig. 5, middle panels) and PGIS–CFP (Fig. 5, right panels) first. Increased FRET intensity is a direct indicator of shorter inter-protein distance (Berney and Danuser, 2003). Then, the separation distances between the co-localized COX-1-RFP and PGIS–CFP, and between COX-2-RFP and PGIS–CFP were measured by FRET using the excitation of the cyanic color (donor) for PGIS–CFP and emission of the red color (acceptor) for COX-RFP. As a result, the intensity of the red color for co-localized COX-2-RFP with PGIS–CFP was twice that of the co-localized COX-1-RFP with PGIS–CFP (Fig. 5, left panels). In general, FRET could only be used to observe the color image if the two molecules are within 50 Å of separation. The earlier results have provided direct evidence, in which: a) the distance between COX-1 or COX-2 and PGIS are within 50 Å; and b) the distance between COX-2-RFP and PGIS–CFP is significantly shorter than that of COX-1-RFP and PGIS–CFP in the cells. In comparison, no FRET signal was observed for the control cells expressing COX-2-RFP (Fig. 5C) and PGIS–CFP (Fig. 5D) alone, respectively.
Fig. 5.
Fluorescence micrographs of COS-7 cells co-expressing PGIS–CFP and COX-1 (A) or COX-2-RFP (B), COX-2-RFP only (C) and PGIS–CFP only (D). The co-localization of the PGIS–CFP (cyanic color, column 3) and COXs-RFP (red color, column 2) in the ER and nucleus membrane (within 50 Å) were observed by the FRET using 440 nm as excitation for CFP and 580 nm as emission for RFP (column 1). The qualitative scales are shown using color columns which represent the integrated fluorescence intensities from the corresponding cell images. Fluorescence intensities were determined using Scion Image software (Frederick, Maryland USA); “Ex” and “Em” represent excitation and emission, respectively.
Discussion
The biosynthesis of the vascular protector PGI2 involved triple catalytic steps and two enzymes (COX-1 or COX-2 coupled to PGIS). The kinetics of the individual COX-1, COX-2 and PGIS has been determined previously (Ruan et al. 2006, 2008a,b). However, to understand how the selective COX-2 inhibitor affects PGI2 biosynthesis more so than that of the nonselective NSAIDs in clinical trials (Vane 2002), the cellular kinetics of COXs coupled to PGIS in biosynthesis of PGI2 is more accurate than that of individual enzymes. Kulmacz (2005) has indicated that the lower Km for PGI2 synthesis and faster initial reaction velocity for COX-2 coupled to PGIS have reflected the higher catalytic efficiency of COX-2 versus COX-1. Based on the information of COX-2 inhibitors increasing myocardial infarction in clinical trials, we have hypothesized that in the presence of COX-1 and COX-2, COX-2 coupled to PGIS might dominate the PGI2 biosynthesis. This study was focused on testing the hypothesis and characterizing the differences in the cellular biosynthesis of PGI2 through the individual COX-1 and COX-2 coupled to PGIS in cells, and demonstrating the mechanisms of the selective COX-2 inhibitor's affect on PGI2 production, which caused a risk for heart disease. To reach the goal, the co-expression of COXs and PGIS in the live cells were used to mimic the catalytic coupling reactions of COXs with PGIS. The enzyme kinetics obtained from the co-expression system allowed us to characterize the PGI2 synthesis by COXs coupled to PGIS in the cellular environment. Our studies have found that the COX-2 coupled to PGIS has a lower Km value than that of COX-1 coupled to PGIS. The initial velocity determined by time course also showed that COX-2 coupled to PGIS was faster than that of COX-1 coupled to PGIS (Table 1). This kinetics information is much closer in reflecting the cellular PGI2 biosynthesis than that of the kinetics studies for the individual enzymes previously determined.
In fact, the biosynthesis of PGI2 through the coupling reaction of COXs with PGIS in the ER membrane, without other molecule involvement, was not directly proven until recently, when we showed that the engineered hybrid enzymes linking COX-1 to PGIS (COX-1-10aa-PGIS) and COX-2 to PGIS (COX-2-10aa-PGIS) could perform triple catalytic reactions and effectively convert AA into PGG2, PGH2 and then PGI2 (Ruan et al. 2006, 2008a). The purified COX-2-10aa-PGIS, without the presence of any other molecules, showed identical triple catalytic activity with that of the hybrid enzymes in cells (Ruan et al. 2008b). The hybrid enzymes of COX-1 or COX-2 linked to PGIS become unique models to study the kinetics of the coupling reactions of the triple catalytic steps from COXs to PGIS in cells. One of the major advantages in using the hybrid enzyme is that it allows us to study kinetics for the ideal coupling reaction at 1:1 molecular ratio of COX and PGIS. Thus, to further confirm the observations that resulted from the co-expression of COXs and PGIS, the hybrid enzymes, COX-1-10aa-PGIS and COX-2-10aa-PGIS were individually expressed in the COS-7 cells. The Km values and initial velocity of the coupling reaction were obtained. Again, the hybrid enzyme which linked COX-2 to PGIS has a lower Km value and faster initial velocity in comparison with that of the hybrid enzyme of COX-1 linked to PGIS (Table 2). It shall be noted that the experimental results from this study were obtained from the co-expressed COX and PGIS with approximately 1:1 (mol/mol) pathophysiological conditions. Thus, the implications of the cell study to clinical heart disease need to be further investigated using animal models with different conditions, something that could be part of our future work.
There are several advantages of using fluorescence fusion enzymes for the topology studies: a) high resolution: the images of COX-RFP and PGIS–CFP shown in Fig. 3 have much higher resolution compared to that of the immunofluorescence-stained wild type COXs and PGIS (Fig. 4); b) instant live cells study: the subcellular localization could be observed under live cell conditions; and c) suitable for FRET: the high resolution fluorescence images are suitable for FRET studies.
FRET is a technique used to measure the distance between two fluorescence molecules (donor and acceptor) with the overlapping emission wavelength from the donor and excitation wavelength of the acceptor. The technology has been extended to measure the protein–protein interactions when the proteins are tagged with fluorescence donor and acceptor (Majoul et al. 2002). In this study, the emission wavelength from PGIS–CFP of 480 nm (±15 nm) is overlapped with the excitation wavelength of 520 nm (±15 nm) of COX-RFP. Thus, the excitation of the PGIS–CFP resulted in an emission of COX-RFP which could be used to measure the distance between the COX and PGIS in the ER membrane. It shall also be indicated that the co-expressed fusion enzymes, COX-RFP and PGIS–CFP could produce a fluorescence signal instantly without any chemical modification. Thus, the intensity of the fluorescence signal is directly related to the concentration of the molecules expressed in the live cells. Consequently, the distance between COX and PGIS could be measured when the cells are still alive under the microscope. As described in the results, the microscopic FRET could only show the emission image when the two molecules are near-by (within 50 Å). Both, the co-expressed COX-1-RFP with PGIS–CFP, and COX-2-RFP with PGIS–CFP showed a FRET signal (Fig. 5, left), which indicated that regardless of the COX-1-RFP or the COX-2-RFP location to the PGIS, the intermolecular distances are less than 50 Å. In addition, the result also showed that the intensity of the FRET signal for COX-2-RFP and PGIS–CFP was much stronger than that of COX-1-RFP and PGIS–CFP. This suggested that the separation between COX-2 and PGIS is shorter than that of COX-1 and PGIS. Because the PGH2 is very unstable in the cytoplasm environment, the shorter distance is favorable to the PGH2 which can move from the COX-2 domain to the PGIS domain with less chances of being degraded. This could be important to the faster catalytic reaction for COX-2 coupled to PGIS than that of COX-1 coupled to PGIS in biosynthesis of PGI2 in the cells. The consequences of directly extrapolating an in vitro modified-protein expression system to clinical significance are needed for further testing.
In conclusion, our studies have provided evidence showing that the interactions between COX-1 or COX-2 and PGIS in the ER membrane are different. In comparison with that of COX-1, COX-2 has priority in coupling with PGIS, therefore dominating PGI2 biosynthesis. Based on the inducible nature of COX-2, these findings have led us to speculate that the preference of COX-2/PGIS coupling over COX-1/PGIS coupling in biosynthesis of PGI2 could be the result of a self-vascular protective event acting in response to the diverse stimulations and stresses involved with vascular ischemia. Selective COX-2 inhibitors, which inactivate COX-2's ability to couple to PGIS, could destroy the self-protection and create a risk that promotes ischemic conditions in the vascular system leading to the triggering of heart diseases.
Acknowledgements
This work was supported by the National Institutes of Health [Grants HL56712 and HL79389 (to KHR), HL100807 (to RD and KHR)] and the American Heart Association [Grant 10GRNT4470042 (to KHR)], T5 DK007676 (for Summer Student Research).
Footnotes
Conflict of interest statement
The authors of this manuscript declare that there are no conflicts of interest.
References
- Berney C, Danuser G. FRET or no FRET: a quantitative comparison. Biophys J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Huang A, So SP, Lin YZ, Ruan KH. Substrate access channel topology in membrane-bound prostacyclin synthase. Biochem J. 2002;362:545–551. doi: 10.1042/0264-6021:3620545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Wu J, So SP, Ruan KH. Identification of the residues in the helix F/G loop important to catalytic function of membrane-bound prostacyclin synthase. Biochemistry. 2003;42:5609–5617. doi: 10.1021/bi026749z. [DOI] [PubMed] [Google Scholar]
- Dogné JM, Hanson J, de Leval X, Pratico D, Pace-Asciak CR, Drion P, et al. From the design to the clinical application of thromboxane modulators. Curr Pharm Des. 2006;12:903–923. doi: 10.2174/138161206776055921. [DOI] [PubMed] [Google Scholar]
- Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- Granstrom E, Diczfalusy U, Hamberg M, Hansson G, Malmsten C, Samuelson B. In: Prostaglandins and the Cardiovascular System. Oates JA, editor. New York: Raven Press; 1982. pp. 15–58. [Google Scholar]
- Kulmacz RJ. Regulation of cyclooxygenase catalysis by hydroperoxides. Biochem Biophys Res Commun. 2005;338:25–33. doi: 10.1016/j.bbrc.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Majerus PW. Arachidonate metabolism in vascular disorders. J Clin Investig. 1983;72:1521–1525. doi: 10.1172/JCI111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoul I, Straub M, Duden R, Hell SW, Soling HD. Fluorescence resonance energy transfer analysis of protein–protein interactions in single living cells by multifocal multiphoton microscopy. J Biotechnol. 2002;82:267–277. doi: 10.1016/s1389-0352(01)00042-3. [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez J, Badimon L. Mechanisms underlying the cardiovascular effects of COX-inhibition: benefits and risks. Curr Pharm Des. 2007;13:2215–2227. doi: 10.2174/138161207781368774. [DOI] [PubMed] [Google Scholar]
- Needleman P, Turk J, Jackschik BA, Morrison AR, Lefkowith JB. Arachidonic acid metabolism. Annu Rev Biochem. 1986;55:69–102. doi: 10.1146/annurev.bi.55.070186.000441. [DOI] [PubMed] [Google Scholar]
- Pace-Asciak CR, Smith WL. Enzymes in the biosynthesis and catabolism of the eicosanoids: prostaglandins, thromboxanes, leukotrienes and hydroxy fatty acids. Enzymes. 1983;16:544–604. [Google Scholar]
- Patrono C, Ciabattoni G, Davi G. Thromboxane biosynthesis in cardiovascular diseases. Stroke. 1990;21:130–133. [PubMed] [Google Scholar]
- Ruan KH, Deng H, Wu J, So SP. The N-terminal membrane domain of the membrane-bound prostacyclin synthase involved in the substrate presentation in the coupling reaction with cyclooxygenase. Arch Biochem Biophys. 2005;435:372–381. doi: 10.1016/j.abb.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Ruan KH. Advance in understanding the biosynthesis of prostacyclin and thromboxane A2 in the endoplasmic reticulum membrane via the cyclooxygenase pathway. Mini Rev Med Chem. 2004;4:639–647. doi: 10.2174/1389557043403710. [DOI] [PubMed] [Google Scholar]
- Ruan KH, Dogné JM. Implications of the molecular basis of prostacyclin biosynthesis and signaling in pharmaceutical designs. Curr Pharm Des. 2006;12:925–941. doi: 10.2174/138161206776055994. [DOI] [PubMed] [Google Scholar]
- Ruan KH, Deng H, So SP. Engineering of a protein with cyclooxygenase and prostacyclin synthase activities that converts arachidonic acid to prostacyclin. Biochemistry. 2006;45:14003–14011. doi: 10.1021/bi0614277. [DOI] [PubMed] [Google Scholar]
- Ruan KH, So SP, Cervantes V, Wu H, Wijaya C, Jentzen RR. (1) An active triple-catalytic hybrid enzyme engineered by linking cyclo-oxygenase isoform-1 to prostacyclin synthase that can constantly biosynthesize prostacyclin, the vascular protector. FEBS J. 2008a;275:5820–5829. doi: 10.1111/j.1742-4658.2008.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan KH, So SP, Wu H, Cervantes V. (2) Large-scale expression, purification, and characterization of an engineered prostacyclin-synthesizing enzyme with therapeutic potential. Arch Biochem Biophys. 2008b;480:41–50. doi: 10.1016/j.abb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson B, Goldyne M, Granstrom E, Hamberg M, Hammarstrom S, Malmsten C. Prostaglandins and thromboxanes. Annu Rev Biochem. 1978;47:997–1029. doi: 10.1146/annurev.bi.47.070178.005025. [DOI] [PubMed] [Google Scholar]
- Smith WL. Prostaglandin biosynthesis and its compartmentation in vascular smooth muscle and endothelial cells. Annu Rev Physiol. 1986;48:251–262. doi: 10.1146/annurev.ph.48.030186.001343. [DOI] [PubMed] [Google Scholar]
- Vane JR. Biomedicine. Back to an aspirin a day? Science. 2002;296:474–475. doi: 10.1126/science.1071702. [DOI] [PubMed] [Google Scholar]