Abstract
The retroviral Gag polyprotein mediates viral assembly. The Gag protein has been shown to interact with other Gag proteins, with the viral RNA, and with the cell membrane during the assembly process. Intrinsically disordered regions linking ordered domains make characterization of the protein structure difficult. Through small angle scattering and molecular modeling we have previously shown that monomeric human immunodeficiency virus (HIV-1) Gag protein in solution adopts compact conformations. However, cryo-electron microscopic analysis of immature virions shows that in these particles, HIV-1 Gag protein molecules are rod-shaped. These differing results imply that large changes in Gag conformation are possible and may be required for viral formation. By recapitulating key interactions in the assembly process and characterizing the Gag protein using neutron scattering, we have identified interactions capable of reversibly extending the Gag protein. In addition, we demonstrate advanced applications of neutron reflectivity in resolving Gag conformations on a membrane. Several kinds of evidence show that basic residues found on the distal N & C-terminal domains enable both ends of Gag to bind to either membranes or to nucleic acid. These results, together with other published observations, suggest that simultaneous interactions of a HIV-1 Gag molecule with all three: protein, nucleic acid and membrane, are required for full extension of the protein.
Keywords: retroviral assembly, neutron reflectivity, SANS, disordered proteins, tethered membranes
Introduction
Expression of a single retroviral protein, termed Gag, is sufficient for virus particle formation in mammalian cells.1; 2 Gag is a multi-domain protein always containing, from N- to C-terminus, a matrix (MA) domain, a capsid (CA) domain, and a nucleocapsid (NC) domain. The particles assembled from Gag are immature virions; in most retroviruses, including lentiviruses such as HIV-1, these particles form as Gag accumulates at the cytoplasmic face of the plasma membrane. Targeting of Gag to the plasma membrane is a function of the MA domain and involves, on the one hand, an electrostatic interaction between basic residues in MA and anionic lipids in the membrane, and on the other hand, a hydrophobic interaction between membrane lipids and myristic acid, a 14-carbon saturated fatty acid at the extreme N-terminus of the protein.3 The protein-protein interactions leading to virus assembly are largely or exclusively a function of the CA domain, while the NC domain plays a principal role in the interactions of Gag with nucleic acids.
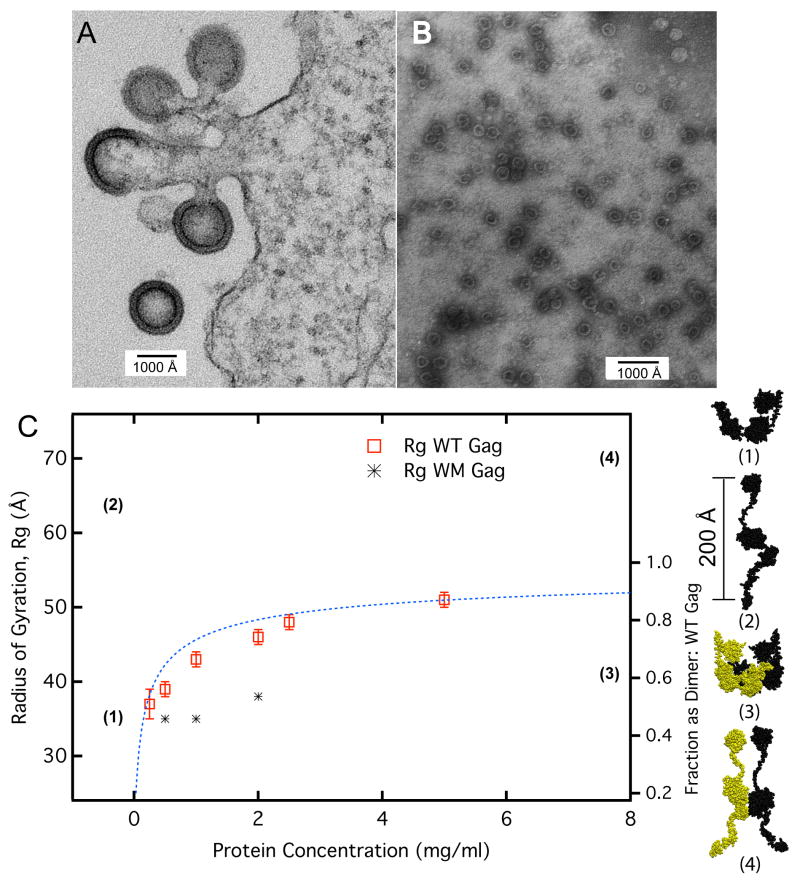
Immature retrovirus particles are roughly spherical, with an average diameter of around 1100 Å. The Gag proteins in these particles are extended rods, approximately 200 Å long.4 They are arranged as radii of the particles, with their N-termini in contact with the lipid bilayer that surrounds the particle and their C-termini projecting into the interior of the particle, presumably in contact with RNA. A transmission electron micrograph of these particles, budding from the plasma membrane of a mammalian cell, is shown in Fig. 1A.
Figure 1.
Gag dimensions. (A) Gag assembly at different stages of immature virion formation imaged by EM as described in supplementary methods. Protein shell in both the spherical virion and incomplete arcs has a thickness of ≈ 200 Å. (B) Recombinant HIV Gag VLPs assembled in vitro with yeast tRNA under standard conditions6 (described in supplementary methods) at physiological salt buffers and visualized by negative stain EM. These VLPs have a diameter 3× smaller than that of native virions and the protein shell is only 70 to Å 80 Å in thickness. (C) Average protein size in solution as a function of protein concentration. Radius of gyration (Rg) was measured by SANS for WT Gag and WM Gag. The percent of WT Gag molecules as dimer was calculated (dashed blue line) using a Kd of 3.9 μM in D2O.14 WM Gag has a binding constant 100× weaker than WT and was assumed to be pure monomer over this concentration range. Models for Gag monomer and dimers are shown on the right based on SANS results for the compact state5 and Cryo EM data for the extended state.4 The dimerization interface solved by solution NMR16 for the CA domain was used as a template for generating the dimer complex. The calculated radius of gyrations for both the compact/extended and monomer/dimer configurations are shown on the plot by their numerical designation. (1), monomer in solution; (2), extended monomer, as in authentic immature virions; (3), dimer of (1); (4), dimer of (2).
We have previously characterized the properties of recombinant HIV-1 Gag protein, purified from bacteria.5 This protein differs from authentic, wild type HIV-1 Gag protein in lacking both the myristic acid modification at its N-terminus and a domain at the extreme C-terminus termed p6. When nucleic acid (NA) is added, this protein assembles into virus-like particles (VLPs); however, these VLPs are far smaller than authentic retrovirus particles, with a diameter of only 250 Å to 300 Å.6 Fig. 1B shows examples of the small VLPs formed with NA alone. These VLPs are too small to be composed of 200 Å rods of Gag. In fact, the shell of protein forming these VLPs appears to be only 70 Å to 80 Å thick. However, correct size VLPs are assembled if inositol pentakisphosphate (IP5) as well as NA is added to purified Gag.7 The striking difference between the particles in Fig. 1A and 1B implies that HIV-1 Gag can adopt alternative conformations, one extended (forming authentic particles) and one bent (as in the VLPs in Fig. 1B).
This conformational freedom presumably reflects the flexibility of the linker regions between the domains in the Gag poly-protein. Solution state NMR demonstrated the MA and CA domains to be rotationally uncoupled by a 26 amino acid linker.8 Structural determination of the CA domain by crystallography revealed five disordered residues linking separate N-terminal and C-terminal domains.9 Furthermore, NC, p6 and p2 domains appear to be largely unstructured, except for the Zn-finger regions within NC.10; 11; 12; 13 Difficulties in characterizing intrinsically disordered domains have led to a lack of high-resolution information for the intact Gag protein.
In previous investigations, structural properties of recombinant Gag protein in solution were probed using hydrodynamic data and small-angle neutron scattering (SANS) together with molecular modeling.5 This study used a Gag mutant, designated WM Gag, that was inhibited from dimerizing. The results indicated that the protein is compact in solution, with its terminal domains situated close in 3-dimensional space. It seems likely that the protein monomer in solution actually adopts an ensemble of inter-converting, relatively compact structures.
In authentic immature particles, each Gag molecule is in contact with other Gag molecules, with the lipid bilayer surrounding the particle, and with the RNA within the particle. The question then arises as to which of these interactions is responsible for the extension of the Gag protein in these particles. In the present work, we have explored the conditions under which HIV-1 Gag protein is compact, as it is in free solution and small VLPs, or extended, as it is in immature virions. We used neutron-scattering methods to dissect the contributions of protein-protein, protein-lipid, and protein-NA interactions to the extension of the protein.
Protein-protein interactions
HIV-1 Gag exists in solution in monomer-dimer equilibrium.14 The interface mediating the dimeric interaction is in the C-terminal portion of the CA domain. In order to determine whether dimerization leads to extension of the wild-type (WT) protein, we used SANS to measure the mean scattering particle size or radius of gyration, Rg, of wild-type Gag over a broad concentration range. Results of this experiment are shown in Fig. 1C. The figure shows the Rg data (left axis) for WT Gag and WM Gag as a function of protein concentration; for the WT Gag, the fraction of protein present in dimers is depicted with blue dashed line and was calculated using a Kd of 3.9μM in D2O buffer.14
As shown in Fig. 1C, the Rg of the WT Gag protein increased as the dimeric fraction increased. At low concentration (0.25 mg/ml, or 5 μM), where 55% of the molecules are in dimers, the weight-averaged Rg is 38 Ǻ, slightly larger than the monomeric Gag value of 35 Ǻ. As the protein concentration increases, the Rg increases, reaching a plateau at ≈ 50 Ǻ; at the maximum concentrations used here (100 μM), > 90% of the molecules are in dimers.
As a frame of reference, models of the compact and extended state of Gag were constructed to provide Rg limits for the monomeric and dimeric protein. The compact Gag conformations were taken from the set of representative structures determined by Datta et al.5 The extended conformation was based on Cryo-EM data of Fuller et al.4 The Cryo-EM results provided distributions of the structured Gag domains arranged radially from the membrane surface in the immature virion. Starting with the compact model, backbone dihedral conformations of the flexible linkers were varied until the Cryo-EM distributions constraints were satisfied. Further details on modeling and defining the structured and flexible regions are given in the Supplementary Material. The software program SASSIE15 was used to perform the conformational search. Even in the extended state multiple linker conformations were possible but the differences in Rg were < 5%. Therefore a representative structure was chosen for experimental comparison. To generate dimers, the Gag monomers were oriented with respect to each other using the dimerization interface between CA domains solved by solution NMR16 as a reference template.
The experimental results indicate that the actual Gag dimer adopts Rg values larger than the compact dimer model but smaller than the extended dimer model. Even at concentrations where 90 % of the population is in dimers, protein dimensions are not consistent with the extended dimer. Rather, dimeric Gag molecules in solution appear to adopt an ensemble of conformations that are shifted towards intermediate, partially extended configurations. Hence protein-protein interactions are not sufficient to fully extend the Gag protein.
Protein-Nucleic Acid Interactions
Within the immature virion the NC domain of Gag is believed to associate with RNA. In vitro studies also demonstrate the importance of Gag-nucleic acid interactions, as assembly of recombinant Gag protein into VLPs can be triggered by addition of RNA or single-stranded DNA.6; 17; 18 However, the effect of nucleic acid on Gag protein conformation has not been previously investigated. The size of recombinant Gag in the presence of single-stranded DNA was examined using SANS. Oligodeoxynucleotides either 5 (TG×2.5) or 14 (TG×7) bases long with the repeating sequence (TG)n were added to Gag, as HIV-1 NC has very high affinity for this sequence.19 Both TG×2.5 and TG×7 bind the NC domain of Gag tightly, but only TG×7 supports assembly, though relatively inefficiently and at low ionic strength.6 Protein and DNA were incubated together in 0.5 M NaCl buffer. Under these conditions, fluorescence anisotropy assays showed that the Kd for protein-DNA binding is below 1 μM (Suppl. Fig. 2), but the binding does not lead to VLP assembly. SANS measurements were performed using a 4 to 12 fold excess of DNA to ensure protein saturation under these conditions.
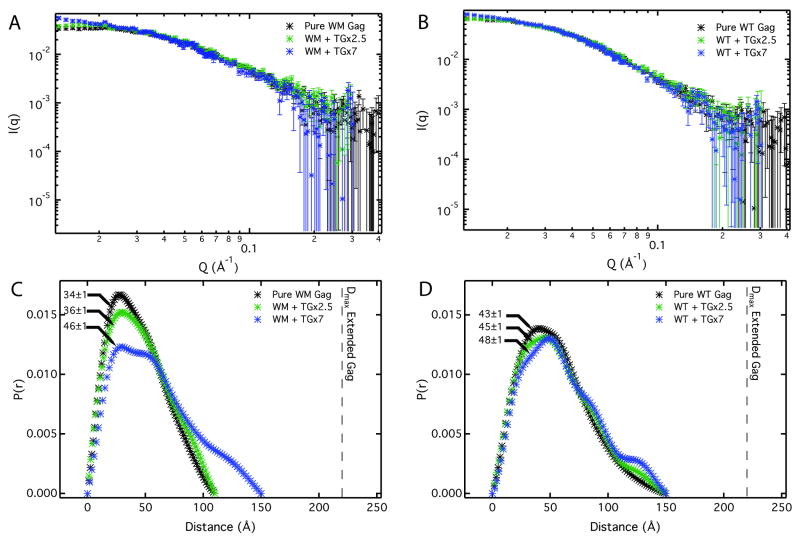
Both WM and WT Gag were studied in order to isolate the contribution of protein-nucleic acid and protein-protein-nucleic acid interactions. The SANS profiles for WM Gag, presumably resistant to protein-protein interactions, are shown in Fig. 2A; little difference is seen between the pure protein (black) and the protein-5-base DNA, TG×2.5 (green) mixture. The sample containing the 14 base, TG×7, (blue) resulted in a slight increase in scattering at low q indicating the presence of a small population of higher order protein oligomers. WT Gag treated similarly was nearly invariant for all protein samples (Fig 2B). The changes in the spectra are due to alteration in protein conformation, as the nucleic acid does not contribute to the observed scattering under the contrast conditions used.
Figure 2.
The influence of single stranded DNA on Gag conformation. WM and WT Gag protein at 20 μM were incubated with short nucleic acid segments containing a TG base sequence repeat. Two different lengths TG×2.5 and TG×7 were used at 240 μM and 80 μM respectively. The SANS spectra of WM Gag and WT Gag are shown in Panel A-B. The pair-distance distribution P(r) determined from the SANS data are given in Panels C & D with Rg values for each condition indicated on the plot. Incubation was performed at 0.5 M NaCl to inhibit protein condensation in the presence of nucleic acid. Gag-TG binding constant, Kd, was determined to be < 1 μM by independent fluorescence anisotropy measurements (see Suppl. Fig. 2). Excess nucleic acid assured complete Gag binding in the SANS experiment.
The pair-distance distribution functions, P(r), shown in Figure 2C and 2D were determined from the SANS profiles and provide both the mean Rg and the maximum dimensions of the protein molecules in solution. For WM Gag, Fig 2C, the pure protein and protein in the presence of short TG×2.5 NA had Rg's of 34 Å ±1 Å and 36 Å ±1 Å, respectively. The resulting molecular sizes are similar to what was found for a compact monomeric protein in solution (Fig. 1C, WM Gag). The complexes formed on TG×7 have an Rg of 46 Å ±1 Å, similar to that determined for WT Gag dimers in solution (Fig 1C). The value of Dmax for WM Gag bound to TG×7 increases to that of WT Gag in solution. It seems plausible that the 14-base strand can bind two Gag molecules via interactions with the NC domain, and thus promote Gag dimerization despite WM Gag's reduced inter-protein affinity. Indeed, WM Gag is capable of assembly into small VLPs in vitro.5 Similar sized complexes were found for WT Gag both as pure protein and in the presence of the TG DNA, where Rg's ranged from 43 Å ±1 Å to 48 Å ±1 Å. NA had little effect on molecular sizes, possibly because WT Gag dimerizes spontaneously in solution. Hence, both WM and WT Gag form similar sized complexes upon binding TG×7. However, as shown in Fig. 2C and 2D, the maximum dimensions observed in both cases were significantly below the expected Dmax calculated from the extended Gag model shown in Fig 1C. These results suggest protein–NA interactions are also not sufficient to extend Gag.
In the presence of longer nucleic acids such as yeast tRNA, ≈ 80 nucleotides in length, Gag protein readily condenses in vitro, forming organized structures like those shown in Fig 1B. These condensed VLPs present a model system for strongly coupled protein-protein-nucleic acid interactions. As discussed previously these VLPs have a diameter and protein shell thickness indicative of a compact Gag protein. Thus, additional interactions are required for Gag protein to adopt extended conformations.
Protein-Membrane interactions
HIV-1 particle assembly occurs at the inner face of the cell membrane. Therefore the membrane itself might play an important role in protein structure. To investigate protein structure and interaction with lipid membranes, a novel system has been engineered as a cell membrane bio-mimetic. We used a tethered bilayer lipid membrane (tBLM) system consisting of a 14-carbon lipid in which the headgroup has been replaced with a short polyethylene glycol linker terminated by a thiol group.20 The thiol anchors the lipid tether to a gold film on a solid Si wafer substrate. The lipid tether sparsely covers the surface and nucleates the formation of a lipid membrane during a rapid solvent exchange procedure.20; 21 The resulting membrane is separated from the support by a 20 Å to 30 Å, highly hydrated layer. We showed recently that the diffusivity of phospholipids in the leaflet distal to the solid support is indistinguishable from that in vesicle membranes.22 The tBLM system by design allows us to control the conditions of protein membrane binding while performing in situ structural measurements.
Lipid membrane and protein structure in the tBLM were investigated by neutron reflectivity (NR). With the technique of NR, the intensity of neutrons reflected off of thin films at increasing grazing angles is measured. At these low angles neutrons penetrate the film surface a few thousand Angstroms. Therefore neutrons that reflect off of buried interfaces (e.g. the interface between the lipid bilayer and the Au substrate or the Au layer and the Cr layer of the substrate) interfere both constructively and destructively, thereby modulating the reflected intensity. The results are interpreted in terms of a 1-D neutron scattering length density (nSLD) profile. The nSLD profile depends on the structure and distribution of lipid groups and protein normal to the membrane plane. NR is one of the few methods that can characterize membrane protein structure in physiologically relevant conditions, that is, on a fluid lipid-bilayer.
Because NR averages over the in-plane organization of molecules on the membrane surface, the nSLD profile is represented using homogenous slabs organized normal to the membrane. The thickness and neutron scattering density of each slab is dependent upon the distribution of chemical groups that the slab represents (lipid chain, lipid headgroup, protein, etc.) Analysis of NR data requires using the slab model and computing a trial reflectivity spectrum by the Parratt formalism.23 Optimization of the slab model parameters is performed to fit the experimental reflectivity data. The resulting nSLD profile is then interpreted in terms of the underlying biological architecture. Uncertainties in our model parameters are rigorously defined using a Monte Carlo error analysis.24 This analysis has been implemented in a customized version of the software suite ga_refl,25 provided by the NIST Center for Neutron Research (NCNR).
Gag binding was studied on a tBLM system composed of 30 % anionic phosphatidylserine (PS) and 70 % neutral lipids. This system has a negative charge density similar to that found in the native viral membrane, but lacks the full complexity of the HIV lipodome.26 The Gag MA domain targets the membrane by a combination of electrostatic interactions originating from a patch of basic residues and hydrophobic interactions originating from an N-terminal myristate group thought to insert into the bilayer. However, the Gag construct in this study lacked the N-terminal myristate group and therefore relied on the contribution of electrostatic interactions to drive membrane binding. Earlier investigations have shown that under certain conditions electrostatics by itself is capable of orienting the MA domain in a manner consistent with proper viral assembly.27
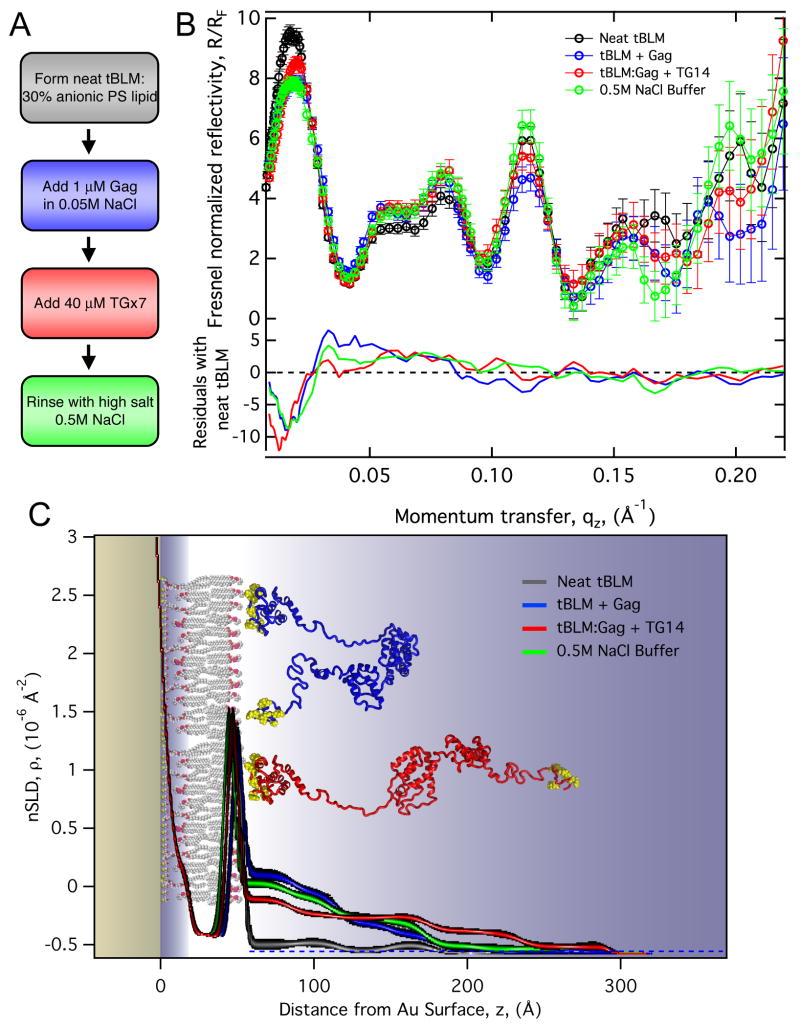
A sequence of experiments outlined in Fig 3A, were conducted to investigate the structure of Gag protein on the model membrane. Initially the neat negatively charged bilayer was formed and measured by NR to ensure a defect-free, > 90 % complete, membrane over the wafer surface. Gag was then introduced into the aqueous phase, where it bound to the membrane. The bound protein was then incubated with the TG×7 to observe the effect of nucleic acid on the membrane-bound protein. Finally the TG×7 construct was removed from the Gag molecules by flushing the sample cell with a high salt (0.5 M NaCl) buffer.
Figure 3.
NR results of WT Gag protein interacting with a tBLM indicates conditions that alter Gag conformation. Panel A, the sequence of measurements performed in situ on the reflectometry instrument. The steps are: 1) Formation of a complete tBLM. 2) Binding WT Gag (buffer: 0.05M NaCl, .001M NaPO4, 5mM TCEP, pH 7.4). 3) Binding of TG×7 DNA to the Gag protein layer. 4) Disassociation of TG×7 using a high ionic strength buffer (same as binding buffer except 0.5 M NaCl). For each experimental condition, three different isotopic aqueous buffer contrasts (mixtures of H2O and D2O) were used. Panel B, the resulting reflectivity spectra for the series of measurements showing the pure H2O buffer data only. Differences in reflectivity from the neat tBLM condition are given as residuals in the bottom of the panel. Panel C, a 1-D nSLD profile of the membrane and Gag determined by fitting the reflectivity data to a “slab model” (see main text for details). Line widths represent the 95 % confidence limits. The inset showing WT Gag cartoons are illustrative models of protein conformations consistent with the overall dimensions determined by reflectivity.
Changes in the reflectivity spectra indicate both binding of Gag and modulation of protein conformation due to the different experimental conditions. Figure 3B shows the reflectivity profiles measured in the H2O aqueous buffer contrast. Differences between the Gag protein conditions and the neat bilayer are shown as residuals in the bottom part of Fig 3B. For each experimental condition reflectivity spectra were measured using three separate isotopic aqueous buffer contrasts (pure H2O, a 1:2 H2O:D2O mixture and pure D2O). The scattering from each contrast provides distinct reflectivity spectra, while the underlying protein and membrane structures are invariant. The reflectivity spectra from the additional buffer contrast conditions are provided in Supplementary Figure S1.
Interpretation of the reflectivity profiles for these complex systems is only possible by the simultaneous fitting of a slab model to the complete set of reflectivity data. A “free-form” interpretation of the protein region (adjacent to outer leaflet headgroups) was used where eight ≈ 30 Å slabs were allowed to vary independently. Hence the best-fit nSLD profile is not based on any assumptions about protein structure. The nSLD profiles are shown in Fig. 3C where line widths are 95 % confidence limits determined by Monte Carlo error analysis. A table of fit parameters used in the slab model that best fit the data and the parameter uncertainties are in Supplementary Table 1.
The nSLD profile for the neat bilayer is shown in Fig 3C (black line) and indicates a nearly defect-free membrane, showing only bulk solvent after the outer-leaflet headgroup region. When Gag was introduced, it formed a protein layer with total dimensions of ≈ 90 Å, Fig 3C (blue line). The subsequent introduction of single stranded TG×7 DNA causes a shift in Gag dimension, extending ≈ 190 Å, Fig. 3C (red line). These dimensions agree well with Gag protein dimensions in the immature virion.4 A nearly complete recovery of the compact state of Gag was induced by the high salt rinse, Fig 3C (green line). The inset showing Gag cartoons structures are illustrative models of possible protein conformations that are consistent with the overall dimensions determined by reflectivity. The SLD profile is likely an average of an ensemble of Gag conformations due to the intrinsically disordered regions between the structured domains. Protein extension was observed under conditions where 30 % surface coverage by Gag was achieved. Additional reflectivity measurements found that below this minimal coverage on the tBLM, Gag protein remained folded (data not shown). Therefore neighboring Gag-Gag interactions are apparently as important as Gag-membrane and Gag-nucleic acid interactions in contributing to the extension of the protein.
Our results show that on their own, neither protein-protein binding, protein-nucleic acid binding, nor protein-membrane binding is sufficient to trigger the rod-shaped conformation that Gag assumes in immature particles. However simultaneously, all three binding interactions extend Gag to dimensions that are consistent with the immature virus particles. Based on the results reported here and the literature of binding interaction studied for the different Gag domains, a consistent model for how Gag conformations are modulated can be formed. This model is depicted schematically in Fig. 4.
Figure 4.
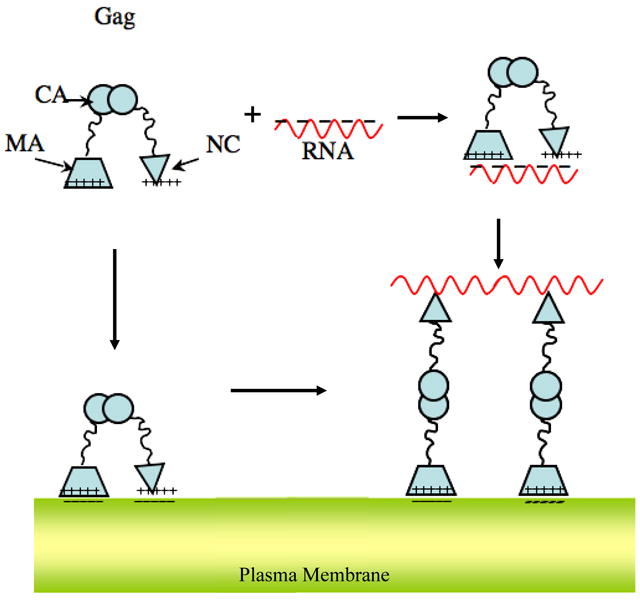
A model for Gag extension consistent with structural characterization and known binding interactions of the protein. The Gag molecule is compact in solution, even when bound to RNA. RNA binds to the NC domain with very high affinity, but evidently can also bind, with lower affinity, to the MA domain. The MA domain targets and anchors Gag to the anionic surface of the plasma membrane. Additionally the NC domain may also associate with the membrane through electrostatic interactions. Only in the presence of all three components i) protein, ii) nucleic acid and iii) membrane, extended Gag is formed. A plausible mechanism is one in which the viral RNA and the membrane separate the terminal domains through their preferential interaction. Cross-linking several membrane bound Gag molecules together by the RNA strand and inter-protein interactions may further stabilize extended Gag.
Salient features of the mechanism are a flexible Gag polyprotein, capable of adopting compact conformations with terminal MA and NC domains in close proximity. The prevalence of positive charges on MA and NC domains allows them to interact simultaneously with polyanionic targets such as nucleic acids or anionic membranes, maintaining the protein in compact conformation(s). Thus compact Gag is observed in the presence of only nucleic acids (Fig.2) or only membranes (Fig.3). However, in the presence of both membranes and nucleic acid, each of the domains binds its preferred target, allowing for extension of the protein – presumably mediated by Gag-Gag interactions. It should be noted that the composition of the membrane might influence this process in vivo.
In fact, a number of disparate observations are all in conformity with this picture. The N-terminal MA domain in Gag has been reported to associate with RNA likely through a patch of basic residues electrostatically attracted to the negatively charged RNA polymer.28; 29 IP5 (which resembles the anionic headgroups in the plasma membrane), together with nucleic acid, induces assembly of correctly sized VLPs7. Interestingly, both ends of the protein participate in stringent binding of IP6 in free solution;14 footprinting shows that both domains can bind either RNA or phosphatidyl inositol (4,5) bisphosphate (a plasma membrane-specific phospholipid);30 free MA as well as free NC can bind RNA; 31; 32 and binding to RNA seems to modulate membrane interactions of Gag, contributing to the specific targeting of Gag to the plasma membrane.31; 33 Furthermore the presence of the MA domain interferes with the ability of Gag to catalyze the annealing of tRNA to a complementary stretch in viral RNA, but this inhibition is relieved by IP6.34
It would appear from these results that HIV-1 Gag is incapable of proper particle assembly in vivo until it reaches the plasma membrane and is in contact with RNA. Further work is needed to dissect the temporal sequence of the events needed in Gag extension and subsequent assembly. Indeed recent observations in live cells seem to indicate that the RNA genome is bound by a small number of HIV Gag molecules and targeted to the PM where more Gag molecules are recruited.35 An understanding of virus assembly intermediates may suggest new leads for the development of therapeutics that inhibit proper virus formation.
Supplementary Material
Acknowledgments
This work utilized facilities supported in part by the National Science Foundation under Agreement No. DMR-0454672. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Siddhartha A.K. Datta, Email: dattasi@mail.nih.gov.
Frank Heinrich, Email: frank.heinrich@nist.gov.
Sindhu Raghunandan, Email: sindhur8902@gmail.com.
Susan Krueger, Email: susan.krueger@nist.gov.
Joseph E. Curtis, Email: joseph.curtis@nist.gov.
Alan Rein, Email: reina@mail.nih.gov.
Hirsh Nanda, Email: hirsh.nanda@nist.gov.
References
- 1.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview: 1997. pp. 263–334. [PubMed] [Google Scholar]
- 2.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou WJ, Parent LJ, Wills JW, Resh MD. Identification of a membrane binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller SD, Wilk T, Gowen BE, Kräusslich HG, Vogt VM. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 5.Datta SAK, Curtis J, Ratcliff W, Clark P, Crist R, Lebowitz J, Krueger S, Rein A. Conformation of the HIV-1 Gag Protein in Solution. J Mol Biol. 2007;365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci USA. 2001;98:10875–10979. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 9.Gamble TR, Yoo SH, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 10.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 11.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 12.Newman JL, Butcher EW, Patel DT, Mikhaylenko Y, Summers MF. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 2004;13:2101–2107. doi: 10.1110/ps.04614804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stys D, Blaha I, Strop P. Structural and functional studies in vitro on the p6 protein from the HIV-1 gag open reading frame. Biochim Biophys Acta. 1993;1182:157–161. doi: 10.1016/0925-4439(93)90137-p. [DOI] [PubMed] [Google Scholar]
- 14.Datta SAK, Zhao Z, Clark PK, Tarasov S, Alexandratos JN, Campbell SJ, Kvaratskhelia M, Lebowitz J, Rein A. Interactions between HIV-1 Gag molecules in solution: An inositol phosphate-mediated switch. J Mol Biol. 2007;365:799–811. doi: 10.1016/j.jmb.2006.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis JE, Raghunandan S, Nanda H, Krueger S. SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput Phys Commun. 2010 submitted. [Google Scholar]
- 16.Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken PZ, Gronenborn AM. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell S, Vogt VM. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu F, Joshi SM, Ma YM, Kingston RL, Simon MN, Vogt VM. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J Virol. 2001;75:2753–2764. doi: 10.1128/JVI.75.6.2753-2764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, McNitt KL, Johnson DG, Hixson CV, Gorelick RJ, Fabris D, Henderson LE, Rein A. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGillivray DJ, Valincius G, Vanderah DJ, Febo-Ayala W, Woodward JT, Heinrich F, Kasianowicz JJ, Losche M. Molecular-scale structural and functional characterization of sparsely tethered bilayer lipid membranes. Biointerphases. 2007;2:21–33. doi: 10.1116/1.2709308. [DOI] [PubMed] [Google Scholar]
- 21.Cornell BA, Braach-Maksvytis VLB, King LG, Osman PDJ, Raguse B, Wieczorek L, Pace RJ. A biosensor that uses ion-channel switches. Nature. 1997;387:580–583. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 22.Shenoy S, Moldovan R, Fitzpatrick J, Vanderah DJ, Deserno M, Loesche M. In-plane homogeneity and lipid dynamics in tethered bilayer lipid membranes (tBLMs) Soft Matter. 2010;6:1263–1274. doi: 10.1039/B919988H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parratt LG. Surface studies of solids by total reflection of x-rays. Phys Rev. 1954;95:359–369. [Google Scholar]
- 24.Heinrich F, Ng T, Vanderah DJ, Shekhar P, Mihailescu M, Nanda H, Lösche M. A new lipid anchor for sparsely-tethered bilayer lipid membranes. Langmuir. 2009;25:4219–4229. doi: 10.1021/la8033275. [DOI] [PubMed] [Google Scholar]
- 25.Kienzle PA, Doucet M, McGillivray DJ, O'Donovan KV, Berk NF, Majkrzak CF. ga_refl, a program for simultaneous fitting of X ray and neutron polarized reflectometry data. 2000 http://www.ncnr.nist.gov/reflpak.
- 26.Brügger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Kräusslich HG. The HIV lipidome: A raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nanda H, Datta SAK, Heinrich F, Lö̲sche M, Rein A, Krueger S, Curtis JE. Electrostatic Interactions and Binding Orientation of HIV-1 Matrix, Studied by Neutron Reflectivity. Biophys J. 2010;99:2516–2524. doi: 10.1016/j.bpj.2010.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purohit P, Dupont S, Stevenson M, Green MR. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA. 2001;7:576–584. doi: 10.1017/s1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hearps A, Wagstaff KM, Piller SC, Jans D. The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry. 2008;47:2199–2210. doi: 10.1021/bi701360j. [DOI] [PubMed] [Google Scholar]
- 30.Shkriabai N, Datta SAK, Zhao ZJ, Hess S, Rein A, Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 31.Alfadhli A, Still A, Barklis E. Analysis of Human Immunodeficiency Virus Type 1 Matrix Binding to Membranes and Nucleic Acids. J Virol. 2009;83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lochrie MA, Waugh S, Pratt DG, Clever J, Parslow TG, Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci USA. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones CP, Datta SAK, Rein A, Rouzina I, Musier-Forsyth K. Influence of IP5 on MA interaction with viral RNA. 2010 communicated. [Google Scholar]
- 35.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci USA. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.