Abstract
RadLex™, the Radiology Lexicon, is a controlled vocabulary of terms used in radiology. It was developed by the Radiological Society of North America in recognition of a lack of coverage of these radiology concepts by other lexicons. There are still additional concepts, particularly those related to imaging observations and imaging observation characteristics, that could be added to the lexicon. We used a free and open source software system to extract these terms from the medical literature. The system retrieved relevant articles from the PubMed repository and passed them through modules in the Apache Unstructured Information Management Architecture. Image observations and image observation characteristics were identified through a seven-step process. The system was run on a corpus of 1,128 journal articles. The system generated lists of 624 imaging observations and 444 imaging observation characteristics. Three domain experts evaluated the top 100 terms in each list and determined a precision of 52% and 26%, respectively, for identification of image observations and image observation characteristics. We conclude that candidate terms for inclusion in standardized lexicons may be extracted automatically from the peer-reviewed literature. These terms can then be reviewed for curation into the lexicon.
Key words: Natural language processing, algorithms, open source
Introduction
RadLex™, the Radiology Lexicon, is a controlled vocabulary representing the terms and concepts of radiology.1 It was developed by the Radiological Society of North America in recognition of a lack of coverage of these radiology concepts by other lexicons.2 RadLex was created and extended by the contributions of committees of radiologists as well as members of other radiology organizations. RadLex is intended to reduce variation and improve clarity in radiology reports and image annotations as well as to provide a standardized means of indexing radiological materials in a variety of settings. Currently, RadLex consists of approximately 12,000 individual terms, organized in a hierarchy with 12 top level categories. Though large, in order to maintain its utility as a standardized tool for the radiology community, RadLex must continue to grow with the field.
RadLex is still missing concepts, particularly those related to imaging observations and imaging observation characteristics: the lingua franca of radiologists. An image observation is something that is seen in the image by a human or machine observer. A “mass” or an “opacity” is an observation. Imaging observation characteristics characterize the observation. “Spiculated” or “diffuse” are examples of image observation characteristics.
While the manual, committee mechanism for extending RadLex, has contributed greatly to the lexicon, it is time consuming, dependent on the schedules of those involved, and costly. As the size of the lexicon increases, this process becomes unsustainable.
Automatic extraction of information from the medical literature is a branch of natural language processing. Many have recognized the potentially valuable content in the peer-reviewed literature and the need to accelerate manual curation of lexicons. Several reviews of text extraction from the medical literature for a variety of purposes have been published.3–5
Our hypothesis is that concepts important for inclusion in RadLex can be automatically extracted from the medical literature at least to the point of facilitating a manual editorial and curation process, using these techniques. This project describes such an automatic term extraction system.
Material and Methods
This work did not involve human subject research. All software was developed in the JAVA language (Sun Microsystems, Mountain View, CA). Documents and data were stored in a MySQL 5.1 database (Sun Microsystems, Mountain View, CA, USA).
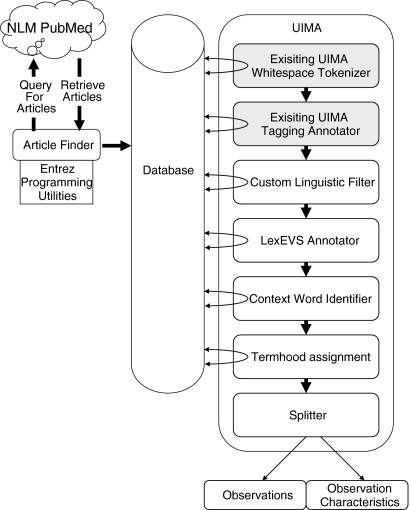
Figure 1 shows an overview of the architecture of the system. An article finder application for gathering a corpus of published, peer-reviewed journal articles was developed using the Entrez Programming Utilities (www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html) from the National Center for Biotechnology Information (NCBI) at the National Library of Medicine (NLM). The article finder used the Apache Axis2 Web service engine (http://ws.apache.org/axis2/) to access the NLM’s PubMed citation index and full text article web service. The article finder searched for articles with the PubMed query syntax. The search string included the terms “imaging findings [Title],” “CT findings [Title],” “MRI findings [Title],” “Xray findings [Title],” and “PET findings [Title].” Using the PubMed ID number of each article, the application located an online version of the article’s text and saved it to local disk. The application parsed the HTML tags to find section headers and segment out the body of the article. The HTML tags were removed, and the text was stored in the database, along with important article meta-data including article title, MeSH headings, journal information, and date of publication.
Fig 1.
The architecture of the automatic extraction system. Shaded modules are existing components of the UIMA architecture.
The term extraction system was written as a combination of standalone Java programs and pipelines for the Apache Unstructured Information Management Architecture (UIMA) framework v2.2.2 (http://incubator.apache.org/uima). UIMA is a system for processing and extracting information from large volumes of documents. UIMA facilitates the creation of processing pipelines, i.e., sets of components applied sequentially to all of the documents in a corpus.
The process consisted of seven steps:
Tokenizer: An existing UIMA whitespace tokenizer was used to identify individual words, punctuation marks, and sentences, marking them as discrete tokens.
Part of Speech Tagging: The UIMA tagger annotator uses hidden Markov models to tag each token as a part of speech.
-
Linguistic Filter: A linguistic filter scans the words of a document and marks sequences of words that fit a defined pattern of parts of speech. We wanted to identify imaging observations and their characteristics. A fully characterized imaging observation is a sequence of zero or more imaging observation characteristics or modifiers, ending in an imaging observation. For example, diffuse interlobular septal thickening consists of the observation (septal thickening) with two imaging observation characteristics (diffuse and interlobular). The pattern we looked for was an adverb, followed by zero or more past tense verbs or adjectives, followed by zero or more nouns, followed by another noun or -ing ending verb. Intervening connector words (“the,” “to,” “of,” etc.) were ignored.
This pattern was determined by trial and error to identify as many fully characterized imaging observations from the text as possible, without being so general as to allow a large number of non-terms onto the list. The frequencies of the phrases in each document were stored in the database.
LexEVS Annotator: A LexEVS (Mayo Clinic, Rochester, MN, USA) annotator was then developed as a module within the UIMA pipeline. LexEVS is a set of programmable interfaces that allows access to controlled terminologies through a client server mechanism. This annotator took the previously identified candidate phrases and looked them up in RadLex and the UMLS Thesaurus. If the term was found in a vocabulary, the category (i.e., its ontological placement, disease, anatomic entity, etc.), term name, vocabulary name, and its entity code (a unique alphanumeric sequence given to each term in a vocabulary) were returned and stored in the database.
-
Context Word Identification: Context words are non-candidate terms that are associated with terms of interest. In our case, positive context words are non-candidate terms that appear near known imaging observations and characteristics. Negative context words are terms that are found not to be associated with known imaging observations and characteristics. RadLex terms that the annotator identified were used to learn context words that surround candidate phrases. The NC-value method 3 finds nouns, adjectives, and verbs within the neighborhood around a known term (in our case a known RadLex term). These words are then given weights based upon how many of the existing RadLex imaging observations each context word is adjacent. To achieve better performance for the radiology domain, we used only verbs and adverbs for both positive and negative context words. We enhanced this NC-value method by distinguishing between preceding and following context words. This is based on the fact that imaging observations almost always appear near other kinds of medical terms. By distinguishing between preceding and following words, the syntactic and semantic relationships that characterize imaging observations can be used to improve identification. For example, consider the sentence “diffuse wall enhancement was seen in T1 weighted images of the gallbladder.” The verb “seen” appears near “diffuse wall enhancement,” an imaging observation, as well as “T1 weighted,” an imaging procedure attribute, and “gallbladder,” an anatomic entity. Without differentiating between whether the verb seen comes before or after a candidate term, there is no way to distinguish that an imaging observation is something that is seen and that the anatomic entity and procedure attribute are attributes of where it is seen.
We also enhanced the method to find both positive and negative context words. Rather than identifying only positive context words which frequently co-occurred with imaging observations, we also identified negative context words which were associated with terms from their ontological meaning as determined by the LexEVS Annotator. This better distinguishes between imaging observations and other kinds of radiology terms. For example, the verb seen is frequently found following imaging observations; it is also frequently found preceding anatomic entities. Following this thinking, our system identifies context words for a variety of categories of RadLex terms found in the documents. Context words found near imaging observations are given positive weights, and those found near other classes, such as anatomic entities or treatments, are given negative weights.
To learn these context words, a pipeline was developed consisting of a single context identification module. All appearances in the articles of existing RadLex terms were located, and the words before and after them within a radius of six words around them were stored as preceding or following context words.
The set of identified context words was then used to generate a context score for every candidate phrase. These context scores represented how similar or dissimilar a candidate phrase’s context was to the context of imaging observations. A UIMA pipeline, consisting only of a context detection module, found all appearances of the candidate phrases and calculated context scores for each one. This context score was based on the weights of the context words that appeared before or after the term within a six word radius.
Termhood Assignment: Termhood is the probability that a term is of particular interest (in our case, an observation or an observation characteristic). The candidate phrases were assigned termhood values which represent how likely each one was to be an imaging observation. This termhood value, a modified version of the NC-value, was calculated based on the context scores, the number of words in the term, and the nesting of the term. The nesting of a term is a representation of how independent that term is. For example, if the phrase “soft tissue mass” appeared 46 times in the documents and the phrase “tissue mass” also appeared only 46 times, it can be said that “tissue mass” itself is not a term, because it only appears within the larger term “soft tissue mass.” The major differences with the NC-value are that we removed frequency as a primary factor, as well as increased the importance of the context scores. Frequency was factored out because most of the newly identified imaging observations did not appear as frequently as other terms. A stop list (a set of words used to filter out some obvious non-terms, like “abstract” and “study”) was also used.
-
Splitter: The list generated by the term ranker consisted of candidate phrases deemed highly likely to contain imaging observations. In order to create one list of observations and another list of characteristics, an application was developed to separate characteristics and observations from these phrases. The application used the ratio between frequencies of words and phrases within a term to distinguish individual characteristics and observations. For example, consider the term “gallbladder wall thickening.” The total frequency of the term “thickening” was 2,108. The total frequency of the term “wall thickening” was 769. This means that 36% of the times that “thickening” appeared in the articles, it was as part of the phrase “wall thickening.” A threshold was set at 30%, because that value gave more accurate results than other thresholds. This frequency ratio was used first to identify the imaging observation of each candidate term. Then, the remainder of the term was divided up, again using frequency ratios, into individual characteristics.
The final score for each observation was the highest termhood value of the candidate phrases in which it appeared. The final score for each characteristic was the sum of the termhood values of the candidate phrases in which it appeared. The top 3,000 ranked candidate phrases were split in this manner, producing the final lists of imaging observations and imaging observation characteristics. Exact match search algorithms to the local LexBIG vocabulary server, by way of the LexEVS services, were used to filter out existing RadLex terms in these lists.
The top 100 imaging observations and the top 100 observation characteristics identified by the system and not already present in RadLex were evaluated by three board certified radiologists all experienced with working with RadLex. Candidate phrases were deemed “valid” if there was consensus among the three reviewers that the term was classified correctly. The precision of the system was calculated as the percentage of valid terms identified in the first 100 candidates.
Results
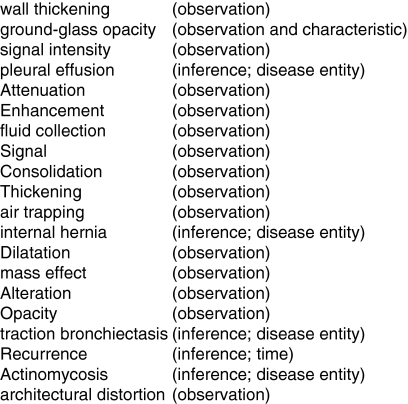
The system was run on a corpus of 1,128 journal articles as identified and retrieved by the article finder program. These articles were processed by the pipeline, and this resulted in two ranked lists, one for imaging observations and another for imaging observation characteristics that were evaluated by the domain experts for inclusion in RadLex. The system generated lists of 624 imaging observations and 444 imaging observation characteristics. Figure 2 lists the top 20 imaging observations identified by the system and their expert classification. Figure 3 lists similar for observation characteristics. Three domain experts evaluated the top 100 terms in each list and agreed that 52 suggested imaging observation characteristics (precision of 52%). From the list of suggested imaging observations, 26 of the top 100 (precision of 26%) new concepts were validated by each of the three experts.
Fig 2.
The top 20 observations identified and their ultimate classification by human experts.
Fig 3.
The top 20 observation characteristics and their ultimate classification by human experts.
Discussion
The automatic term extraction system we developed was able to identify new imaging observations and imaging observation characteristics from journal articles for inclusion in RadLex. Medical journal articles were selected as the input for our system because they are credible, peer-reviewed, semi-structured, rich sources of terms of interest. Articles collected from older issues of journals can be used to help find fundamental terms used across time, whereas recent articles contain many new terms reflecting current technologies and advances in the field. The process is also low cost in that all software used is open source and freely available.
The system provides a mechanism for accelerated expansion of RadLex by analyzing large collections of documents. Unlike manual methods, this system reduces demand on the committees of domain experts and committees. The domain experts can focus on the review and validation of proposed terms. Adding concepts without this validation would limit the utility of the lexicon. Additionally, once a term has been approved, the domain expert can then focus on determining relationships surrounding the new term as well as its location within the category hierarchy.
There is still room for improvement in the ranked lists created by the system. As evidenced by the precision values, many terms on the lists were either not classified correctly or not appropriate for inclusion in RadLex.
LexEVS is a powerful tool for exploiting a variety of controlled vocabularies. In this case, it was used as a part of the LexEVS annotator, checking whether or not terms were already in RadLex and where they belonged. This was essential for context processing as well. The tools for accessing, managing, distributing, and storing vocabularies and vocabulary information are not limited to RadLex but can be applied to many other vocabularies as well. This system can be modified by using any of the available vocabularies as a part of the annotator and, additionally, by adjusting context and processing, other categories of phrases within RadLex.
Conclusion
Candidate terms for inclusion in standardized lexicons may be extracted automatically from the peer-reviewed literature. Doing so has the potential for growing these vocabularies much faster than manual curation by committees of experts. While the sensitivity of the current process is limited, there is evidence to suggest that this could be improved with modification of the algorithms. In addition, a similar process could be developed to perform automatic relationship identification and hierarchical placement. Combined with a user-friendly method of distributing and displaying lists of potential phrases and relationships for domain expert validation, these tools could enhance standardized vocabularies going forward. These vocabularies will be all the more critical as structured reporting an image annotation advance.
Acknowledgments
RH and AVE were funded through the National Science Foundation (NSF) Research Experience for Undergraduates (REU) program. We wish to thank Daniela Raicu, PhD and Jacob Furst, PhD for leading the NSF REU MedIX program. Also, a special thanks to Emily Kawaler and Francis Ferraro for their support and contributions throughout the project.
References
- 1.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26(6):1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 2.Langlotz CP, Caldwell SA. The completeness of existing lexicons for representing radiology report information. J Digit Imaging. 2002;15(Suppl 1):201–205. doi: 10.1007/s10278-002-5046-5. [DOI] [PubMed] [Google Scholar]
- 3.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Brief Bioinform. 2005;6(1):57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 4.Erhardt RA, Schneider R, Blaschke C. Status of text-mining techniques applied to biomedical text. Drug Discov Today. 2006;11(7–8):315–325. doi: 10.1016/j.drudis.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Brief Bioinform. 2007;8(5):358–375. doi: 10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]