Abstract
Multi-site imaging research has several specialized needs that are substantially different from what is commonly available in clinical imaging systems. An attempt to address these concerns is being led by several institutes including the National Institutes of Health and the National Cancer Institute. With the exception of results reporting (which has an infrastructure for standard reports, albeit with several competing lexicons), medical imaging has been largely standardized by the efforts of DICOM, HL7, and IHE. What are not well developed in this area are the tools required for multi-site imaging collaboration and data mining. The goal of this paper is to identify existing clinical interoperability methods that can be used to harmonize the research and clinical worlds, and identify gaps where they exist. To do so, we will detail the approaches of a specific multi-site trial, point out the current deficiencies and workarounds developed in that trial, and finally point to work that seeks to address multi-site imaging challenges.
Key words: Clinical information systems, clinical trial, controlled vocabulary
Introduction
The rise of multi-site collaborative science is a well-recognized trend among both scientists and funding bodies. As a result, groups such as the National Institutes of Health (NIH) and the private sector are increasingly promoting grants and contracts that leverage the large accrual and statistical power that can be gained via multi-site trials. However, there are many challenges in supporting such trials: data integrity, data confidentiality, regulated access, quality assurance of data transmission/storage, and result validation. To appreciate this, it is useful to construct usage scenarios that point out the missing functionality of current state of the art tools for multi-site, collaborative imaging research. For example, consider a radiologist, participating in one or more clinical trials, who in the course of the day uses an ideal clinical/collaborative-research software suite as described below.
Ideal Situation
8 am
Dr. Rad logs into a workstation for the day. All relevant applications auto-start, the worklists for the clinical assignment of the day pop-up, and Dr. Rad begins reading with all applications synched to the same patient context. All clinical exams and their relevant prior studies display the way the radiologist likes them (with associated reports and history), regardless of where they were performed. Series identification, series alignment, co-registration, and change maps have been generated. Any 3D/CAD (computer-aided diagnosis) processing that can be scripted has already occurred automatically.
10 am
An email alerts Dr. Rad that one or more exams are awaiting his attention in worklist “Research 1”. Dr. Rad enters the worklist and selects the first case. The same session certificate that authenticated Dr. Rad for his clinical work has been registered for this research site, and checked against the access control lists maintained by the research image archive administrator. Dr. Rad is permitted access to this worklist, and can view all available compares on this patient depending on the research protocol, but in the process of doing so the back end image-manager routes this exam to a research worklist. Since Dr. Rad is a blind reader in this study, the exam has been mapped to an alias in place of the patient's real name and other identifiers (similarly for compares, reports and histories if allowed in the research protocol). Any exam specific processing has been done automatically via plug-ins that functions with the PACS application.
10:03 am
Dr. Rad begins reporting on the case using the same structured reporting tool that he uses in his clinical work. The tool determines, based on the performing location and Accession number, that results should be sent to a different results repository, rather than Dr. Rad's home radiology information system (RIS). The Digital Image Communications in Medicine (DICOM) structured report (SR) object is data mined at the research core lab for elements of interest to the research protocol.
10:06 am
Dr. Rad returns to his clinical work at the exact point he interrupted it, wondering, “Wasn't research among sites always like this?”
Reality Today
In the world of legacy PACS and information systems in which our practice currently lives, the description above is a remote dream. Typically, the radiologist has to manually open and log into six applications to begin the clinical day. Synchronization of the patient context among those applications also has to be done manually. Comparison exams may even reside on a different PACS. It is typically four or five in the afternoon before the radiologist can think of their research, and at that point they have to log into research dedicated tools that have a different user interface then what they are accustomed to.1 The specific details of how this is done at our site will be outlined in the “Current Efforts” section.
Take-Home Points
In addition to the radiologist, there are more stakeholders in the scenarios above. Two critical ones are the IT administrators and the funding agencies. In the Ideal Situation, the administrator is happy because she can create a single image archive with scalable user access controls that verify that only IRB approved users can access specific cases. The overhead costs to manage image data in Reality Today are high because: it is a manual process, data is often replicated on numerous systems, and the storage costs may not be tiered appropriately based on performance needs. Data sent from study participants should automatically be anonymized with an audit trail of exam accesses (as opposed to sharing clinical data wherein the data would require encryption).2 Standardized research tools would also reduce redundant development efforts; often data mining tools are reinvented for every new study because standard reporting constructs are not used. Finally, consolidating data from numerous research projects could lead to a common database farm for meta-analysis by permitted users.
From the funding point of view, the Ideal Situation means that NIH (or others) does not have to pay each award winner to reinvent viewing, archiving, anonymizing, analysis and other software. The manual process of de-identification, study transmission, form entry and other tedious tasks performed by study coordinators and staff can be minimized with a more automated and standardized system that performs commodity functions.
Current Efforts
In the past 5–8 years, the rise of multi-site imaging trials has progressed from nearly non-existent to several major new NIH sponsored projects per year. Some current projects exemplify best practices, and in particular better tools for data mining.3 In particular, the authors are familiar with the following 5 year project.
Mayo Clinic and LTRC Radiology Core Lab
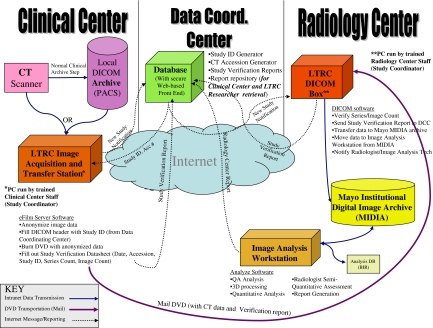
The Lung Tissue Research Consortium (LTRC) is an NIH/NHLBI-sponsored multi-site project. The goal of the LTRC is to enable better management of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis diseases by creating a data and tissue repository including lung tissue and bodily fluid specimens, volumetric high-resolution CT data of the chest and extensive clinical questionnaire and laboratory studies from donors with and without disease. The tissue and data repositories formed by the LTRC will serve as a resource for any researcher who can apply for samples and data through the NIH/NHLBI. The LTRC participating Clinical Centers (CC) collect lung tissues, The Data Coordinating Center (DCC), Radiology Core Laboratory (RCL), and Pathology Core Laboratory (PCL) provide a standardized method of collecting the samples, de-identifying the information, transferring data to the core laboratories, analyzing samples, reporting results and preparing the stored tissue samples, results, and image data for re-distribution to other researchers (Fig. 1).
Fig 1.
The data flow required by the Lung Tissue Consortium Radiology Data Coordinating Center. The flows provide feedback loops to assure that scans acquired at participating sites are received, processed, and archived. A variety of protocols including Web, DICOM, and mail are used to carry all the required messages and data payloads.
The imaging goals of this scientific endeavor include application of proven computer-assisted analysis algorithms and development of new effective and efficient tools to quantify pulmonary disease. The LTRC RCL systems and reporting for the LTRC are established to define the value of characterizing specific features of pulmonary disease through automated texture analysis, histogram analysis, vascular and bronchial attenuation patterns, regional pulmonary elasticity, architectural distortion, total and functional lung parenchymal volumes, and semi-automated segmentation of pathology.
The imaging informatics aspect of this project required establishment and maintenance of a computer system to receive clinically acquired and LTRC-specific CT datasets at each participating CC institution, de-identify these studies to LTRC-specific codes provided by the DCC, and transfer the image data to the RCL. A tracking system for the receipt and analysis CT images which matches the number and type of images in the clinical center to those stored in the RCL was developed, to assure that no data was lost in the transfer process and that blinded radiology results stored at the DCC were appropriate for the data acquired at the CC.
The specific systems developed for the image acquisition and transfer from the CC’s to the RCL include a generic DICOM receiver and viewer (eFilm Workstation version 2.9, Merge eMed corporation) on a standard PC at each clinical center. This DICOM receiver is configured as a destination for each CC's PACS and the CT scanners used to acquire the LTRC-specific CT protocols. Therefore, any studies performed for the LTRC or historical imaging that is to be used by the LTRC can be transferred to the LTRC CC workstation electronically through standardized DICOM transmission. The PC at each institution has unique password-protected access for each LTRC study coordinator, a firewall that limits incoming traffic to only DICOM information and automated antivirus and system patch updates in place.
The receipt of images on the CC workstation is tracked by an internal database and the series/image counts are utilized for study tracking by the DCC through a web-based form completed by the CC staff. The images on the CC workstation are de-identified by the CC staff using new LTRC ID’s provided by the DCC. A custom “scrubbing” utility parses the eFilm database for study information, loads DICOM images for a patient, removes and/or replaces identifying information in the DICOM headers of image data, and re-writes a de-identified copy of the DICOM files. Dual-entry of the LTRC ID into the scrubbing utility assures a low error rate for the de-identification process.
To minimize the potential of data loss, each CC is required to store the original identified/clinical image data acquired by the LTRC protocols either to a local archive or to archive media approved by the CT scanner manufacturer for the duration of the LTRC study. The transmission of the de-identified LTRC CT data from the CC’s to the RCL can be performed by electronic transfer (via a DICOM send) or through creation of media and physical transfer of the media to the RCL. However, given the very large size of standardized LTRC datasets (1 Gb) and limited bandwidth available at some clinical sites, the standard method of transfer to the RCL is DICOM-format images on DVD media packaged in damage-resistant mailers. This is a cost-effective and reliable way to transfer large datasets in the setting of non-urgent timeline for results reporting in the LTRC. The same software and mechanisms would be amenable to electronic methods, with error checking and correction to assure reliable transfer.
Upon exam receipt at the RCL, the DVD media are checked by a simple DICOM header parser and upload utility on a RCL PC that checks all DICOM data on the DVD and provides a study description and series/image count that is provided to the DCC through a web-based form entry system. This information is verified against the information provided from the CC to the DCC and an error report is generated if an exact match is not made. In the event of errors, reconciliation can be made through the same web-based system and requests for re-sending of data or resolution of errors is tracked by the DCC database and transferred to the appropriate CC.
The RCL utility transfers the DICOM image data to a research-specific storage pool within a highly available multi-tiered institutional DICOM archive. This archive includes multiple DICOM functions necessary for the storage of and transfer of medical imaging data. An internal database and rule-based management software allows this archive to perform the actions necessary in the Integration of the Health Care Enterprise (IHE) image management actor profile. Additionally, as for the LTRC archive, incoming data can be assigned to a logically separate storage pool within the entire archive. This allows clinical data from different areas and different research project data to be managed, verified by external systems and stored in different methods without necessitating separate systems for each project or clinical need. This storage pool is also configured to automatically forward new DICOM data from the archive to a research data processing server specific to the LTRC. Received image datasets are loaded on a dedicated LTRC 3D analysis workstation, by a 3D processing technologist. Software on this workstation is not designed for general image interpretation, but is optimized for display and volumetric analysis of lung CT data. This application is based on more generalized tools in the Analyze 7.0 and AVW image analysis software libraries (Mayo Biomedical Imaging Resource, Mayo Clinic, Rochester, MN) and the TCL/TK open source image analysis and visualization toolkits.
A board-certified radiologist views the CT image data, segmentation, and image analysis results and completes a structured report that includes both regional semi-quantitative reporting of specific pulmonary findings and coded diagnosis from a pre-defined list. This structured report is transmitted to the DCC through the same web-based forms utilized for RCL study tracking and other CC, and PCL results.
The DCC developed and maintains the database and computer software necessary to process, store, and analyze data furnished by the CCs, PCL, and RCL. The DCC controls data access by the investigators, study coordinators and research staff through a central database with specific access privileges for each registered staff member based on their role in the study and the site where they work. For a public resource, the DCC prepares a subset of public use data sets corresponding to tissue collections prepared by the PCL. The DCC will also direct the TCL and the RCL to provide specified LTRC resources and data sets to investigators approved by the NIH/NHLBI for access.
For any researchers applying for LTRC data, the DCC will coordinate the complication of relevant clinical/laboratory data and instruct the RCL and PCL regarding which tissue samples and image datasets are to be extracted and shipped to requesting investigators. At the RCL, this will be accomplished in an automated fashion through a batch of C-Move requests based on LTRC identifiers from a workstation to the LTRC DICOM storage pool. If additional changes in the LTRC identifiers are required before transfer to another investigator, the same scrubbing utility used for removal of patient identifiers could be utilized.
While the preceding works well enough in this particular case, there are some limitations:
Suppose a given participant of LTRC wants to read the cases from a remote site, there is currently no easy way to do this
Consider an LTRC participant who also is a member of another grant/contract. That person will need to exit the viewer they are using for the LTRC work, and remotely logon to a different computer to use a different viewer, if that is even possible.
Consider the difficulties of administering access rights on the image archive to dozens or hundreds of researchers, particularly if there is more than one archive.
Recent Developments: IHE and caBIG
Before delving into the details of imaging informatics, it is useful to consider just what informatics is. In general, the study of informatics is a body of facts and knowledge related to a domain of study, and how it is acquired, stored, transmitted, represented and mined for meaning. In the case of medical informatics there are several zones that scale up in size: bio-informatics (cell size and below), imaging informatics (organ systems), clinical informatics (patient sized systems) and health informatics (concerned with populations).4
Excepting imaging informatics, large multi-site trials have been adept for some time at conducting all types of informatics research. A recurring theme of this work is adoption of commonly agreed to testing procedures, clinical questionnaires and standard lexicons (such as RadLex) to describe results.5,6 However, even when a common vocabulary is defined, it becomes possible to share experimental protocols and results only when a common protocol is used to transmit such data. Often, the protocols used in the trial are very different form the normal clinical protocols. For many health care transactions, particularly clinical systems, that protocol is HL7 (Health Level 7).7 Using HL7, it is possible for laboratory, pathology, radiology and other departmental database systems to communicate their results to the electronic medical record (EMR) systems that are commonly referred to by clinicians in their daily practice. In its current widely used form (HL7 V2.5) the protocol uses standard message delimiting characters to frame commonly agreed to data elements. However, the protocol also permits large blobs of free text to be encapsulated in a transmission to other systems. While this makes the standard flexible, it also challenges researchers who must try to parse natural language reports in the HL7 stream for findings in a reproducible manner.
While HL7 enabled multi-site research trials to proceed apace with data that could be encoded and exchanged textually, imaging collaborations have been relatively sparse until recently. There are several reasons for this; not least is that the magnitude of data produced by imaging systems and unstructured text reports has made it very difficult to share results among sites until the wide availability of both broadband networks and universal protocols. Paradoxically, imaging informatics is in many ways both more, and less, advanced then its general health care informatics siblings. Less advanced in that standard lexicons exist, but are sparsely used making data mining very difficult; more advanced in that image features, acquisition parameters, and methods of communicating and sharing images are very standard. The primary reason for that latter is DICOM.
DICOM is the result of a partnership between the American College of Radiology and National Electrical and Manufacturing Association. The closest analog to the version in use today, DICOM 3.0, made its debut at the 1993 Radiological Society of North America (RSNA), InfoRAD exhibit where an open source implementation from the Mallinckrodt Institute of Radiology at Washington University known as the Central Test Node was debuted.8 In general, DICOM is based on the concept of service-object pairings. That is, there are objects (CT images, CR images, Structured reports, etc.) and operations that can be performed on them (FIND, GET, PRINT, STORE, etc.). DICOM has been a powerful unifying force in imaging informatics, and it no longer requires programming or soldering skills to interface a CT scanner to a laser filming device. However, when imaging equipment has to communicate with health care databases, HL7 enters the picture and as we have seen above, HL7 is a rather loose protocol. Because of this, the “simple” act of interfacing a RIS to a PACS is a major undertaking for vendors (and customers) every time a new combination is attempted.
Taken together, DICOM and HL7 have enabled great strides in imaging systems to communicate with each other, and even to share some non-image information with the other text based medical informatics systems commonly used such as the EMR. However, there have continued to be areas that have not been well served by merely addressing communication protocols, and multi-site imaging research is one of these. To address these implementation domain issues, it is useful to construct “use cases” that conceptually represent the actors in a transaction, and how they must interact to accomplish the goal of the use case. There are two different large scale initiatives that have taken this approach and they are described below.
IHE
In 1997 RSNA, Healthcare Information and Management Systems Society, several academic centers and a number of medical imaging vendors embarked on a program to solve integration issues across the breadth of health care informatics.9,10 IHE is an international initiative that rather than defining new protocols like HL7 or DICOM starts with several use cases and defines Integration Profiles that are implemented by health care Actors to accomplish the use case goal(s). The integration profiles are ultimately rendered as HL7 or DICOM messages, but IHE defines the correct behavior and message content, thereby removing much of the ambiguity in the standards. There are IHE Frameworks (profiles and actors) defined for Radiology, Cardiology, Lab, IT Infrastructure, Patient Care Coordination and Trial Implementations.11
Version (V6.0) of the IHE Radiology Framework defines 14 integration profiles, implemented by 25 actors: Scheduled Workflow, Patient Information Reconciliation, Consistent Presentation of Images, Presentation of Grouped Procedures, Access to Radiology Information, Key Image Note, Simple Image and Numeric Report, Basic Security (now Audit Trail), Charge Posting, Post Processing Workflow, Reporting Workflow, Evidence Document, Portable Data for Imaging, and NM Image.
The actors are: Acquisition Modality, ADT Patient Registration, Audit Record Repository, Charge Processor, Department System, Scheduler/Order Filler, Display, Enterprise Report Repository, Evidence Creator, External Report Repository, Image Archive, Image Display, Image Manager, Master Patient Index, Order Placer, Performed Procedure Step Manager, Portable Media Creator, Portable Media Importer, Post-processing Manager, Print Composer, Print Server, Report Creator, Report Manager, Report Reader, Report Repository, Secure Node, Time Server.
Clearly, much of the capability implied by these lists overlaps with the functionality needed to perform, transfer data, report, store and track charges related to a research examination. Some of the functionality needed for clinical practice may not be needed for a multi-site project, however, there is some functionality missing in this list too—notably de-identification tools and more sophisticated access controls based on IRB member lists.
caBIG
Bringing together the combined efforts of numerous workers, the National Cancer Institute created the caBIG (Cancer Bioinformatics Grid) consortium in 2004 to create standard lexicons to describe treatment protocols, and then promote standard analytic tools and result sharing in widely accessible formats. To date, caBIG defines four Workspaces with a plethora of applications under each: Clinical Trial Management Systems, Integrative Cancer Research, In vivo Imaging, and Tissue Banks and Pathology Tools.12
Resources built under the caBIG architecture know of each other’s existence and can share data using a web framework known as the Globus Toolkit, a grid computing toolkit developed by the physics community to perform distributed computing on non-heterogeneous networks.13 Message content is passed among nodes via the Extensible Markup Language protocol, which is the protocol used by HL7 V3.0. Furthermore, authentication is performed with government approved standards (X.509 certificates or assertion based authentication).14 Once authenticated, user rights (authorizations) are retrieved from LDAP (Lightweight Directory Access Protocol) servers that permit system administrators to create authorization rules for specific caBIG modules (for example the Protein Management System may require users to be Medical Doctors with 10 years practice experience before granting access).
A key point to note is, caBIG does not specify data exchange using DICOM, so while healthcare systems that use HL7 V3.0 will be caBIG capable, the vast majority of PACS and imaging modalities will require intervening computers to broker the DICOM to caBIG translation.
Discussion
While the IHE Radiology Framework provides a comprehensive framework for integrating a clinical enterprise, only a subset of those Actors are required to construct a useful multi-site, clinical imaging trials system. Conversely, the caBIG approach eschews the complexity of the clinical world and has developed fine tools that concentrate solely on research needs, but this also has drawbacks. Ideally, one should not require radiologists to use two different platforms to accomplish both their clinical and research duties. It therefore comes down to the following question, “What path will vendors be more likely to implement in their largely clinically focused products; a platform developed by the medical research arm of one nation’s government, or a standard that is international in scope and already leverages other international standards?” Assuming the latter, this Discussion will study those IHE concepts most helpful to overcome the difficulties outlined in the “Current Practice” section, and the relevant IHE implementation profiles to a research trial. As was shown in the “Current Practice” section, the primary workflows in multi-site imaging research are:
image the patient with research protocols and transmit the patient identified exam to the exam anonymizer
anonymize the studies and substitute the IRB approved Study ID before sending to the Imaging core lab's archive
at the core lab, acknowledge the receipt of the images and verify that the total study content was received
store the exam under its study ID in the core lab image archive
pull the exams to the research image processor, perform processing, present it to the DICOM General Purpose Physician Worklist where the researcher reads and reports findings to the Core Results Repository.
Archive the imaging Study and Results for long term access to the trial participants and to allow researchers who will perform retrospective data mining on the results
Point A is addressed via the current generation of IHE/DICOM compliant acquisition devices that can perform reproducible scan protocols and share the results to external systems. These devices are described by the IHE Acquisition Modality Actor and must support at least the Scheduled Workflow Implementation Profile, Consistent Presentation of Images, Presentation of Grouped Procedures and Key Image Notes. The advantage to the researcher of these profiles is that they are assured that if the image displays device they use supports these profiles as well, they are seeing the exam exactly the way the performing technologist saw it as the patient was scanned. Further, the Grouped Procedures support assures that the right images are profiled to the right exam orders and thus will display correctly against other exams of that same body part.
Point B was not well represented by IHE until V6.0 via a new supplement. Named “Teaching File and Clinical Trials Export Profile”, this implementation profile seeks to achieve the ability to “… select images, series or studies (which may also contain key image notes, reports, evidence documents and presentation states) that need to be exported for teaching files or clinical trials”[15-04-22]. The supplement also “… defines an actor for making the export selection, which would typically be grouped with an Image Display or Acquisition Modality, and an actor for processing the selection, which is required to support a configurable means of de-identifying the exported instances”. These new actors are named Export Selector and Export Manager, respectively. These actors will typically be grouped with an Image Display Actor which also provides the ability to receive DICOM exams over a network via the push model.
Point C is addressed by the IHE Image Manager/Archive Actors by supporting IHE Scheduled Workflow (and in particular the DICOM Store Commit function) between the archive and the Image-Display/Export Actors from Point B. The benefit of this workflow is that it guarantees correct study demographics, profiles to the proper worklist, validates the accuracy of data transmission on the sending and receiving sides, and de-identifies the exam for HIPAA compliance.
Point D is served by the Image Archive Actor that will store the exam (until the scheduled deletion time) awaiting fetch requests from authenticated and authorized agents. By combining the Archive Actor with other features implemented by the Authentication Actor, it is possible for a single archive to contain exams from many unrelated clinical trials, and enforce access restrictions on users so that they only see those exams for trials where they are a member.
Point E is covered by the Post-Processing Workflow Manager Actor that schedules (or honors queries for) exams between the Image Archive/Manager Actors and the Image Display Actor. There are numerous profiles that would need to be covered in a fully IHE compliant clinical system, but for trials the relevant implementation profiles are: Scheduled workflow, Post-processing workflow, Consistent Presentation of Images, Key Images, and Access to Radiology information. Together these profiles assure that Dr. Rad sees the research exams in a known worklist, with relevant priors (and histories if permitted by the research protocol), and that the images have the same quality and presentation state they were prepared with by the technologist.
Finally, point F is serviced via sending the final imaging exam to yet another Image Archive Actor, whilst results are created and stored by Report Creator/Manager/Repository Actors that implement Reporting, Evidence Documents, and Simple Image and Numeric Workflow Profiles. As in other cases, by judicious use of Authentication Actor functions, a single Report Repository can store results from many trials, and restrict Trial member A from seeing results that were stored under Trial B. However, there remains the issue of structured formatting of the report that can foster later data mining; this is likely where caBIG efforts could combine with existing DICOM standards such as DICOM SR to facilitate both a constrained vocabulary and a strict data format.16
For contrast it is useful to examine a teaching file project developed by the RSNA. It has been suggested that the RSNA Medical Imaging Resource Center (MIRC) teaching-file project could be pressed into multi-site clinical imaging trials service.17 Indeed recent features have been added with this in mind. MIRC provides the ability to create trusted federations of MIRC sites which can cross query each other’s image stores. In addition, MIRC supports centralized user/role management tools and the Principle Investigator in charge of a MIRC store can anonymize data coming in from each contributor to the store.18,19 In many ways, MIRC realizes several of the requirements for a pure clinical trials system. What separates it from the ideal system described in the Introduction are several features:
It is a separate system from the clinical viewer/reporting system (i.e., no IHE/HL7 reporting support) requiring the Radiologist to break out of their normal workflow
MIRC uses a separate viewing application with a different user interface
There is no support for physician interpretation worklists (no automatic comparison exams)
There is no support for automated hanging protocols to present studies to the radiologist with a consistent look and field regardless for where the exams were acquired.
Nevertheless, for a project that began as a teaching file, MIRC has made significant inroads into the requirements of a multi-site imaging trials tool as listed in the Discussion under points A–F.
Conclusions
From the preceding, it is clear that there is great overlap in the functional requirements of multi-site imaging trials, teaching applications and the clinical workflow challenges already addressed by IHE and other standards to deal with enterprise imaging. While the full set of IHE Actors is not needed to fulfill imaging trial objectives, a moderate set consisting of the following actors would likely prove useful:
Image Acquisition
Image Display plus Export Selector/Manager
Image Manager/Archive
Post Processing Manger
Image Display with the research image processing tools
Report Creator/Manager/Repository
Authentication (to perform authentication and authorization of the above)
A modern clinical PACS should already implement the prior list, so with the simple addition of the IHE Teaching File and Clinical Trials Export profile, most institutions with a recent PACS could participate in multi-site trials. However, the data mining potential of such trials would be extended if the structured lexicons of caBIG were somehow married to the structured data formats of DICOM (i.e., DICOM SR)
References
- 1.Langer S. Radiology speech recognition: workflow, integration, and productivity issues. Curr Probl Diagn Radiol. 2002;31(3):95–104. doi: 10.1067/cdr.2002.125401. [DOI] [PubMed] [Google Scholar]
- 2.Langer SG, Stewart BK. Computer security: a primer. J Digit Imaging. 1999;12(3):114–131. doi: 10.1007/BF03168630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MS, Sumit SK, Pais RC, Lee YZ, McNitt-Gray MF, Goldin JG, Cardenas AF, Aberle DR. Database Design and Implementation for Quantitative Image Analysis Research. IEEE Transactions on Information Technology in Biomedicine. 2005;9(1):99–108. doi: 10.1109/TITB.2004.837854. [DOI] [PubMed] [Google Scholar]
- 4.Blois MS, Shortliffe EH. The computer meets medicine: Emergence of a discipline. In: Shortliffe EH, Perreault LE, editors. Medical informatics: computer applications in health care. Reading, MA: Addison–Wesley; 1990. p. 20. [Google Scholar]
- 5.Rubin DL. Creating and curating a terminology for radiology: ontology modeling and analysis. J Digit Imaging. 2008;21(4):355–362. doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langlotz CP, Caldwell SA. The completeness of existing lexicons for representing radiology report information. J Digit Imaging. 2002;15(Suppl 1):201–205. doi: 10.1007/s10278-002-5046-5. [DOI] [PubMed] [Google Scholar]
- 7.Hammond WE. HL7—more than a communications standard. Stud Health Technol Inform. 2003;96:266–271. [PubMed] [Google Scholar]
- 8.Bidgood WD, Jr, Horii SC. Introduction to the ACR-NEMA DICOM Standard. Radiographics. 1992;12(2):345–355. doi: 10.1148/radiographics.12.2.1561424. [DOI] [PubMed] [Google Scholar]
- 9.Channin DS. M:I-2 and IHE: integrating the health care enterprise, year 2. Radiographics. 2000;20(5):1261–1262. doi: 10.1148/radiographics.20.5.g00se391261. [DOI] [PubMed] [Google Scholar]
- 10.Channin DS, Parisot C, Wanchoo V, Leontiev A, Siegel EL. Integrating the health care enterprise, a primer: part 3, what does IHE do for me?”. Radiographics. 2001;21(5):1351–1358. doi: 10.1148/radiographics.21.5.g01se401351. [DOI] [PubMed] [Google Scholar]
- 11.Carr CD. IHE: a model for driving adoption of standards. Comput Med Imaging Graph. 2003;27(2–3):137–146. doi: 10.1016/S0895-6111(02)00087-3. [DOI] [PubMed] [Google Scholar]
- 12.Kakazu KK. The Cancer Biomedical Informatics Grid (caBIG): pioneering an expansive network of information and tools for collaborative cancer research. Hawaii Med J. 2004;63(9):273–275. [PubMed] [Google Scholar]
- 13.Liu BJ, Zhou MZ. Utilizing data grid architecture for the backup and recovering of clinical image data. Comput Med Imaging Graph. 2005;29(2–3):95–102. doi: 10.1016/j.compmedimag.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Lin K, Daemer G et al (2006) caBIG security technology evaluation white paper. an NCI caBIG Architecture Workspace Project
- 15.TI 2005-04-22 TCE Rev. 1.1. The IHE Teaching File and Clinical Trial Export Draft, copyright ACC/HIMMS/RSNA.
- 16.Clunie DA. DICOM structured reporting and cancer clinical trials results. Cancer Inform. 2007;4:33–56. doi: 10.4137/cin.s37032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegel E, Channin D, Perry J, Carr C, Reiner B. Medical image resource center: an update on the RSNA's medical image resource center. J Digit Imaging. 2002;15(1):2–4. doi: 10.1007/s10278-002-1000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel E, Reiner B. The radiological society of North America's medical image resource center: an update. Digit Imaging. 2001;14(2 Suppl 1):77–79. doi: 10.1007/BF03190302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MIRC Site. http://mircwiki.rsna.org/index.php?title=Main_Page. Last viewed September 2009.