Abstract
OBJECTIVE
IKZF1 encoding Ikaros, an essential regulator of lymphopoiesis and immune homeostasis, has been implicated in the development of childhood acute lymphoblastic leukemia (C-ALL). Because recent genome-wide association (GWA) studies have linked a region of the 3′-UTR of IKZF1 with C-ALL susceptibility, we tested whether IKZF1 is associated with the autoimmune disease type 1 diabetes.
RESEARCH DESIGN AND METHODS
rs10272724 (T>C) near IKZF1 at 7p12 was genotyped in 8,333 individuals with type 1 diabetes, 9,947 control subjects, and 3,997 families of European ancestry. Association was tested using logistic regression in the case-control data and by the transmission disequilibrium test in the families. Expression data for IKZF1 by rs10272724 genotype were obtained using quantitative PCR of mRNA/cDNA generated from peripheral blood mononuclear cells from 88 individuals, whereas expression data for five other neighboring genes were obtained from the online Genevar dataset.
RESULTS
The minor allele of rs10272724 (C) was found to be protective from type 1 diabetes (odds ratio 0.87 [95% CI 0.83–0.91]; P = 1.1 × 10−11). rs10272724 was not correlated with levels of two transcripts of IKZF1 in peripheral blood mononuclear cells.
CONCLUSIONS
The major susceptibility genotype for C-ALL confers protection from type 1 diabetes. Our finding strengthens the link between autoimmunity and lymphoid cancers. Further investigation is warranted for the genetic effect marked by rs10272724, its impact on IKZF1, and the role of Ikaros and other family members, Ailios (IKZF3) and Eos (IKZF4), in autoimmunity.
IKZF1 encodes Ikaros, a zinc-finger transcription factor with a regulatory role in lymphopoiesis and the maintenance of cytokine expression in mature lymphocytes (1,2). Mice with a null mutation in IKZF1 (Ikaros−/−) display complete penetrance of leukemia, increased CD4+ to CD8+ T-cell ratios, and defects in T-cell maturation, including decreased T-cell receptor signaling thresholds during central tolerance (1). Mice heterozygous for the null mutation in IKZF1 (Ikaros+/−) exhibit milder features, but abnormalities including reduced B-cell precursors, a highly increased proliferative response to T-cell receptor engagement, and high rates of T-cell leukemia and lymphomas have been observed (3).
Human studies suggest a specific role for Ikaros in childhood acute lymphoblastic leukemia (C-ALL). Development of C-ALL is linked to somatically acquired mutations that add to inherited cancer susceptibility alleles (4). In 2009, deletions of IKZF1 were reported in 28.6% of C-ALL affected adults (5) and in 83.7% of BCR-ABL1 C-ALL patients examined (6). Combined, the human and murine studies suggest that diminished expression of IKZF1 interrupts lymphocyte development, creating conditions that maintain the rapidly dividing lymphoblasts that characterize ALL.
Two recent genome-wide association (GWA) studies have identified a C-ALL susceptibility locus near IKZF1 (7,8). The identified C-ALL-associated single nucleotide polymorphisms (SNPs), rs4132601 (T>G) and rs11978267 (A>G), as well as two others near IKZF1, rs11980379 (T>C) and rs10272724 (T>C), showed evidence of association with type 1 diabetes in a separate GWA study (P = 2.0 × 10−6, 2.6 × 10−6, 2.5 × 10−6, and 1.4 × 10−6, respectively; www.t1dbase.org), although not at the level the authors required for follow-up (P < 1 × 10−6) (4). All four SNPs are in high linkage disequilibrium (LD, r2>0.98) in the British 1958 Birth Cohort (B1958) controls (http://www.b58cgene.sgul.ac.uk), suggesting they mark the same potential causal variant for type 1 diabetes. Type 1 diabetes is a disease of abnormal immunity, wherein autoimmune destruction of pancreatic β-cells results in a state of insulin dependency (8,9). Type 1 diabetes results from the influence of environmental factors acting on an underlying genetic susceptibility (9). Although no direct link between IKZF1 and type 1 diabetes has been reported, the previously reported, but not highlighted or followed up, association of the chromosome 7p12.2 region containing IKZF1 with type 1 diabetes (10; www.t1dbase.org), the immunologic importance of IKZF1 (11), and the association of IKZF1 with susceptibility to C-ALL (7,8) prompted us to select the SNP most associated with type 1 diabetes, rs1027274 (T>C) (P = 1.4 × 10−6) near IKZF1,for follow-up genotyping to assess association with type 1 diabetes and to determine whether a common genetic component exists for C-ALL and type 1 diabetes.
RESEARCH DESIGN AND METHODS
Samples.
A total of 8,333 type 1 diabetes cases, 9,947 controls, and 3,997 families were genotyped at rs10272724 using TaqMan (Supplementary Methods). All samples were of white European ancestry. Cases were diagnosed with type 1 diabetes before 17 years of age (mean age at diagnosis = 7.8 years) and were from the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory Genetic Resource Investigating Diabetes study (http://www.childhood-diabetes.org.uk/grid.shtml). Controls were obtained from the British 1958 Birth Cohort (n = 6,899; http://www.cls.ioe.ac.uk/studies.asp?section=000100020003) and the WTCCC UK Blood Service (UKBS) sample collection (n = 3,048) (10,12).
Statistical analysis.
STATA version 10 (StataCorp LP, College Station, TX) was used to perform association analyses (http://www.stata.com). rs10272724 was in Hardy-Weinberg equilibrium in unaffected parents and control subjects (P > 0.05). Case-control data were modeled using logistic regression, with disease status as the outcome variable and counts of the minor allele (coded 0, 1, and 2) as the independent variable, assuming a multiplicative allelic effects model. Geographic region was included as a covariate in the model (Supplementary Methods). Families were analyzed using the transmission disequilibrium test (13) (Supplementary Methods). The interdependency of rs10272724 and rs4948088 in the 7p12 region was examined by stepwise logistic regression.
Peripheral blood mononuclear cell isolation and preparation of DNA and RNA.
Peripheral blood mononuclear cells (PBMCs) were purified from heparinized blood diluted 1:1 in PBS (without Ca2+ and Mg2+, GIBCO, Invitrogen, Carlsbad, CA), and 15 mL aliquots were layered onto 10 mL aliquots of Lympholyte (Cedarlane Laboratories Ltd., Burlington, Ontario) followed by centrifugation at 800g for 20 min at room temperature. The harvested PBMC layer was washed twice with ice-cold PBS and centrifuged at 300g for 10 min at 4°C. Pellets were resuspended in TRIzol (Invitrogen) and stored at −80°C in aliquots of 10 × 107 cells/mL. Total RNA from 1 × 107 PBMCs in TRIzol was prepared using chloroform extraction followed by purification with the RNeasy Mini kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. RNA quality was assessed using an Agilent 2100 Bioanalyser, and concentration was evaluated by Nanodrop (Thermo Scientific, Waltham, MA). First strand DNA synthesis was carried out on 1 μg of RNA using Superscript III RT kit and oligo-dT (Invitrogen).
Quantitative PCR evaluation of IKZF1 expression.
Quantitative (q)PCR primers and probe were designed to two transcripts of IKZF1, herein referred to as “Isoform 1” and “Isoform 2” (Supplementary Methods). Probes were labeled with the fluorochrome FAM and quenched using TAMRA. qPCR assays contained 5 μL of 1:10 dilution of oligo-dT primed cDNA, prepared as described above, in 20 μL assays. Cycle threshold (Ct) values were measured using a 7900HT ABI prism (Applied Biosystems, Carlsbad, CA) and analyzed using SDS v2.1 software (ABI). qPCR reactions were run in triplicate, and the ΔCt for the transcript level qPCRs was calculated using the IKZF1 Isoform 1 or Isoform 2 qPCR Ct value minus the single copy gene β2 microglobulin qPCR Ct value. Expression values were compared via one-way ANOVA using Prism software (GraphPad Software Inc., La Jolla, CA).
mRNA expression of genes near rs10272724.
Correlation between rs10272724 and expression in three types of cell lines (primary fibroblasts, Epstein–Barr virus-immortalized lymphoblastoid cell lines, and phytohemagglutinin-stimulated primary T-cells) derived from umbilical cord samples of 75 newborns of Western European origin via the GenCord project was examined in silico (14) using the publicly available HapMap online GENe Expression VARiation (Genevar) resource (http://www.sanger.ac.uk/resources/software/genevar/). Nine probes that passed quality control assessment (Supplementary Table 1) were evaluated for correlation of mRNA expression with rs10272724 genotype by linear regression using the Genevar 2.0.1 Java tool (15) (Supplementary Methods). A P < 0.0056 significance threshold was used within each cell type based on a Bonferroni correction for testing nine transcripts.
Donors.
Blood for qPCR experiments was collected from 88 nondiabetic donors (rs10272724: 44 TT, 33 CT, 11 CC) selected from a pregenotyped bioresource and processed within 4 hours (www.cambroidgebioresource.org.uk).
Study ethics.
All DNA samples were collected with ethical approval from the National Health Service Cambridgeshire Research Ethics Committee. Written consent was obtained from all individuals.
RESULTS
The minor allele of rs10272724 (T>C) near IKZF1 was protective from type 1 diabetes (odds ratio [OR] 0.87 [95% CI 0.83–0.91], P = 4.8 × 10−9; Table 1, Supplementary Table 2). No evidence of association of rs10272724 with sex or age-at-diagnosis was obtained (P > 0.1). Some samples overlapped with those used in the previous GWA study for type 1 diabetes (10). However, in each of the sample sets used by Barrett et al. (10) and in our unique samples, the effect is in the same direction and the overall significance is increased, suggesting the effect is not solely attributable to the original samples (Table 1). The minor allele of rs10272724 was also protective in the families with type 1 diabetes, i.e., under-transmitted to affected offspring (Table 1; relative risk 0.87 [95% CI 0.81–0.93], P = 7.6 × 10−5). Combined, these results indicate that the minor allele, C, of rs10272724 at 7p12.2 is convincingly associated with a reduced risk of type 1 diabetes (P = 1.1 × 10−11). Barrett et al. (10) reported a replicated association with rs4948088 (C>A) with type 1 diabetes in the 7p12.1 region, 554 kb downstream of IKZF1. However, this SNP, marking a confirmed type 1 diabetes locus (http://www.t1dbase.org/page/Regions), is not in LD with rs10272724 in IKZF1 (D’ = 0.02, r2 = 0.0001), and we found by regression analysis that the two effects were independent as both SNPs improve a model with the other SNP included (P < 5.5 × 10−5).
TABLE 1.
Association of rs10272724 (T>C) near IKZF1 in 8,333 type 1 diabetes cases, 9,947 control subjects, and 3,997 families with type 1 diabetes
| Sample set | No. cases | No. controls | MAF cases | MAF controls | OR (95% CI) | P* |
|---|---|---|---|---|---|---|
| Full case-control set |
8,333 |
9,947 |
0.25 |
0.28 |
0.87 (0.83–0.91)† |
4.8 × 10−9 |
| Analysis of genotypes generated in the current report, within sample sets used by published GWA studies‡ | ||||||
| Set 1 (T1DGC) |
3,850 |
3,772 |
0.25 |
0.28 |
0.85 (0.79–0.91)† |
1.6 × 10−5 |
| Set 2 (WTCCC) |
1,827 |
1,507 |
0.25 |
0.28 |
0.84 (0.76–0.92) |
0.0014 |
| Set 3 (Swafford) |
2,666 |
4,668 |
0.26 |
0.28 |
0.90 (0.83–0.98) |
0.011 |
| No. families |
MAF unaffected parents |
No. transmitted |
No. not transmitted |
RR (95% CI) |
P |
|
| Families with type 1 diabetes |
3,997 |
0.27 |
1,423 |
1,642 |
0.87 (0.81–0.93) |
7.6 × 10−5 |
| Overall | 1.1 × 10−11 | |||||
MAF, minor allele frequency; RR, relative risk.
*No evidence of deviation from a multiplicative allelic effects model was obtained (P = 0.33), so P values assuming multiplicative allelic effects are reported.
The overall P value was obtained by combining the P value from the case-control sets and the family transmission disequilibrium test using Fisher’s method for combined probability.
†This OR is subject to winner’s curse. ‡Barrett et al. (10) analyzed three datasets in their GWA study, two of which overlap with the samples in the current study. Barrett et al. used the Illumina 550 K SNP chip to genotype rs10272724 in set 1 (4). Genotypes were 99.6% concordant between the Illumina platform and TaqMan in the 3,850 cases and 3,772 controls genotyped using both technologies. Affymetrix 500 K Mapping Array genotypes for neighboring SNPs were used to impute rs10272724 genotypes in set 2 by Barrett et al. because rs10272724 was not genotyped by Affymetrix. T1DGC, Type 1 Diabetes Genetics Consortium; WTCCC, Wellcome Trust Case-Control Consortium.
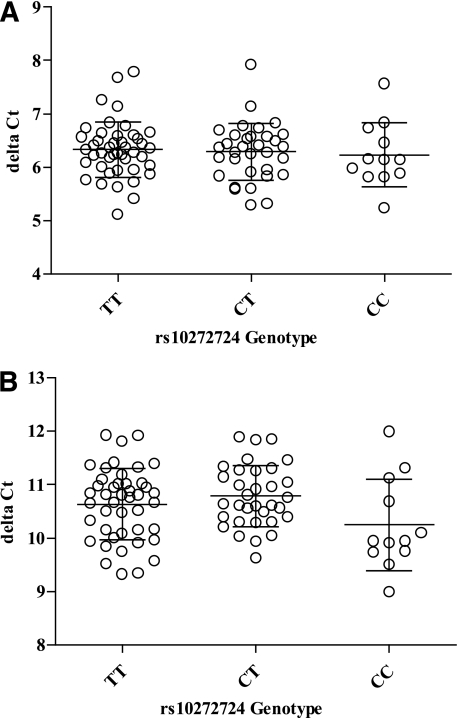
The relative abundance of two different sets of IKZF1 transcripts in mRNA extracted from PBMCs isolated from 88 nondiabetic individuals was assessed using qPCR to determine the association of rs10272724 alleles with gene expression. No evidence of allele-specific expression was obtained (P > 0.1; Fig. 1). Others have reported a correlation of rs4132601 with IKZF1 expression in Epstein–Barr virus-transformed lymphoblastoid cell lines from the HapMap GENEVAR resource ([8]; P = 0.0017). However, this is likely to be a false effect. The probe originally used (Illumina ID ILMN_1676575) contains two high-frequency SNPs in its target, rs62447207 (G>T) and rs62447208 (C>G) in LD with rs10272724 (r2 = 0.76), rendering it unreliable, as evidenced by the absent IKZF1 probe in the latest Genevar data from the study by Dimas et al. (14). In results from 13 eQTL datasets displayed at http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/, there is no reliable evidence for an IKZF1-associated eQTL. Next, correlation of rs10272724 genotype and expression of nine transcripts from other genes within 1 Mb of rs10272724 in three types of cell lines was tested using the latest Genevar dataset by Dimas et al. (14). No correlation was observed between rs10272724 and expression of any of the five neighboring genes ([14], Supplementary Fig. 1).
FIG. 1.
Association of IKZF1 expression with rs10272724 (T>C) genotype in 88 unaffected individuals. A: qPCR analysis of IKZF1 Isoform 1 transcript expression. B: qPCR analysis of IKZF1 Isoform 2 transcript expression. δCt were calculated using the IKZF1 Isoform 1 or Isoform 2 qPCR Ct value minus the single copy gene β2 microglobulin (B2M) qPCR Ct value. Expression values were compared via one-way ANOVA using Prism software.
DISCUSSION
We provide evidence that the minor allele of rs10272724 (C), in near-perfect LD with the minor, C-ALL–susceptible, allele of rs4132601 (G), is protective from type 1 diabetes. Of the nearby genes in the region, IKZF1 appeared to be the most likely candidate causal gene for type 1 diabetes in this region given its causal role in C-ALL and its known role in lymphocyte development. However, our analysis revealed that the previously reported correlation of IKZF1 mRNA expression with rs4132601 may be inaccurate given complications with the probe used in the data accessed by Papaemmanuil et al. (8). Furthermore, an IKZF1 genotype-phenotype correlation at rs10272724 was not confirmed by our qPCR analysis of PBMCs. IKZF1 is now an attractive candidate causal gene in type 1 diabetes, and a functional link between rs10272724 and IKZF1 may yet be elucidated on examining specific isoforms of Ikaros. We note that our qPCR assay evaluated the expression of two transcripts conserved across 10 splice variants of IKZF1, spanning six of the seven identified active and dominant-negative isoforms of the protein. In addition, many allele-specific effects on gene expression are known to be tissue or cell-type specific and may even be restricted to particular phases of development. Another ALL-related gene, AF4/FMR2 family member 3 (AFF3), has been associated with type 1 diabetes (10) (J. Cooper, C.W., and J.A.T., unpublished observations), so our report also confirms a second genetic link between these diseases. AFF3 has been implicated as a susceptibility allele in rheumatoid arthritis (rs10865035 and rs9653422) (16), further highlighting the connection between autoimmunity and lymphoid cancers.
Our finding with IKZF1 marks the third Ikaros family member to be associated with type 1 diabetes, the others being the transcription factors Ailios (IKZF3) on chromosome 17q21.2 and Eos (IKZF4) on chromosome 12q13.2 (10,17), whose targets include BCL-2 and FOXP3, respectively. Ikaros family members interact to coordinate functions of immunologic development and homeostasis (18), so it will be important to explore the effect of these interactions in disease etiology. The more than 30 reported interaction partners of the Ikaros family members (http://www.t1dbase.org) suggest the role of IKZF1 in type 1 diabetes could be subtle and yet far reaching. Interactions with the histone deacetylase and chromodomain-helicase-DNA-binding families of proteins suggest chromosome remodeling events could be involved (11,19). Ikaros interactions with the Notch (18) and STAT (20) protein families also suggest that signaling events that affect T-cell activation and maturation could affect the ultimate development of a diabetogenic, leukemia-protected or nondiabetogenic, leukemia-susceptible T-cell repertoire. Thus, further investigation is warranted to elucidate the phenotypic effect of the genetic feature marked by rs10272724, its impact on IKZF1, and the role of Ikaros in autoimmunity.
ACKNOWLEDGMENTS
This work was funded by the Juvenile Diabetes Research Foundation International (JDRF), the Wellcome Trust, the National Institute for Health Research Cambridge Biomedical Research Centre, the National Science Foundation, and the National Institutes of Health Division of Intramural Research. A.D.-E.S. is a National Institutes of Health Cambridge Health Science Research Scholar and a National Science Foundation Graduate Research Fellow. L.J.D is supported by a Wellcome Trust Intermediate Clinical Fellowship. C.W. is supported by the Wellcome Trust (089989). The Cambridge Institute for Medical Research is in receipt of a Wellcome Trust Strategic Award (079895). The authors thank the U.K. Medical Research Council and Wellcome Trust for funding the collection of DNA for the British 1958 Birth Cohort (MRC Grant G0000934, Wellcome Trust Grant 068545/Z/02). They also acknowledge use of DNA from The U.K. Blood Services collection of Common Controls (UKBS collection), funded by the Wellcome Trust Grant 076113/C/04/Z, the Wellcome Trust/JDRF Grant 061858, and the National Institutes of Health Research of England. The collection was established as part of the Wellcome Trust Case-Control Consortium (funding for the project was provided by the Wellcome Trust under award 076113). This research uses resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development, and Juvenile Diabetes Research Foundation and supported by U01 DK062418. Funding for the project was provided by the Wellcome Trust under award 076113.
No potential conflicts of interest relevant to this article were reported.
A.D.-E.S., J.M.M.H., L.J.D., and J.A.T. researched data, contributed to discussion, wrote the article, and reviewed and edited the article. C.W. and M.J.L. contributed to discussion, wrote the article, and reviewed and edited the article. D.S., M.M.-A., T.M., and H.S. researched data and reviewed and edited the article.
The authors thank all of the patients, control subjects, and family members for participation and the Human Biological Data Interchange and Diabetes UK for U.S. and U.K. multiplex families, respectively; the Norwegian Study Group (Dag Undlien and Kjersti Ronningen) for Childhood Diabetes for the collection of Norwegian families; Constantin Ionescu-Tirgoviste and Cristian Guja, Institute of Diabetes, Nutrition and Metabolic Diseases, Bucharest for Romanian families; and the Academy of Finland and the JDRF for the Finnish families. DNA samples from the British 1958 Birth Cohort were prepared and provided by S. Ring and W. McArdle, Avon Longitudinal Study of Parents and Children (ALSPAC) Laboratory, University of Bristol; R. Jones, Department of Community Based Medicine, University of Bristol; M. Pembrey, Clinical and Molecular Genetic Unit, Institute of Child Health, London; D. Strachan, Division of Community Health Sciences, St. George’s, University of London; and P. Burton, Department of Health Sciences, University of Leicester. The authors thank David Dunger and Barry Widmer, Department of Paediatrics, University of Cambridge, Addenbrooke’s Hospital, and the British Society for Paediatric Endocrinology and Diabetes for the type 1 diabetes case collection and the Cambridge BioResource for nondiabetic donors. The authors also thank H. Stevens, P. Clarke, G. Coleman, S. Duley, D. Harrison, and S. Hawkins, JDRF/Wellcome Trust Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research, University of Cambridge, for preparation of DNA samples.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-0446/-/DC1.
REFERENCES
- 1.Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med 1999;190:1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umetsu SE, Winandy S. Ikaros is a regulator of Il10 expression in CD4+ T cells. J Immunol 2009;183:5518–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol 2002;2:162–174 [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med 2004;350:1535–1548 [DOI] [PubMed] [Google Scholar]
- 5.Martinelli G, Iacobucci I, Storlazzi CT, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol 2009;27:5202–5207 [DOI] [PubMed] [Google Scholar]
- 6.Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008;453:110–114 [DOI] [PubMed] [Google Scholar]
- 7.Treviño LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet 2009;41:1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet 2009;41:1006–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Concannon P, Chen WM, Julier C, et al. Type 1 Diabetes Genetics Consortium Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the Type 1 Diabetes Genetics Consortium. Diabetes 2009;58:1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Sif S, Jones B, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 1999;10:345–355 [DOI] [PubMed] [Google Scholar]
- 12.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 1993;52:506–516 [PMC free article] [PubMed] [Google Scholar]
- 14.Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 2009;325:1246–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang TP, Beazley C, Montgomery SB, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 2010;26:2474–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton A, Eyre S, Ke X, et al. YEAR Consortium. BIRAC Consortium Identification of AF4/FMR2 family, member 3 (AFF3) as a novel rheumatoid arthritis susceptibility locus and confirmation of two further pan-autoimmune susceptibility genes. Hum Mol Genet 2009;18:2518–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd JA, Walker NM, Cooper JD, et al. Genetics of Type 1 Diabetes in Finland. Wellcome Trust Case Control Consortium Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007;39:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkenschlager M. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett 2010;584:4910–4914 [DOI] [PubMed] [Google Scholar]
- 19.Koipally J, Georgopoulos K. A molecular dissection of the repression circuitry of Ikaros. J Biol Chem 2002;277:27697–27705 [DOI] [PubMed] [Google Scholar]
- 20.Yap WH, Yeoh E, Tay A, Brenner S, Venkatesh B. STAT4 is a target of the hematopoietic zinc-finger transcription factor Ikaros in T cells. FEBS Lett 2005;579:4470–4478 [DOI] [PubMed] [Google Scholar]