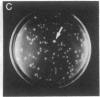
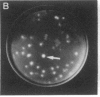
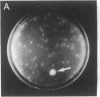
Abstract
Genomic clones were isolated that code for three glycosidases proposed to be involved in the catabolism of cell wall components in Saccharomyces cerevisiae. alpha-Mannosidase (AMS1), exoglucanase (BGL1), and endochitinase (CTS1) genes were isolated with the aid of filter assays based on the hydrolysis of 4-methylumbelliferyl glycosides, which permitted the in situ monitoring of these glycosidase activities in yeast colonies. Uracil prototrophs resulting from transformation with a multicopy YEp24 yeast genomic library were screened, leading to the identification of transformants possessing high levels of glycosidase activity. Restriction maps of plasmids from multiple isolates were used to localize glycosidase-overproduction genes, which were subcloned into a Schizosaccharomyces pombe/S. cerevisiae shuttle vector. Transformation of Sch. pombe with BGL1 and CTS1 subclones resulted in the appearance of these activities in this organism, and an AMS1 plasmid caused a 2-fold increase in endogenous alpha-mannosidase levels. Insertion of the marker gene LEU2 into putative AMS1 sequences disrupted plasmid-encoded alpha-mannosidase overproduction. S. cerevisiae strains that incorporated a restriction fragment containing ams1::LEU2 into their chromosomal DNA by homologous recombination expressed no detectable alpha-mannosidase activity in either the haploid or homozygous recessive diploid states, whereas heterozygous and wild-type cells exhibited levels proportional to AMS1 gene dosage. No readily apparent phenotype was associated with the alpha-mannosidase deficiency; however, labeling experiments utilizing [2-3H]mannose suggest that alpha-mannosidase may function in mannan turnover.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Cabib E., Bowers B. Chitin and yeast budding. Localization of chitin in yeast bud scars. J Biol Chem. 1971 Jan 10;246(1):152–159. [PubMed] [Google Scholar]
- Correa J. U., Elango N., Polacheck I., Cabib E. Endochitinase, a mannan-associated enzyme from Saccharomyces cerevisiae. J Biol Chem. 1982 Feb 10;257(3):1392–1397. [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Elango N., Correa J. U., Cabib E. Secretory character of yeast chitinase. J Biol Chem. 1982 Feb 10;257(3):1398–1400. [PubMed] [Google Scholar]
- Hien N. H., Fleet G. H. Separation and characterization of six (1 leads to 3)-beta-glucanases from Saccharomyces cerevisiae. J Bacteriol. 1983 Dec;156(3):1204–1213. doi: 10.1128/jb.156.3.1204-1213.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien N. H., Fleet G. H. Variation of (1 leads to 3)-beta-glucanases in Saccharomyces cerevisiae during vegetative growth, conjugation, and sporulation. J Bacteriol. 1983 Dec;156(3):1214–1221. doi: 10.1128/jb.156.3.1214-1221.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek-Kelly S., Akiyama T., Saunier B., Tkacz J. S., Herscovics A. Characterization of a specific alpha-mannosidase involved in oligosaccharide processing in Saccharomyces cerevisiae. J Biol Chem. 1985 Feb 25;260(4):2253–2257. [PubMed] [Google Scholar]
- Kaya T., Aikawa M., Matsumoto T. Purification and properties of alpha-mannosidase from bakers' yeast. J Biochem. 1977 Nov;82(5):1443–1449. doi: 10.1093/oxfordjournals.jbchem.a131832. [DOI] [PubMed] [Google Scholar]
- Olivero I., Hernandez L. M., Larriba G. Regulation of beta-exoglucanase activity production by Saccharomyces cerevisiae in batch and continuous culture. Arch Microbiol. 1985 Nov;143(2):143–146. doi: 10.1007/BF00411037. [DOI] [PubMed] [Google Scholar]
- Opheim D. J. alpha-D-Mannosidase of Saccharomyces cerevisiae. Characterization and modulation of activity. Biochim Biophys Acta. 1978 May 11;524(1):121–130. doi: 10.1016/0005-2744(78)90110-9. [DOI] [PubMed] [Google Scholar]
- Pastor F. I., Herrero E., Sentandreu R. Metabolism of Saccharomyces cerevisiae envelope mannoproteins. Arch Microbiol. 1982 Aug;132(2):144–148. doi: 10.1007/BF00508720. [DOI] [PubMed] [Google Scholar]
- Reichelt B. Y., Fleet G. H. Isolation, properties, function, and regulation of endo-(1 leads to 3)-beta-glucanases in Schizosaccharomyces pombe. J Bacteriol. 1981 Sep;147(3):1085–1094. doi: 10.1128/jb.147.3.1085-1094.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sánchez A., Villanueva J. R., Villa T. G. Saccharomyces cerevisiae secretes 2 exo-beta-glucanases. FEBS Lett. 1982 Feb 22;138(2):209–212. doi: 10.1016/0014-5793(82)80443-2. [DOI] [PubMed] [Google Scholar]
- del Rey F., Villa T. G., Santos T., Garcia-Acha I., Nombela C. Purification and partial characterization of a new, sporulation specific, exo-beta-glucanase from Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1982 Apr 29;105(4):1347–1353. doi: 10.1016/0006-291x(82)90935-4. [DOI] [PubMed] [Google Scholar]