Abstract
BACKGROUND
African American men have the highest rates of prostate cancer worldwide, and immunogenetic studies suggest that people of African descent have increased susceptibility to diseases of inflammation. Since genetic susceptibility is an etiological factor in prostate cancer, we hypothesize that sequence variants in the promoter region of the CD14 gene that regulate inflammation may modify individual susceptibility to this disease.
METHODS
The CD14 promoter was screened for single-nucleotide polymorphisms (SNPs) using dHPLC. One variant, −260 C>T (rs2569190), was genotyped via restriction digest in all study participants (264 cases and 188 controls). The association of disease status and the polymorphism was analyzed by unconditional logistic regression. Odds ratios with 95% confidence intervals were calculated, stratifying by ethnicity and adjusting for age. Two-sided P-values of ≤0.05 were considered as statistically significant.
RESULTS
Eleven variants (four novel) were identified in the promoter region of CD14. A marginal association between the C genotypes (C/C + C/T) and prostate cancer was found (P =0.07). When stratified by age, among men ≥55 years of age, the C genotypes were significantly associated with prostate cancer (P <0.05). When stratified by self-reported ethnicity, African American males who had the C genotypes were at a higher risk for prostate cancer (P <0.05).
CONCLUSIONS
This is the first study to show an association between the C genotypes of the CD14 (−260) variant and prostate cancer which supports the hypothesis that genetic variation in the inflammatory process can contribute to prostate cancer susceptibility in African American men.
Keywords: CD14, prostate cancer, chronic inflammation, innate immunity
INTRODUCTION
The incidence of prostate cancer varies significantly across ethnic groups, with African American men having a higher prevalence and greater morbidity than in most other populations [1]. Most of the literature on population differences in disease frequencies for common complex diseases involving gene–environment interactions, as is true for most health disparities, have highlighted social, cultural, environmental, and economic causal factors, while obscuring potential underlying gene-based biological factors [2]. The underlying genetic heterogeneity of human biological groups classified stereotypically by “racial”/ethnicity or continental origin compromises the power of genomic studies to dissect the biological determinants in health disparities. The broad spectrum of data emerging on genome-wide associations of single-nucleotide polymorphisms (SNPs), non-randomly distributed in the human genome, plays a critical but as yet largely uncharacterized role in health disparities. Advances in molecular and genetic studies of complex diseases have demonstrated a causal relationship between infection, inflammation, and complex diseases, such as asthma and cancer [3]. It remains to be determined whether genome variation in African Americans in regulation of the inflammatory response to pathogens may contribute to the higher prevalence and greater morbidity of prostate cancer in African American men.
As the hallmark of innate immunity, inflammation is an ancient, universal first line of host defense against pathogens [4]. Evolutionarily conserved germ line pattern recognition receptors (PRRs) are the sentinels of the primary host alarm system in response to bacterial and other pathogen infections [5]. As a PRR, CD14 plays a major role in pathogen-activated signal transduction pathways in the production of inflammatory cytokines [6,7]. Our laboratory is particularly interested in assessing the biomedical significance of DNA polymorphisms in African Americans (www.genomecenter.howard.edu), with particular attention to (a) the evolutionary impact of pathogen environment on alleles and haplotypes in innate and adaptive immune response genes, and (b) significance of genomic heterogeneity among natural populations of African descent in dissecting the biological determinants of gene–environment interactions in health disparities.
The CD14 (−260) polymorphism is located near the Sp1 transcription factor binding site, which is known to have a major influence on CD14 expression [8]. CD14 is a PRR of particular interest because it is thought to play a critical role in enhancing the dose-dependent sensitivity of cells to microbial products through tethering of certain pathogen-associated molecular patterns (PAMPs) to the plasma membrane and facilitating their interaction with downstream TLRs signaling of inflammatory cytokines [9]. Lin and Karin [10] recently reported a cytokine-mediated link between innate immunity, inflammation, and cancer. In this report we demonstrate an association of a polymorphism in the CD14 gene (−260C>T), which encodes a PRR, with prostate cancer in African American men.
MATERIALS ANDMETHODS
Study Population
Unrelated men of African descent (i.e., self-reported African Americans, Afro-Caribbean, African-Latino, East and West Africans) were recruited from the Washington, DC, area through the Division of Urology at the Howard University Hospital and/or prostate cancer screening at the Howard University Cancer Center. Prostate cancer cases were between 40 and 85 years of age. All the cases were diagnosed within 1 year of enrollment. The study population of African descent consisted of 254 prostate cancer patients and 188 male controls. Unaffected male volunteers were enrolled among individuals undergoing regular physical exams in the Division of Urology at Howard University Hospital and/or men participating in screening programs for prostate cancer at the Howard University Cancer Center. The screening program was demographically similar to the patient population seen in the Division of Urology clinics. The recruitment of controls occurred concurrently with the recruitment of prostate cancer patients. Blood samples were collected from each subject. Clinical characteristics including Gleason grade, prostate-specific antigen (PSA), and diagnosis of prostatitis were obtained from medical records. The mean age of cases 68.95 ± 9.73 was significantly greater than controls 57.33 ± 11.37, as was also the frequency of prostatitis. All control subjects had PSA levels <4.0 ng/ml and normal digital rectal exams. The clinical profile of the study population is shown in Table I. Individuals diagnosed with benign prostatic hyperplasia were not considered in this analysis. Prostate cancer cases were diagnosed by transrectal ultrasound-guided biopsy using standard saturation technique [11]. Biopsy cores were reviewed by members of the Department of Pathology Howard University College of Medicine. Prostate cancer cases were classified according to well-established parameters of the Gleason Scoring System [12,13]. Prostatitis cases were diagnosed based on the following criteria: (a) clinical presentation of perineal or scrotal discomfort, (b) irritative voiding symptoms with or without associated lower back pain, (c) the presence of leukocytes with or without bacteria in expressed prostatic secretions, (d) in patients who underwent prostate biopsy, the presence of focal or diffuse inflammatory infiltrates. Howard University Institutional Review Board approved the study and written consent was obtained from all participants.
TABLE I.
Clinical Profile for Men of African Descent in the Study Population
| Controls (total eligible in %) | Cases (total eligible in %) | P-value | |
|---|---|---|---|
| Total eligiblea | 188 | 254 | |
| Mean age | 57.33 ± 11.37 | 68.95 ± 9.73 | <0.01 |
| Age with family history | |||
| <60 | 11 (5.9) | 9 (3.5) | |
| 61–70 | 5 (2.7) | 16 (6.3) | |
| >71 | 2 (1.1) | 11 (4.3) | |
| Prostatitis | 7 (3.7) | 61 (24.0) | <0.01 |
| Gleason score | |||
| <4 | 18 (7.1) | ||
| 5–7 | 120 (47.2) | ||
| 8–10 | 26 (10.2) | ||
| Unknown | 90 (35.4) | ||
| PSA | |||
| <4 | 13 (5.1) | ||
| 4–10 | 79 (31.1) | ||
| 11–20 | 27 (10.6) | ||
| 21–50 | 18 (7.1) | ||
| 51–100 | 8 (3.1) | ||
| >101 | 23 (9.1) | ||
| Unknown | 86 (33.9) | ||
Self-reported men of African descent.
SNP Discovery
Genomic DNA was obtained from isolated lymphocytes using cell lysis, proteinase K-treatment, protein precipitation, and DNA precipitation. A total of 2.5 kb upstream of the transcription start site of CD14 gene was amplified using custom-designed primers. Forty-eight randomly selected DNA samples were amplified in a 25 μl PCR reaction containing 1× PCR buffer II, 30 ng of whole genome amplified DNA, 10 pmol of each forward and reverse primers, 1.5 mM MgCl2, 0.3 mM dNTP (ABI), 0.5U Platinum Taq DNA polymerase (Invitrogen). PCR reactions were performed as follows: denaturation at 95°C for 15 sec, annealing for 15 sec at 50°C, and extension at 72°C for 25 sec for 35 cycles followed by 10 min at 72°C in PE thermal cycler. The fragments, size from 430 to 600 bp, were amplified. Before loading the PCR samples into the dHPLC instrument (WAVE DNA Fragment Analysis System, Transgenomic) for variant detection; the samples were heated at 95°C for 4–8 min and removed from the thermal cycler. The samples were then cooled at room temperature for 20 min, loaded into the dHPLC instrument, and run with the start gradient of 45% buffer A (0.1 M triethylammonium acetate (TEAA) solution, pH 7.0) and 55% buffer B (0.1 M TEAA containing 25% acetonitrile, pH 7.0). The stop gradients of 36% buffer A and 64% buffer B were used, respectively, acquisition time 8.7 min. Those samples demonstrating heteroduplex peaks were sequenced (Big Dye Terminator v.1.1, ABI 377) in both directions to identify any variants within the PCR fragment.
Genotyping
Restriction fragment length polymorphism was used for genotyping CD14 (−260) variant (rs2569190) located on chromosome 5q31.1. The PCR fragment was amplified using CD14-260 forward 5′-TGAGTCATCAGGACACTGCC-3′ and CD14-260 reverse 5′-TCACCTCCCCACCTCTCTTC-3′. Genotyping was performed using restriction digestion under the following conditions: 10 μl PCR product, 2 μl 10× buffer 2, 7.5 μl water, and 0.5 μl enzyme (5 U) in a total volume of 20 μl. The HaeIII digests were incubated at 37°C for 6 hr with expected fragment sizes of 70 and 80 bp for CC, 150, 70, and 80 bp for CT, and 150 bp for TT. The resultant fragments were electrophoresed on a 3% agarose gel containing ethidium bromide. The bands were then visualized by UV transillumination (Fig. 1).
Fig. 1.
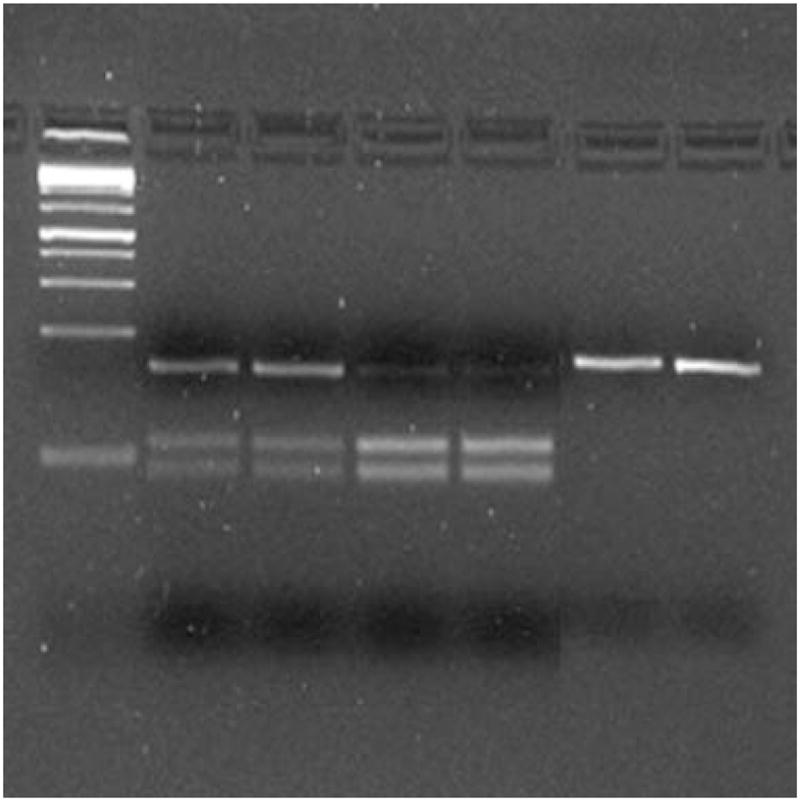
RFLP analysis ofrs2569190 using HaeIII. Samples1and 2 represent heterozygotes whereas samples 3 – 6 represent homozygotes for the wild type and variant, respectively.
Statistical Analysis
The statistical analyses for the case–control study were done with the SAS/STAT® software, version 9.1 (SAS Institute, Inc., Cary, NC). Allele frequencies in cases and controls were tested for Hardy–Weinberg equilibrium (Graphpad Software, http://www.graphpad.com/quickcalcs/index.cfm). The association of disease status and polymorphism was analyzed by unconditional logistic regression. Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated, stratifying by ethnicity and adjusting for age. Two-sided P-values of ≤0.05 were considered as statistically significant. Genetic modeling was performed using the statistical analysis package R (Lucent Technologies, Murray Hill, NJ). Quality control was performed for each assay by repeating 20% of samples in a blinded fashion.
Electronic Database Information
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP/ (for human SNP allele frequencies).
HapMap: http://www.hapmap.org/ (for human SNP allele frequencies).
RESULTS
SNP Discovery in the Promoter Region of CD14
Eleven variants were identified in the promoter region of CD14 (Table II). Of these, seven including the (−260) variant (rs2569190) were found in dbSNP and HapMap (NCBI build 36, dbSNP b126) websites. The remaining four variants were classified as novel. The (−260) variant was chosen for genotyping because of (a) the allele frequency difference in the SNP between the European-based CEPH population and African-based Yoruba population from the International Haplotype Project data (Table II) and (b) possible effect of the SNP on the biology of prostate cancer.
TABLE II.
Putative Promoter Variants Identified in the Screening of CD14
| Positiona | Variant | Effectb | dbSNP rs# | CEU | YRI | AA-1c | AA-2d |
|---|---|---|---|---|---|---|---|
| −135 | A>G | No change | Novel | N/A | N/A | N/A | N/A |
| −260 | C>T | Loss of Sp1 and ETF sites | rs2569190 | T: 47.4%; C: 52.6% | T: 29.3%; C: 70.7% | T: 33.3%; C: 66.7% | T: 33.3%; C: 66.7% |
| −618 | A indel | Loss of GATA-1 site | Novel | N/A | N/A | N/A | N/A |
| −651 | C>T | No change | rs5744455 | C: 70%; T: 30% | N/A | C: 89.1%; T: 10.9% | N/A |
| −911 | A>C | Elf-1>GATA-1 | rs5744454 | A: 70%; C: 30% | N/A | A: 77.1%; C: 22.9% | N/A |
| −1247 | C>T | Loss of NF-1 and Oct-1 sites | rs2569191 | C: 52.6; T: 47.4% | N/A | C: 50%; T: 50% | N/A |
| −1347 | C>T | No change | Novel | N/A | N/A | N/A | N/A |
| −1461 | G>T | Addition of Sp1 site | Novel | N/A | N/A | N/A | N/A |
| −1721 | C>T | HNF3, MEB1, GLO>Oct-1, HB, HNF3B, TBP | rs2915863 | C: 40%; T: 60% | N/A | C: 22.9%; T: 77.1% | N/A |
| −1957 | A>G | Loss of E1, MyoD, Sp1, ER sites | rs3138076 | A: 70%; G: 30% | N/A | A: 77.1%; G: 22.9% | N/A |
| −2253 | C>A | No change | rs5744448 | A: 0.9%; C: 99.1% | A: 10.9%; C: 89.1% | A: 10.4%; C: 89.6% | N/A |
Number of base pairs from the ATG start codon.
Transcriptional binding site change.
African American frequency data taken from dbSNP.
African American frequency data taken from our control population.
Frequency Analysis of CD14 (−260) Variant in Cases and Controls
Genotypic and allelic analyses of the (−260) variant showed a marginal difference between cases and controls. The frequency of those with the C genotypes, 90.59%, was higher, but not significantly different from those without C (T/T) 83.72% in case–control analyses of the variant. Although the latter was not statistically significant, it was noted that the homozygous TT genotype at a frequency of 16.28% among controls was higher than the 9.42% among cases, whereas the frequency of the heterozygous C/T genotype for cases, 43.50%, was higher than the 34.30% heterozygous genotype for controls. Chi-square analysis showed a marginal association of the C genotypes with prostate cancer (P =0.07). There was no association between the C allele and prostate cancer among this population.
Association of CD14 (−260) Variant and Prostate Cancer Risk
Although the association between the C genotypes and prostate cancer cases in the general population was marginal, when the population was stratified according to age, the 90.78% frequency of those with the C genotypes among cases was significantly greater than the 80.61% of those with this genotype among the controls. Chi-square and OR analyses showed that the C genotypes were significantly associated with prostate cancer (OR: 2.18, 95% CI: 1.07–4.44; P <0.05) indicating that in the 55 years and older age range, males with the C genotypes have approximately twice the risk of developing prostate cancer (Table III). We also found that when the population was stratified according to ethnicity, the C genotypes were again significantly associated with prostate cancer among self-identified African Americans (OR: 2.40, 95% CI: 1.20–4.70; P <0.05) (Table III). Lastly, there was not an association found between the C allele and prostate cancer when the population was stratified according to age or self-reported ethnicity.
TABLE III.
CD14 Genotypic and Allelic Analyses in Prostate Cancer
| Controls | % | Cases | % | ORa | 95% CI | P-valueb | |
|---|---|---|---|---|---|---|---|
| African descent | n =172 | n =223 | |||||
| TT | 28 | 16.28 | 21 | 9.42 | 1.00 | ||
| CT | 59 | 34.30 | 97 | 43.50 | 1.91 | 0.92–3.97 | 0.19 |
| CC | 85 | 49.42 | 105 | 47.09 | 1.82 | 0.89–3.72 | 0.26 |
| CC/CT | 144 | 83.72 | 202 | 90.59 | 1.86 | 0.95–3.67 | 0.07 |
| C allele | 229 | 66.57 | 307 | 68.83 | 1.10 | 0.82–1.50 | 0.49 |
| Age ≥55 years old | n =98 | n =206 | |||||
| TT | 19 | 19.39 | 19 | 9.22 | 1.00 | ||
| CT | 37 | 37.76 | 91 | 44.17 | 2.17 | 1.01–4.69 | |
| CC | 42 | 42.86 | 96 | 46.60 | 2.18 | 1.02–4.66 | 0.10 |
| CC/CT | 79 | 80.61 | 187 | 90.78 | 2.18 | 1.07–4.44 | <0.05 |
| C allele | 121 | 61.73 | 283 | 68.69 | 1.36 | 0.95–1.94 | 0.09 |
| African Americans | n =139 | n =204 | |||||
| TT | 25 | 17.99 | 17 | 8.33 | 1.00 | ||
| CT | 45 | 32.37 | 89 | 43.63 | 2.40 | 1.09–5.30 | 0.08 |
| CC | 69 | 49.64 | 98 | 48.04 | 1.10 | 1.07–1.13 | 0.23 |
| CC/CT | 114 | 82.01 | 187 | 91.67 | 2.40 | 1.20–4.70 | <0.05 |
| C allele | 183 | 69.85 | 285 | 65.83 | 1.20 | 0.87–1.67 | 0.27 |
Adjusted for age.
χ2 P-values.
Genetic Modeling of the CD14 (−260) Variant in Cases and Controls
Various genetic models were tested for the (−260) variant taking the population as a whole, as well as, stratifying the population based on age and self-reported ethnicity. We found the best-fitting model to be the recessive model (Table IV). The results indicated that in men of African descent, older men (≥55), and African Americans, respectively, the TT genotype was associated with a decreased risk of prostate cancer (OR: 0.53, 95% CI: 0.29–0.98, P =0.04; OR: 0.42, 95% CI: 0.21–0.84 (P =0.015); OR: 0.41, 95% CI: 0.21–0.80 (P =0.008), respectively).
TABLE IV.
Association of the CD14 (−260) Variant With Prostate Cancer
| ORa | 95% CI | P-valueb | Best-fitting genetic model | |
|---|---|---|---|---|
| African descent | ||||
| CC/CT | 1.00 (ref.) | |||
| TT | 0.53 | 0.29–0.98 | 0.041 | Recessive |
| Age ≥55 years old | ||||
| CC/CT | 1.00 (ref.) | |||
| TT | 0.42 | 0.21–0.84 | 0.015 | Recessive |
| African Americans | ||||
| CC/CT | 1.00 (ref.) | |||
| TT | 0.41 | 0.21–0.80 | 0.008 | Recessive |
Adjusted for age.
χ2 P-values.
DISCUSSION
This is the first report of an association between the C genotypes of the CD14 (−260) variant and prostate cancer which supports the hypothesis that genetic variation in the inflammatory process can contribute to prostate cancer susceptibility in populations of African descent. When subjects were stratified first on age, the C genotypes were found to be significantly associated with prostate cancer in men 55 years of age and older, whereas when age was treated as a continuous variable the association with the C genotypes was only marginal. This is consistent with other reports that show that the risk for developing major cancers in women and men, prostate cancer included, increases with age [14–16].
The latest SEER statistics show that African American men have the highest incidence, as well as, the highest mortality rate of prostate cancer when compared to men of other ethnicities [17]. This significantly higher frequency of prostate cancer among African American men shows that ethnic diversity within the larger study population of African descent can be used to assess the impact of population substructure on prostate cancer susceptibility. With the C genotypes conferring a greater than two times risk for prostate cancer in African American men demonstrates the potential value of utilizing genetic diversity within the African Diaspora to stratify the population in research on the biology of health disparities. The significant difference in prostate cancer frequency found with the subset of self-reported African American men in contrast to the ethnically more heterogeneous total sample population of African descent raises intriguing and provocative questions about the impact of population history and environment in susceptibility to complex diseases, like health disparities. Of particular interest is the relationship between the CD14 variant, innate immunity, and continental origin of populations in response to environmental factors, such as pathogens, in health disparities. The finding reported here of an association between the C genotypes of CD14 with prostate cancer in African Americans raises intriguing questions about the biology of CD14. The frequency of the (−260) variant in populations of African descent is known to differ significantly from European populations which may reflect natural selection in allele frequency due to pathogen-related environmental differences between these two disparate populations. In our studies on the genetics of innate immunity in health disparities, we use significant differences in the SNP frequencies between CEPH/European and Yoruban/African populations, taken from HapMap data, to identify a subset of natural variants that we term “environmentally informative markers.” For this reason, the CD14 (−260) variant was chosen for this study.
The functionality of the CD14 (−260) variant has been linked to diseases of chronic inflammation, specifically allergy and asthma [18,19]. The homozygous TT genotype has an increase in serum levels of soluble CD14 (sCD14) and a decrease in total serum IgE compared to carriers of the C genotypes (CC and CT), which suggests that CD14 may play a role in the regulation of IgE synthesis as well as IgE-mediated conditions such as allergy and asthma [8]. Also, Benhnia et al. [20] demonstrated that CD14 signaling is responsible for maintaining a complex balance of pro- and anti-inflammatory mediators to produce disease resistance or susceptibility to the spirochetal bacterium, Borrelia burgdorferi in Lyme disease, a chronic inflammatory disorder.
It is noteworthy that TT homozygotes for the (−260) variant had significantly lower levels of total serum IgE as well as significantly higher serum levels of sCD14 when compared to both CC and CT genotypes [13]. It is expected that decreased serum levels of sCD14 and increased total serum IgE levels would be advantageous in an environment where extracellular parasitic disease, specifically schistosomiasis is endemic. In April of 2008, there were 207 million cases of schistosomiasis reported in Sub-Saharan Africa and Eastern Brazil [21]. Hagan [22] has associated the production of very high concentrations of specific and non-specific IgE in response to helminth antigen with a protective advantage against infection. Thus, in an environment where effective immunity to schistosomiasis is important to survival, carriers of the C genotypes (CC/CT) would have a selective advantage over those carrying the TT genotype. This is consistent with evidence of the higher frequency of the C allele in the West African Yoruba population and populations of African descent like African Americans. It is possible that this variant is an example of antagonistic pleiotropy, which refers to the expression of a gene that can have both beneficial and detrimental effects on the host [23]. As stated above, in an environment where helminth infection is rampant, the ability to mount an effective inflammatory response to the pathogen is advantageous to host survival. However, this “adaptive advantage” may be a risk factor for disease in an environment where induction of a comparable innate inflammatory response is not associated with host defense to a physical stressor, such as a pathogen, but rather to “perceived (i.e., psychosocial) stressors,” that are well established risk factors in health disparities like prostate cancer.
In this regard, it is also noteworthy that the expression of membrane bound CD14 (mCD14) in bladder cancer cell lines has been shown to induce interleukin-8 (IL-8) in response to the PAMP, lipopolysaccharide (LPS). Shimizu et al. [24] found that treatment of human uroepithelial cancer cell lines, T24 and 5637, with phosphatidyl inositol-specific phospholipase C to remove mCD14 severely suppressed the induction of IL-8. Transfection of UM-UC-3, another human uroepithelial cancer cell line, with CD14 cDNA-expressing mCD14, resulted in a more efficient induction of IL-8 in response to LPS stimulation, indicating that mCD14 and not sCD14 is essential in the induction of IL-8 in bladder epithelial cells in response to LPS. It is notable that IL-8, a member of the CXC chemokine family, is secreted by leukocytes and tumor cells [25]. IL-8 is also known to enhance angiogenesis in tumors and stimulate endothelial cell growth via increased secretion of vascular growth factors and basic fibroblast growth factor [26–28]. Interestingly, IL-8 levels were increased in prostate cancer tumors advanced to the stage where they no longer respond to anti-androgens [29,30]. Lee et al. [31] demonstrated that IL-8 is involved in the activation of the androgen receptor and confers androgen-independent growth. The tyrosine kinases Src and FAK (focal adhesion kinase) were found to be involved in the IL-8-induced signaling pathway. Lentschat et al. [32] and Solomon et al. [33] found that mCD14 resides in cholesterol-rich lipid microdomains that contain src kinases. It remains to be determined whether populations of African descent with a higher frequency of the C allele for the (−260) variant also have more mCD14 than T allele carriers, suggesting that CD14 in prostate cancer cells may have an effect similar to that found in bladder cancer cell lines.
The association between the CD14 (−260C>T) polymorphism and prostate cancer reported here has interesting implications for the role of PRRs in regulation of the inflammatory response in health disparities. In addition, this finding is instructive in highlighting the added value of genetic variation in innate immune response genes to dissecting the biology of complex diseases, such as prostate cancer in African American men. The importance of cooperation between innate and adaptive immune mechanisms for effective tumor immunotherapy and tumor immune surveillance is widely recognized.
Acknowledgments
The authors wish to first thank all of the men who volunteered to participate in this genetic study and to acknowledge that this research was funded in part by the National Institutes of Health (S06 GM08016 and RR03048) and the Department of Defense (DAMD W81XWH-07-1-0203 and DAMD W81XWH-06-1-0066).
Grant sponsor: National Institutes of Health; Grant numbers: S06 GM08016, RR03048; Grant sponsor: Department of Defense; Grant numbers: DAMD W81XWH-07-1-0203, DAMD W81XWH-06-1-0066.
References
- 1.American Cancer Society. Cancer facts & figures for African Americans 2005–2006. Atlanta: American Cancer Society; 2005. pp. 1–28. [Google Scholar]
- 2.Royal CD, Dunston GM. Changing the paradigm from ‘race’ to human genome variation. Nat Genet. 2004;36:S5–S7. doi: 10.1038/ng1454. [DOI] [PubMed] [Google Scholar]
- 3.Correa P. Human gastric carcinogenesis: A multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 4.Podolsky DK. Mucosal immunity and inflammation. V. Innate mechanisms of mucosal defense and repair: The best offense is a good defense. Am J Physiol. 1999;277:G495–G499. doi: 10.1152/ajpgi.1999.277.3.G495. [DOI] [PubMed] [Google Scholar]
- 5.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, Lowry SF. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R, Janeway C. The Toll receptor family and microbial recognition. Trends Microbiol. 2000;8:452–456. doi: 10.1016/s0966-842x(00)01845-x. [DOI] [PubMed] [Google Scholar]
- 7.Medzhitov R, Janeway C. Innate immunity. N Engl J Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 8.LeVan TD, Bloom JW, Bailey TJ, Karp CL, Halonen M, Martinez FD, Vercelli D. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol. 2001;167:5838–5844. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 9.Triantafilou M, Miyake K, Golenbock DT, Triantafilou K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J Cell Sci. 2002;115:2603–2611. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- 10.Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raja J, Ramachandran N, Munneke G, Patel U. Current status of transrectal ultrasound-guided prostate biopsy in the diagnosis of prostate cancer. Clin Radiol. 2006;61:142–153. doi: 10.1016/j.crad.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Gleason DF. Histologic grading of prostate cancer: A perspective. Hum Pathol. 1992;23:273–279. doi: 10.1016/0046-8177(92)90108-f. [DOI] [PubMed] [Google Scholar]
- 13.Gleason DF. Classification of prostate carcinomas. Cancer Chemother Rep. 1966;50:125–128. [PubMed] [Google Scholar]
- 14.Yancik R. Cancer burden in the aged: An epidemiologic and demographic overview. Cancer. 1997;80:1273–1283. [PubMed] [Google Scholar]
- 15.Yancik R, Ries LA. Aging and cancer in America. Demographic and epidemiologic perspectives. Hematol Oncol Clin North Am. 2000;14:17–23. doi: 10.1016/s0889-8588(05)70275-6. [DOI] [PubMed] [Google Scholar]
- 16.Ries LA, Reichman ME, Lewis DR, Hankey BF, Edwards BK. Cancer survival and incidence from the surveillance. Epidemiology and End Results (SEER) Program. Oncologist. 2003;8:541–552. doi: 10.1634/theoncologist.8-6-541. [DOI] [PubMed] [Google Scholar]
- 17.Horner MJ, Ries LAG, Krapho M, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinch Comb DG, Edwards BK, editors. SEER Cancer Statistics Review, 1975–2006. Bethesda, MD: National Cancer Institute; 2008. Available at: http://seer.cancer.gov/csr/1975_2005. [Google Scholar]
- 18.Baldini M, Lohman C, Halonen M, Erickson RP, Holt PG, Martinez FD. A polymorphism* in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20:976–983. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- 19.Barnes KC. Genetic epidemiology of health disparities in allergy and clinical immunology. J Allergy Clin Immunol. 2006;117:243–254. doi: 10.1016/j.jaci.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 20.Benhnia MR-E-I, Wroblewski D, Akhtar MN, Patel R, Lavezzi W, Gangloff SC, Goyert SM, Caimano MJ, Radolf JD, Sellati TJ. Signaling through CD14 attenuates the inflammatory response to Borrelia burgdorferi, the agent of Lyme disease. J Immunol. 2005;174:1539–1548. doi: 10.4049/jimmunol.174.3.1539. [DOI] [PubMed] [Google Scholar]
- 21.Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J. Helminth infections: The great neglected tropical diseases. J Clin Invest. 2008;118:1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagan P. IgE and protective immunity to helminth infections. Parasite Immunol. 1993;15:1–4. doi: 10.1111/j.1365-3024.1993.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 23.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 24.Shimizu T, Shin-ichi Y, Takahashi S, Kunishima Y, Takeyama K, Masumori N, Takahashi A, Matsukawa M, Itoh N, Tsukamoto T, Fujii N. Membrane-anchored CD14 is important for induction of interleukin-8 by lipopolysaccharide and peptidoglycan in uroepithelial cells. Clin Diagn Lab Immunol. 2004;11:969–976. doi: 10.1128/CDLI.11.5.969-976.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie K. Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 2001;12:375–391. doi: 10.1016/s1359-6101(01)00016-8. [DOI] [PubMed] [Google Scholar]
- 26.Huang S, Mills L, Mian B, Tellez C, McCarty M, Yang XD, Gudas JM, Bar-Eli M. Fully humanized neutralizing antibodies to interleukin-8 (ABX-IL8) inhibit angiogenesis, tumor growth, and metastasis of human melanoma. Am J Pathol. 2002;161:125–134. doi: 10.1016/S0002-9440(10)64164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto J, Sakaguchi H, Aoki I, Tamaya T. Clinical implications of expression of interleukin 8 related to angiogenesis in uterine cervical cancers. Cancer Res. 2000;60:2632–2635. [PubMed] [Google Scholar]
- 28.Fasciani A, D’Ambrogio G, Bocci G, Monti M, Genazzani AR, Artini PG. High concentrations of the vascular endothelial growth factor and interleukin-8 in ovarian endometriomata. Mol Hum Reprod. 2000;6:50–54. doi: 10.1093/molehr/6.1.50. [DOI] [PubMed] [Google Scholar]
- 29.Aalinkeel R, Nair MP, Sufrin G, Mahajan SD, Chadha KC, Chawda RP, Schwartz SA. Gene expression of angiogenic factors correlates with metastatic potential of prostate cancer cells. Cancer Res. 2004;64:5311–5321. doi: 10.1158/0008-5472.CAN-2506-2. [DOI] [PubMed] [Google Scholar]
- 30.Lehrer S, Diamond EJ, Mamkine B, Stone NN, Stock RG. Serum interleukin-8 is elevated in men with prostate cancer and bone metastases. Technol Cancer Res Treat. 2004;3:411. doi: 10.1177/153303460400300501. [DOI] [PubMed] [Google Scholar]
- 31.Lee LF, Louie MC, Desai SJ, Yang J, Chen HW, Evans CP, Kung HJ. Interleukin-8 confers androgen-independent growth and migration of LNCaP: Differential effects of tyrosine kinases Src and FAK. Oncogene. 2004;23:2197–2205. doi: 10.1038/sj.onc.1207344. [DOI] [PubMed] [Google Scholar]
- 32.Lentschat A, Karahashi H, Michelsen KS, Thomas LS, Zhang W, Vogel SN, Arditi M. Mastoparan, a G protein agonist peptide, differentially modulates TLR4- and TLR2-mediated signaling in human endothelial cells and murine macrophages. J Immunol. 2005;174:4252–4261. doi: 10.4049/jimmunol.174.7.4252. [DOI] [PubMed] [Google Scholar]
- 33.Solomon KR, Kurt-Jones EA, Saladino RA, Stack AM, Dunn IF, Ferretti M, Golenbock D, Fleisher GR, Finberg RW. Heterotrimeric G proteins physically associated with the lipopolysaccharide receptor CD14 modulate both in vivo and in vitro response to lipopolysaccharide. J Clin Invest. 1998;102:2019–2027. doi: 10.1172/JCI4317. [DOI] [PMC free article] [PubMed] [Google Scholar]


