ABSTRACT
Colorectal cancer (CRC) is the second leading cause of death from cancer in the United States. Aggressive research in the last decade has led to a wealth of information about this disease; for example, we now know that more than 80% of sporadic colon tumors contain mutations in the Wnt and TGFβ signaling pathways. The latest avenue of research is revealing the existence of and role for the cancer stem cell (CSC) model, which promotes the idea that malignancies originate from a small fraction of cancer cells that show self-renewal and multi- or pluripotency. The model also endorses that CSCs are capable of initiating and sustaining tumor growth. The body of evidence in favor of the CSC model is rapidly growing and includes analyses from flow cytometry of numerous CSC biomarkers, abnormal signaling pathways, symmetric division, dietary augmentation, and analysis of the behavior of these cells in spheroid culture formation. Although the incidence of death from CRC remains high, fervent research, both basic and translational, is beginning to improve patient outcomes. This paper focuses on stem cell biology in the context of CRC to help understand the mechanisms leading to tumor development and therapy resistance, with possible therapeutic indications.
Colorectal cancer (CRC) is the second leading cause of death from cancer in the United States. In 2009 there will have been an estimated 147,000 newly diagnosed cases of CRC and nearly 50,000 deaths associated with this disease.1 The age-adjusted incidence in the United States is 61.2 CRC cases per 100,000 population among men and 44.8 per 100,000 population among women.1 These incidences, while relatively high, have been slowly declining since 1985.2
A growing body of evidence supports the idea that human cancers can be considered a stem cell disease. According to the cancer stem cell (CSC) model, malignancies originate from a small fraction of cancer cells that show self-renewal and pluripotency and are capable of initiating and sustaining tumor growth.3 The cancer-initiating cells, or “cancer stem cells,” were first identified in hematologic malignancies and most recently in several solid tumors, including CRC.
The hypothesis of stem cell–driven tumorigenesis in colon cancer raises questions as to whether current treatments are able to efficiently target the tumorigenic cell population that is responsible for tumor growth and maintenance. This review will focus on several aspects of stem cell biology in the context of CRC to help understand the mechanisms that give rise to tumor development and therapy resistance. It will briefly review current knowledge on normal intestinal stem cells in the context of intestinal crypt biology, which has led to a new theory on the origins of colon adenomas and cancers, followed by a summary of the current status of colon CSC markers, signaling pathways, and prospective therapeutic options.
COLONIC STEM CELLS AND CRYPT ORGANIZATION
Colonic Crypt Organization
The colon is organized into four histologically distinct layers. The epithelial layer, at the luminal surface, consists of a single sheet of columnar epithelial cells folded into finger-like invaginations that are supported by the lamina propria to form the functional unit of the intestine, called crypts of Lieberkühn. There are four epithelial cell lineages. The terminally differentiated cells (enterocytes, goblet cells, and endocrine cells), which are found in the top third of the crypt, are derived from multipotent stem cells located at the bottom of the crypt. During asymmetric division, these multipotent cells undergo self-renewal and generate a population of transit amplifying cells that, upon migration upward through the crypt, proliferate and differentiate into one of the epithelial cell types of the intestinal wall. The fourth type of cells, the Paneth cells, differentiate during a downward migration to the base of the crypt, where they reside below the stem cell population.4
A variety of functions have been attributed to Paneth cells. These functions include modulation of the intestinal microflora and maintenance of mucosal defense barriers through production of antimicrobial peptides (cryptdins, lysozyme). The location of Paneth cells at the crypt base, as well as their production of growth factors and other regulatory molecules,5–7 suggests that they may also contribute to the stem-cell niche through short-circuit paracrine loops and/or regulate the proliferation and differentiation programs of other cell lineages.
A normal human crypt contains roughly 2,000 cells and is believed to have approximately 19 stem cells. Analyses of mitochondrial DNA mutations in these crypt cells have revealed that normal human colon crypts expand by fission, providing evidence that crypt structure and function are derived from the expansion of stem cells.8
Colon Stem Cells
Stem cells are defined by two functional properties: the ability to perpetuate themselves throughout an extended time period (self-renewal) and the potential to generate differentiated cells of the tissue of origin (multipotency). Despite the significant recent progress in the field of stem cell biology, the identification, isolation, and characterization of stem cells of the colonic crypt remain elusive. A more recent model, “the stem cell zone,” is based on the identification of small undifferentiated cycling cells interspersed within the Paneth cells, termed crypt base columnar cells (CBCs), which are believed to be the true intestinal stem cells.9
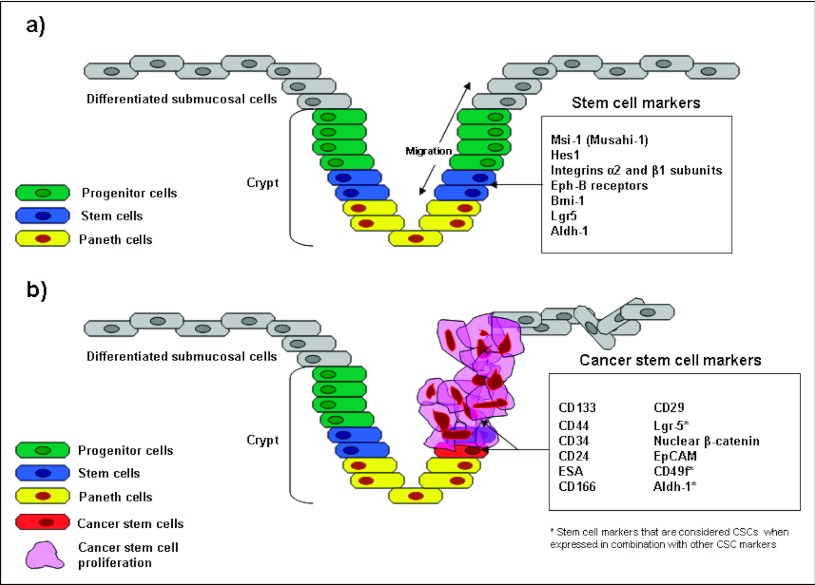
Several molecules have been proposed as markers of stem cells in the intestine (Table 1), including Musashi-1 (Msi-1), Hes1, integrins α2 and β1 subunits, EphB receptors, Bmi-1, Lgr5, and Aldh1 (Figure 1). The RNA-binding protein Msi-1, originally found in Drosophila melanogaster, is thought to be involved in asymmetric division during neuronal development.10 Immunohistochemical analysis performed in normal human colon crypts reveals that the majority of cells expressing Msi-1 reside in the lower region of the crypt, which corresponds to the expected position of the colonic stem cells.11 However, immunoreactivity is observed above the bottom of the crypt, suggesting that Msi-1 is still expressed by early transient-amplifying progenitor cells. Similarly, the expression of Hes1, a transcriptional repressor transactivated by Msi-1, has been evaluated in the mouse small intestine epithelium.12 Hes1 and Msi-1 were coexpressed by the putative stem cells at the crypt base, although Hes1 was expressed by a broader population of cells. Other putative biomarkers have been evaluated to distinguish the stem cell population within the colon, such as members of the integrin superfamily of transmembrane glycoproteins, including α2 and β1 subunits.13 More recently, EphB receptors were described as important regulators of migration and proliferation in the intestinal epithelium. The expression of both EphB2 and EphB3 tyrosine kinase receptors was reported at the bottom of the crypt in the mouse colon.14 Inhibition of EphB2/EphB3 signaling was shown to reduce the number of proliferating cells without altering the stem cell number, suggesting that EphB receptors are unlikely to be an independent biomarker of colonic stem cells. Conversely, a more promising intestinal stem cell marker might be Bmi-1, a factor involved in the self-renewal of hematopoietic and neural stem cells. Bmi-1 was recently reported to be expressed within the bottom crypts in the small intestine predominantly by the cells at the +4 position.15 G protein–coupled receptor 5 (Lgr5), also known as Gpr49, is proposed as a biomarker of colonic stem cells.16 Lgr5 is predicted to encode a seven-transmembrane protein with a large extracellular domain for ligand binding and a short cytoplasmic tail for coupling to G proteins. In the mouse colon, Lgr5 expression is restricted to cycling columnar cells at the crypt base, and it has been demonstrated that Lgr5-expressing cells differentiate into the expected functional lineages of the colonic epithelium.16 More recently, it has been described that single sorted Lgr-5 positive stem cells can also initiate long-term culture by generating crypt–villus organoids in which all differentiated cell lines are present.
Table 1.
Markers that have been proposed to characterize normal intestinal SCs and used to isolate colon CSCs
| Marker | Function | |
|---|---|---|
| Normal intestinal SCs | Musahi-1 | RNA binding protein |
| Hes-1 | Transcriptional repressor | |
| EphB receptors | Cell surface receptors | |
| Bmi-1 | Policomb-repressor protein | |
| Lgr-5 | Unknown, Wnt target gene | |
| Aldh-1 | Enzyme | |
| Colon CSCs | CD133 | Unknown |
| CD44 | Hyaluronic acid receptor | |
| CD166 | Adhesion molecule | |
| Aldh-1 | Enzyme |
Figure 1.
This represents the crypt of Lieberkühn, found in the colon wall, which continues into projecting villi (not shown here). Wnt signals are turned on in crypt stem and progenitor cells, and they are off in differentiated cells present in the villi, hence establishing the crypt–villus boundary. (a) Normal intestinal epithelium showing the crypt stem cells (blue) migrating/differentiating in both directions to produce enterocytes, goblet cells, enteroendocrine cells, as well as Paneth cells. Some common colonic stem cell markers are presented. (b) Tumorigenic intestinal epithelium, in which a colonic stem cell has acquired certain mutations to become a cancer stem cell (CSC), proliferating aberrantly and disrupting the characteristics of the adjacent cells. Some common CSC markers are presented.
Emerging new data also reveal a role for transforming growth factor-β (TGF-β) signaling in the development of gut endoderm17 and the transition of stem cells to a more differentiated phenotype. The signaling mechanisms that regulate development and cancer in most organ systems can be similar. In the gut, the TGF-β signaling pathway is a prominent player for both situations. Studies have localized the TGF-β-receptor-2 (TBR2) to both differentiated cells of the villus and undifferentiated cells near the bottom of the crypt18 during gut development. Recently, it was suggested that TGF-β and Wnt pathways synergistically promote CRC tumorigenesis. In this case, compound heterozygous ApcΔ716/+/Smad4± mice developed larger colon polyps that could progress to malignant adenocarcinoma.19 In addition, Apc+/N1638 mice20 developed increased tumor multiplicity when expressed on a Smad4 heterozygote background. Furthermore, inactivation of TBR2 in intestinal epithelial cells of Apc+/N1638 mice induces transformation and invasion of tumors, which are initiated by the APC mutation. These tumors are dramatically increased in a cell-autonomous manner.21 In addition, current studies indicate epigenetic regulation of the TGF-β pathway members, specifically β2–general spectrin (β2SP), a scaffolding protein that functions as a potent TGF-β–signaling Smad3/4 adaptor, may play a key role in generating transitional stem cells and inducing epithelial cell differentiation.17,22–24 The results of these studies confirm that disruption of both TGF-β and Wnt signaling cooperate to drive tumor initiation, likely in colonic stem cells, and progression in vivo.
Manipulating Stem Cell Marker Genes Reveals Function
Manipulating specific genes that are considered markers for crypt stem cells has resulted in two outcomes: identifying the function of that gene, and helping to identify the stemness of crypt cells. The following is a summary of results for down-regulated stem cell markers: (1) Msi-1: siRNA-mediated reduction of Msi-1 leads to mitotic catastrophe in tumor cells. Knockdown of Msi-1 also results in tumor growth arrest in xenografts, reduced cancer cell proliferation, and increased apoptosis alone and in combination with radiation injury. Moreover, there is inhibition of Notch-1 and up-regulation of p21WAF1 upon knockdown of Msi-1.25 (2) Hes-1: Targeted disruption of the Hes1 gene increases enteroendocrine and goblet cells and reduces enterocytes in the fetal mouse intestine,26 suggesting that Hes1 normally functions to negatively regulate specification of enteroendocrine goblet cell lineages and to positively regulate specification of enterocytes.27 (3) Lgr-5: LGR5 deficiency deregulates Wnt signaling and leads to precocious Paneth cell differentiation in the mouse fetal intestine. This deregulation is associated with overexpression of Wnt target genes in the intervillus epithelium. Transcriptional profiling of mutant mice ileums reveals that LGR5 function is associated with expression of stem cell and stem cell niche markers.28
COLON CANCER AND THE CRC STEM CELL THEORY
Colorectal Carcinogenesis
From a molecular point of view, CRC is one of the best-characterized cancers, mainly because studies of hereditary cases, which account for about 15% of CRCs, have revealed many biologic aspects of this neoplasm. The genetic events include increases in cellular proliferation and the silencing of genes involved in inhibition of proliferation and apoptosis. The accumulation of mutations involving oncogenes and tumor suppressor genes follows the progression of the disease along the adenoma–carcinoma sequence, in which the neoplastic process, initiated by APC or β-catenin mutations and tumor progression, results from the sequential mutation of other genes, such as K-Ras and p53, in the context of a growing genomic instability. This model has been further refined, and studies performed on relatively rare inherited cases, such as Beckwith Wiedemann Syndrome, led to the identification of genetic alterations in pathways such as TGF-β that play a major role in the development of sporadic CRC.
CRC Stem Cells
CSCs are defined by characteristics similar to those of normal colonic stem cells, mainly their abilities to self-renew, a characteristic that drives tumorigenesis, and to (aberrantly) differentiate, a property that generates the bulk of cells within a tumor. These self-renewing “cancer stem cells” might constitute only a small fraction of the tumor cells, with the bulk of the tumor composed of more differentiated cells that lack self-renewal capacity. According to the CSC hypothesis, it can be assumed that the first mutational hit occurs in a colonic stem cell located at the crypt bottom that, being long lived, can accumulate oncogenic mutations over years or decades. Once transformed, mutated stem cells can divide symmetrically and asymmetrically giving rise to other CSCs and progenitors, which in turn generate other cancer cells devoid of self-renewal ability. Eventually, the entire niche will be colonized by mutant stem cells, and the crypt will be filled with their progeny. The proliferating cancer cells will be subjected to further changes that may result in the progression of cancer.
Colon CSC Markers
Specific markers have been correlated to the cancer stem cell phenotype (Table 1). Experiments using flow cytometric analysis and spheroid culture formation to identify cells with surface markers that might correlate with the stem cell tumorigenic phenotype have shown the involvement of molecules including CD133,29–31 CD44,30–33 CD34,34 CD24, epithelial-specific antigen (ESA),32,33,35 CD166,31 CD29,31 Lgr5,31 nuclear β-catenin,31 EpCAM,35 CD49f,35 and ALDH 1.36
Several studies have demonstrated the expression of specific cell surface biomarkers. In the first two of these studies, CD133 was used to identify a colon cancer–initiating cell (CC-IC) population in human tumors.37,38 The tumorigenic potential of CD133+ CC-IC was evaluated by sorting freshly dissociated tumor cells and injecting them into immunocompromised mice. CD133+ cells, which account for approximately 2.5% of the bulk tumor cells, were shown to be devoid of the intestinal epithelial differentiation marker cytokeratin 20 (CK20), while expressing the epithelial adhesion molecule BerEp4 (also known as EpCAM). Although this suggests that CD133+ cells signify colon CSCs, it has also been reported that only 1 in 262 CD133+ cells would be a true CSC.38 The study also demonstrated the expression of CD133 in normal colon tissue, although at lower frequency, suggesting that CD133+ CC-ICs in cancer samples might result from oncogenic transformation of normal colonic stem cells.
Cell surface proteins other than CD133 have been reported to mark colon CSCs. For example, CD166 combined with CD4435 or CD24 combined with CD29 (R. Fodde, PhD unpublished data, Dec.1, 2007) may define the colorectal CSC population.
Recent evidence indicates that spheroid cultures of primary cancer cells are superior to “regular” adherent grown cultures in medium containing serum37,39 because xenotransplanted tumors derived from such spheroid cultures more faithfully preserve the original gene expression profiles and tumor morphology,31 CD24, CD29, CD44, and CD166, which have all been described to enrich for CSCs in CRCs (R. Fodde, PhD unpublished data, Dec.1, 2007),35 were also expressed on a subpopulation in those spheroid cultures. However, immunohistochemical analysis of normal colonic epithelium showed that CD44 expression occurs not only in the stem cell compartment at the crypt bottom but also in cells within the proliferative compartment: thus, the specificity of CD44 for colonic stem cells remains to be determined.
In other studies, in six of six human CRCs tested, the ability to engraft in vivo in immunodeficient mice was restricted to a minority subpopulation of epithelial cell adhesion molecule EpCAMhigh/CD44+ epithelial cells.35 Tumors originating from EpCAMhigh/CD44+ cells maintained a differentiated phenotype and reproduced the full morphologic and phenotypic heterogeneity of their parental lesions. Analysis of the surface molecule repertoire of EpCAMhigh/CD44+ cells led to the identification of CD166 as an additional differentially expressed marker, useful for CSC isolation in three of three CRCs tested.35 To better characterize the colon-CSC surface marker repertoire and evaluate whether colon-CSCs could be further enriched by subfractionation of the EpCAMhigh/CD44+ population, a systematic evaluation was started of markers already described as differentially expressed in other stem cell models, such as CD49f.35,37,38,40–42 Analysis of CD49f expression revealed a consistent and reproducible pattern, similar to that of EpCAM.35
More recently, aldehyde dehydrogenase 1 (ALDH1) has been proposed as a promising new marker for normal and malignant human colonic stem cells.36 Flow cytometric isolation of ALDH1+ cancer cells and implantation of as few as 25 cells in NOD/SCID mice generate tumor xenografts. Further isolation of cancer cells using a second marker (CD44 or CD133 serially) only modestly increased enrichment based on tumor-initiating ability.
THE TGF-β SIGNALING PATHWAY AND COLON CANCER
The TGF-β signaling pathway is involved in the control of cell proliferation, differentiation, migration, and apoptosis and is one of the most commonly altered pathways in human cancers.43,44 It is also important for stem cell maintenance, function, and carcinogenesis. TGF-β pathway signals are conveyed through serine/threonine kinase receptors to specific intracellular mediators known as Smad proteins.45 To date, eight Smad proteins have been found, and they are classified into three functional classes: (1) receptor-activated Smads (R-Smads): Smad1, Smad2, Smad3, Smad5, and Smad8; (2) comediator Smads: Smad4 and Smad10; and (3) inhibitory Smads: Smad6 and Smad7. Smad proteins function through adaptor proteins such as SARA and β2SP and by interacting with multiple other signal transduction pathways.22,46 Downstream targets of TGF-β signaling are key cell-cycle checkpoint genes including CDKN1A (p21), CDKN1B (p27), and CDKN2B (p15). In most cases, their activation leads to growth arrest.
Belonging to a ligand-receptor family that also includes bone morphogenetic protein and activin.47–49 TGF-β serves as a tumor suppressor in normal intestinal epithelium by inhibiting cell proliferation and inducing apoptosis. Many CRCs escape the tumor-suppressor effects of TGF-β and are resistant to TGFβ-induced growth inhibition.50 In fact, TGF-β is also often excessively produced in colorectal cancers, presumably owing to loss of feedback inhibition with disruption of its intracellular SMAD signaling pathway.51 Also, the autocrine activity from elevated secretion of TGF-β ligand has further consequences in that signaling through SMAD-independent pathways, unmasked with interruption of SMAD-dependent signaling, enhances cell proliferation and cell motility, two phenotypes consistent with tumorigenesis and metastatic behavior.52
Moreover, the TBR2 gene contains microsatellite sequences prone to replication errors, especially in the presence of MMR gene inactivation.53 Frameshift mutations of TBR2 are found in >80% of CRCs that demonstrate microsatellite instability.54 Mutations in the type I receptor (TBR1) have also been identified in human CRC cell lines, and reconstitution of TBR1 expression has been shown to reduce tumorigenicity.55 Smad4 mutatations have been identified in 16% to 25% of CRCs, and Smad2 alterations in approximately 6% of CRCs.55 Similarly, mice with a homozygous deletion of Smad3 have been shown to develop aggressive CRCs at an early age, depending on the genetic background of the mice.56
A relatively recent discovery that has revealed a novel underlying mechanism of CRC is the specific correlation in human CRC specimens of Smad4 expression and β2SP, a mediator of Smad3/4 nuclear translocation. When CRC specimens were compared with normal human colon tissue, distinct patterns of β2SP and Smad4 expression at the tips and crypts of colonic mucosa were observed. Further analysis demonstrated reduced β2SP in Dukes stage B1 tissues. However, the mouse genetic studies revealed a bigger picture. In these studies, 3 of 19 6-to-8-month-old β2SP±/Smad4± mutant mice developed colonic adenomas compared with none of the wild-type controls or Smad4± mutant mice. Based on accumulating evidence, β2SP functions by conferring cell polarity and maintaining cell architecture. Its loss is associated with the transition from hyperplasia to adenoma. Down-regulation of β2SP combined with loss of Smad4 is also seen in advanced and metastatic CRC.57 These data indicate a strong role for β2SP in TGF-β signaling and in the suppression of early CRC and, later, in metastatic disease with Smad4.
THE CANONICAL WNT SIGNALING PATHWAY AND COLON CANCER
In adult tissues, Wnt signaling is essential for the regulation of self-renewal, proliferation, and differentiation of pluripotent stem cells.58–63 Wnt signaling also appears to take on the same roles in several differentiated somatic cell types and in cancer cells.58–63 Consistent with these observations, different components of the Wnt signaling pathway are linked to tumorigenesis, including adenomatous polyposis and colon carcinoma.
The canonical Wnt signaling pathway is referred to as the Wnt/β-catenin pathway, because it can regulate β-catenin protein levels to control the activation of Wnt-responsive target genes. All Wnt signaling pathways are initiated by interaction of Wnt proteins with Frizzled (Fzd) receptors, but in this pathway the Wnt signaling will activate only if the binding of the Wnt protein to the Fzd receptor takes place in the presence of the coreceptor Lrp5/6 (low-density lipoprotein receptor-related protein 5/6) resulting in the formation of a Wnt:Fzd:Lrp5/6 trimolecular complex.64,65 This complex provides a favorable environment for the recruitment of Dishevelled (Dvl).66 The formation of the Wnt-Fzd-Lrp5/6 complex also promotes the Lrp5/6-mediated degradation of axin.65 The inhibition of glycogen-synthase kinase 3β (Gsk3β) activity by Wnt with the degradation of axin blocks the formation of the protein complex consisting of Gsk3β, axin, and adenomatous polyposis coli (Apc) tumor suppressor protein. If the formation of the protein complex of Gsk3β, axin, and Apc tumor suppressor protein does not occur, accumulation of free β-catenin results in its translocation to the nucleus. Once positioned in the nucleus, the free β-catenin acts as a transcription factor and activates T-cell factor (Tcf) and lymphoid enhancer factor (Lef) by forming nuclear complexes with members of the Tcf/Lef transcription factor family.67 This leads to the transcription and expression of a variety of Wnt-responsive target genes such as Myc, Ccnd1 (cyclin D1), and Axin2. In addition, the complexes of Tcf/Lef and β-catenin may cooperate with factors activated by other signaling pathways to alter cellular remodeling processes. The canonical Wnt signaling pathway is also activated by several other cellular mechanisms. The shifting of proteins from the cadherin-bound pool to the cytoplasmic pool can increase the amount of available free β-catenin for the activation of target genes.68
The Wnt cascade is essential in establishing cell fate along the crypt–villus axis of the intestinal epithelium. Accumulation of nuclear β-catenin, the hallmark of active Wnt signaling, is evident in the crypt cells of the normal intestine, whereas differentiated villus cells present β-catenin at their basolateral membrane, where it is important to ensure cell adhesion. Deletion of the transcription factor Tcf4, the most prominent effector of Wnt signaling in the gastrointestinal tract, produces a severe intestinal phenotype accompanied by neonatal lethality. Although the villus epithelial compartment is practically unaffected in these mice, intestinal crypts are completely absent and no proliferating cells are observed, indicating that Wnt signals are required for the maintenance of the crypt proliferative compartment. Thus, in physiologic conditions, the β-catenin:Tcf4 complex drives the transcription of a set of target genes that determine the characteristics of the intestinal crypt cells. Wnt signals are turned off in differentiated cells present in the villi, hence establishing the crypt–villus boundary (Figure 1). In rare cases in which APC is not inactivated, human intestinal tumors usually show activating mutations in β-catenin itself, or loss-of-function mutations in Axin2, a protein that cooperates with APC in β-catenin degradation.68
The Wnt signaling pathway was first causally associated with carcinogenesis when it was found to be permanently activated in familial adenomatous polyposis (FAP)69,70 and spontaneous forms of colon cancer.71,72 Chronic activation of Wnt signaling in these intestinal tumors results from inactivating mutations in the APC locus and, to a much lesser extent, in β-catenin and AXIN2/conductin genes. Although it appears that mutations of Wnt pathway components are sufficient to generate constitutive activation of Wnt signaling in CRC, recent evidence suggests that additional autocrine mechanisms involving stimulation of the pathway by secreted WNT proteins also have a role. Thus, it was shown that CRC cells frequently express WNT, and that treatment with secreted Fzd-related protein (SFRP) via interference with Wnt receptor binding. Importantly, SFRP genes are subject to inactivation by hypermethylation in CRC cells, indicating that an epigenetic mechanism leads to a boost of the Wnt signal.73
CROSS-TALK BETWEEN TGF-β AND WNT SIGNALING PATHWAYS
Do the Wnt and TGF-β pathways cooperate for colorectal tumor progression? The answer appears to be yes. Another mechanism to silence TGF-β signaling, in addition to genetic ablation of its receptor, is deployment of the BMP and activin membrane–bound inhibitor (BAMBI), a pseudoreceptor that is related to TGF-β receptor type I but lacks an intracellular kinase domain that is important in activating intracellular SMADs.74 BAMBI resembles the homodimerization domain of TGF-β receptor type I to act as a decoy to prevent the formation of receptor complexes between the type I and type II receptors after TGF-β ligand binding.74 BAMBI is aberrantly elevated in most CRCs when compared with matched normal colon tissue from the same patient, and Wnt signaling, as evidenced by experimental interruption of β-catenin or T-cell factor nuclear transcriptional activity, induces the expression of BAMBI.75,76 Thus, Wnt signaling through β-catenin activation transcriptionally activates BAMBI as a mechanism to block TGF-β signaling. Additionally, there is evidence for positive feedback regulation by BAMBI on Wnt signaling.75 BAMBI interacts with the Wnt receptor Frizzled5, and its coreceptor LRP6 promotes the nuclear localization of β-catenin; and overexpression of BAMBI promotes Wnt/β-catenin transcriptional activity, including the expression of c-myc and cyclin D1, two of Wnt/β-catenin's transcriptional targets.77 Last, perhaps as a countermeasure to negatively regulate its own pathway, TGF-β–SMAD signaling can also induce BAMBI expression, because the BAMBI gene contains SMAD-binding elements in its promoter.76 Thus, BAMBI can be induced by both Wnt signaling and TGF-β–SMAD signaling to positively regulate Wnt signaling and negatively regulate TGF-β signaling. The overall effect of BAMBI would be to increase cellular growth through enhancement of Wnt proliferative signaling and inhibition of TGF-β–SMAD suppressive signaling.78
Evidence that β-catenin is responsible for the aberrant expression of BAMBI in colorectal tumor cells comes from experiments using a dominant-negative mutant of Tcf4 or by using an inhibitor of β-catenin-Tcf interaction; results showed repression of BAMBI in CRC cell lines. Furthermore, overexpression of BAMBI inhibits the tumor cell response to TGF-β signaling. These results suggest a mechanism by which β-catenin interferes with TGF-β–mediated growth arrest by inducing the expression of BAMBI, and thus contributing to colorectal and hepatocellular tumorigenesis.75
THERAPEUTICS TARGETING CRC STEM CELLS
The identification and potential targeting of colon CSCs and their signaling pathways, as well as understanding their surrounding environment, might lead to more effective, early diagnosis of cancer and focused treatment options; yet, many challenges remain.
The identification of these cells is not clear cut; hence, more specific stem cell markers are needed to enable targeting of “malfunctioning” cells at a premalignant level. Such targeting may promote differentiation and/or block the self-renewal pathway, possibly leading to a less chemoresistant cancer. Although epithelial-specific antigen (ESA), CD44, CD133, CD166, and Lgr5 are relatively good markers, they may also be potential targets for attacking CSCs in the colon. Expression of ESA, a cell surface marker, is associated with increased proliferation and decreased differentiation. CD44 is normally expressed at the base of dividing crypts in the proliferative zone; during neoplastic conditions, the distribution of CD44 expression extends to the luminal surface. CD133 expression seems to be restricted to undifferentiated cells, including endothelial progenitor cells, hematopoietic stem cells, prostatic epithelial stem cells, and leukemias. In a previous mouse model of acute myeloid leukemia, CD44 was targeted with a monoclonal activating antibody,79 which resulted in reversal of differentiation blockade, and prevention of normal homing of the cells to both bone marrow and spleen. A possible negative consequence of targeting CSCs is that the normal “stem cells” might also be affected. However, recent studies suggest differential sensitivity of normal and malignant stem cells to these agents.80
Another possibly beneficial treatment strategy is to target “cancer-initiating” signaling pathways such as the Wnt, Hedgehog, Notch, and TGF-β pathways, with a goal of suppressing self-renewal and promoting differentiation. β2-spectrin is decreased in colon tumor samples, and this information could potentially be applied as a screening method in the clinic to increase suspicion, or possibly confirm the presence, of colon cancer. Furthermore, the inhibition of CDK4, a gene target that is normally suppressed by the TGFβ-signaling pathway, shows promise in decreasing proliferation in colon cancer cells. Hence, further research is needed to identify specific targets in these pathways.
The stem cell niche is composed of fibroblasts, endothelia, and inflammatory cells and plays an important role in the maintenance and promotion of CSCs into more invasive and metastatic potentials.81 This niche is an anchoring site for stem cells and adhesion molecules. Cytokines facilitate communication between the stromal elements and the stem cells. Factors that are members of major developmental pathways, including the Wnt, bone morphogenic, Notch, and TGF-β pathways, are constantly secreted into the stromal microenvironment.82 This suggests that strategies aimed at the stem cell microenvironment might prove effective. A recent report described the autocrine production of IL-4 by CD133+ colon cancer stem-like cells.83 Though their cell isolates were relatively resistant to 5-fluorouracil or oxaliplatin, pretreatment of the cells with anti-IL-4 significantly increased ability of the chemotherapeutic agents to decrease tumorigenic growth. These findings indicate that IL-4 protects CD133+ cells and could thus dictate therapy refractoriness.83 In xenografts, the addition of IL-4 antibodies significantly reduced tumor growth after chemotherapy. In vivo, this enhancement was expressed as a substantial slowing of tumor growth, and normal tumor growth resumed upon withdrawal of the chemotherapeutic agent, showing that IL-4 exerts a protective effect on the CD133+ stem-like colon cancer isolates. Furthermore, interpretation of this microenvironment will allow better understanding of the relatively quiescent nature of CSCs as well as their innate multidrug resistance, which has been a major concern in chemotherapy failure and disease relapse.
Symmetric stem cell division can contribute to tumor growth.84 This finding suggests that systemic therapies for CRC and other cancers need to control or eliminate symmetric CSC division in tumors, while minimally affecting normal stem cell division in nontumoral tissues.44
There is extensive evidence that dietary supplementation reduces colon cell proliferation, the extent of the crypt's proliferative zone, and colon carcinogenesis in humans and rodents.85 Decreased sensitivity of colon cells to Ca2+ and disruption of their apoptotic mechanisms is seen in malignant transformation of colon cells. In fact, Ca2+ supplementation has been reported to stimulate colon cell proliferation in adenomatous polyps. This apparent antagonistic response is likely mediated by disrupted Wnt signaling in colorectal tumors and demonstrates the tightly linked and regulated process of colonic stem cell self-renewal, proliferation, and differentiation. Targeting of Wnt-driven stem cell proliferation along with dietary Ca2+ loading to restrain APC and the calcium-sensing receptor (CaSR) and promote adherens junctions, however, may prove an effective strategy. This also highlights the need for novel combinations targeting the multifaceted nature of CRC stem cells.
CONCLUSIONS
The identification of CRC stem cells has had a significant effect on CRC research, prevention, and therapy. New potential biomarkers and signaling pathways could have important roles in the specificity, identification, and understanding of CSCs and may represent targets to suppress self-renewal or promote differentiation. Other potential targets involve the tumor microenvironment, which is emerging as a crucial stem cell/CSC regulator, as well as stem cell symmetric division. Substantial additional research in this field is needed and requires use of cutting-edge materials and methods including induced pluripotent stem cells, which are capable of differentiating into definitive endoderm,86–88 pluripotent human, monkey, and mouse ES cells; gut primary cells and cell lines; mouse knockout/knockin technology; and sound translational/clinical approaches. It is hoped that results of these synergistic research efforts will eventually reduce the number of CRC cases, currently at 61.2 and 44.8 CRC cases/100,000 men and women, respectively, to near 0.
REFERENCES
- 1. Jemal A, Siegel R, Ward E, et al. : Cancer statistics, 2009. Cancer J Clin 59(4):225–249, 2009 [DOI] [PubMed] [Google Scholar]
- 2. Lieberman DA: Clinical practice. Screening for colorectal cancer. N Engl J Med 361(12):1179–1187, 2009 [DOI] [PubMed] [Google Scholar]
- 3. Boman BM, Wicha MS: Cancer stem cells: a step toward the cure. J Clin Oncol 26(17):2795–2799, 2008 [DOI] [PubMed] [Google Scholar]
- 4. Troughton WD, Trier JS: Paneth and goblet cell renewal in mouse duodenal crypts. J Cell Biol 41(1):251–268, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raaberg L, Nexo E, Damsgaard J, et al. : Immunohistochemical localisation and developmental aspects of epidermal growth factor in the rat. Histochemistry 89(4):351–356, 1988 [DOI] [PubMed] [Google Scholar]
- 6. de Sauvage FJ, Keshav S, Kuang WJ, et al. : Precursor structure, expression, and tissue distribution of human guanylin. Proc Natl Acad Sci U S A 89(19):9089–9093, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mulherkar R, Desai SJ, Rao, et al. : Expression of enhancing factor gene and its localization in mouse tissues. Histochemistry 96(4):367–370, 1991 [DOI] [PubMed] [Google Scholar]
- 8. Greaves LC, Preston SL, Tadrous PJ, et al. : Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A 103(3):714–719, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ricci-Vitiani L, Fabrizi E, Palio E, et al. : Colon cancer stem cells. J Mol Med 87:1097–1104, 2009 [DOI] [PubMed] [Google Scholar]
- 10. Nakamura M, Okano H, Blendy JA, et al. : Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 13(1):67–81, 1994 [DOI] [PubMed] [Google Scholar]
- 11. Nishimura S, Wakabayashi N, Toyoda K, et al. : Expression of Musashi-1 in human normal colon crypt cells: a possible stem cell marker of human colon epithelium. Dig Dis Sci 48(8):1523–1529, 2003 [DOI] [PubMed] [Google Scholar]
- 12. Kayahara T, Sawada M, Takaishi S, et al. : Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett 535(1–3):131–135, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Fujimoto K, Beauchamp RD, Whitehead RH: Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology 123(6):1941–1948, 2002 [DOI] [PubMed] [Google Scholar]
- 14. Holmberg J, Genander M, Halford MM, et al. : EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 125(6):1151–1163, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Sangiorgi E, Capecchi MR: Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40(7):915–920, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barker N, van Es JH, Kuipers J, et al. : Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449(7165):1003–1007, 2007 [DOI] [PubMed] [Google Scholar]
- 17. Mishra L, Derynck R, Mishra B: Transforming growth factor-beta signaling in stem cells and cancer. Science 310(5745):68–71, 2005 [DOI] [PubMed] [Google Scholar]
- 18. Barnard JA, Beauchamp RD, Coffey RJ, et al. : Regulation of intestinal epithelial cell growth by transforming growth factor type beta. Proc Natl Acad Sci U S A 86(5):1578–1582, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takaku K, Oshima M, Miyoshi H, et al. : Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 92(5):645–656, 1998 [DOI] [PubMed] [Google Scholar]
- 20. Alberici P, Jagmohan-Changur S, De Pater E, et al. : Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene 25(13):1841–1851, 2006 [DOI] [PubMed] [Google Scholar]
- 21. Munoz NM, Upton M, Rojas A, et al. : Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res 66(20):9837–9844, 2006 [DOI] [PubMed] [Google Scholar]
- 22. Tang Y, Katuri V, Dillner A, et al. : Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science 299(5606):574–577, 2003 [DOI] [PubMed] [Google Scholar]
- 23. Tang Y, Kitisin K, Jogunoori W, et al. : Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci U S A 105(7):2445–2450, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin L, Amin R, Gallicano GI, et al. : The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene 28(7):961–972, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sureban SM, May R, George RJ, et al. : Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology 134(5):1448–1458, 2008 [DOI] [PubMed] [Google Scholar]
- 26. Jensen J, Pedersen EE, Galante P, et al. : Control of endodermal endocrine development by Hes-1. Nat Genet 24(1):36–44, 2000 [DOI] [PubMed] [Google Scholar]
- 27. Mills JC, Gordon JI: The intestinal stem cell niche: there grows the neighborhood. Proc Natl Acad Sci U S A 98(22):12334–12336, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia MI, Ghiani M, Lefort A, et al. : LGR5 deficiency deregulates Wnt signaling and leads to precocious Paneth cell differentiation in the fetal intestine. Dev Biol 331(1):58–67, 2009 [DOI] [PubMed] [Google Scholar]
- 29. Singh SK, Clarke ID, Hide T, et al. : Cancer stem cells in nervous system tumors. Oncogene 23(43):7267–7273, 2004 [DOI] [PubMed] [Google Scholar]
- 30. Collins AT, Berry PA, Hyde C, et al. : Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 65(23):10946–10951, 2005 [DOI] [PubMed] [Google Scholar]
- 31. Vermeulen L, Todaro M, de Sousamelloqq F, et al. : Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A 105(36):13427–13432, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. : Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100(7):3983–3988, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li C, Heidt DG, Dalerba P, et al. : Identification of pancreatic cancer stem cells. Cancer Res 67(3):1030–1037, 2007 [DOI] [PubMed] [Google Scholar]
- 34. Bonnet D, Dick JE: Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3(7):730–737, 1997 [DOI] [PubMed] [Google Scholar]
- 35. Dalerba P, Dylla SJ, Park IK, et al. : Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 104(24):10158–10163, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang EH, Hynes MJ, Zhang T, et al. : Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 69(8):3382–3389, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. : Identification and expansion of human colon-cancer-initiating cells. Nature 445(7123):111–115, 2009 [DOI] [PubMed] [Google Scholar]
- 38. O'Brien CA, Pollett A, Gallinger S, et al. : A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445(7123):106–110, 2007 [DOI] [PubMed] [Google Scholar]
- 39. Lee J, Kotliarova S, Kotliarov Y, et al. : Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9(5):391–403, 2006 [DOI] [PubMed] [Google Scholar]
- 40. Singh SK, Hawkins C, Clarke ID, et al. : Identification of human brain tumour initiating cells. Nature 432(7015):396–401, 2004 [DOI] [PubMed] [Google Scholar]
- 41. Shackleton M, Vaillant F, Simpson KJ, et al. : Generation of a functional mammary gland from a single stem cell. Nature 439(7072):84–88, 2006 [DOI] [PubMed] [Google Scholar]
- 42. Storms RW, Trujillo AP, Springer JB, et al. : Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci U S A 96(16):9118–9123, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Massague J, Blain SW, Lo RS: TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103(2):295–309, 2000 [DOI] [PubMed] [Google Scholar]
- 44. Boman BM, Huang E: Human colon cancer stem cells: a new paradigm in gastrointestinal oncology. J Clin Oncol 26(17):2828–2838, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Shi Y, Massague J: Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113(6):685–700, 2003 [DOI] [PubMed] [Google Scholar]
- 46. Tsukazaki T, Chiang TA, Davison AF, et al. : SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell 95(6):779–791, 1998 [DOI] [PubMed] [Google Scholar]
- 47. Beck SE, Jung BH, Fiorino A, et al. : Bone morphogenetic protein signaling and growth suppression in colon cancer. Am J Physiol Gastrointest Liver Physiol 291(1):G135–G145, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jung B, Doctolero RT, Tajima A, et al. : Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology 126(3):654–659, 2004 [DOI] [PubMed] [Google Scholar]
- 49. Jung BH, Beck SE, Cabral J, et al. : Activin type 2 receptor restoration in MSI-H colon cancer suppresses growth and enhances migration with activin. Gastroenterology 132(2):633–644, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoosein NM, McKnight MK, Levine AE, et al. : Differential sensitivity of subclasses of human colon carcinoma cell lines to the growth inhibitory effects of transforming growth factor-beta 1. Exp Cell Res 181(2):442–453, 1989 [DOI] [PubMed] [Google Scholar]
- 51. Grady WM, Carethers JM: Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135(4):1079–1099, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chow JY, Cabral JA, Chang J, et al. : TGFbeta modulates PTEN expression independently of SMAD signaling for growth proliferation in colon cancer cells. Cancer Biol Ther 7(10):1694–1699, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ilyas M, Efstathiou JA, Straub J, et al. : Transforming growth factor beta stimulation of colorectal cancer cell lines: type II receptor bypass and changes in adhesion molecule expression. Proc Natl Acad Sci U S A 96(6):3087–3091, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takayama T, Miyanishi K, Hayashi T, et al. : Colorectal cancer: genetics of development and metastasis. J Gastroenterol 41(3):185–192, 2006 [DOI] [PubMed] [Google Scholar]
- 55. Xu Y, Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet 16(spec. no. 1):R14–R20, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thiagalingam S, Lengauer C, Leach FS, et al. : Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet 13(3):343–346, 1996 [DOI] [PubMed] [Google Scholar]
- 57. Tang Y, Katuri V, Srinivasan R, et al. : Transforming growth factor-beta suppresses nonmetastatic colon cancer through Smad4 and adaptor protein ELF at an early stage of tumorigenesis. Cancer Res 65(10):4228–4237, 2005 [DOI] [PubMed] [Google Scholar]
- 58. Nusse R: An ancient cluster of Wnt paralogues. Trends Genet 17(8):443, 2001 [DOI] [PubMed] [Google Scholar]
- 59. Grigoryan T, Wend P, Klaus A, et al. : Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev 22(17):2308–2341, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang H, He X: Wnt/beta-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol 20(2):119–125, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ling L, Nurcombe V, Cool SM: Wnt signaling controls the fate of mesenchymal stem cells. Gene 433(1–2):1–7, 2009 [DOI] [PubMed] [Google Scholar]
- 62. Nusse R: Wnt signaling and stem cell control. Cell Res 18(5):523–527, 2008 [DOI] [PubMed] [Google Scholar]
- 63. Mosimann C, Hausmann G, Basler K: Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10(4):276–286, 2009 [DOI] [PubMed] [Google Scholar]
- 64. Wehrli M, Dougan ST, Caldwell K, et al. : arrow encodes an LDL-receptor-related protein essential for Wingless signaling. Nature 407(6803):527–530, 2000 [DOI] [PubMed] [Google Scholar]
- 65. Mao J, Wang J, Liu B, et al. : Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell 7(4):801–809, 2001 [DOI] [PubMed] [Google Scholar]
- 66. Kishida M, Hino S, Michiue T, et al. : Synergistic activation of the Wnt signaling pathway by Dvl and casein kinase Iepsilon. J Biol Chem 276(35):33147–33155, 2001 [DOI] [PubMed] [Google Scholar]
- 67. Ishitani T, Ninomiya-Tsuji J, Matsumoto K: Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol 23(4):1379–1389, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Espada J, Calvo MB, Diaz-Prado S, et al. : Wnt signaling and cancer stem cells. Clin Transl Oncol 11(7):411–427, 2009 [DOI] [PubMed] [Google Scholar]
- 69. Kinzler KW, Nilbert MC, Su LK, et al. : Identification of FAP locus genes from chromosome 5q21. Science 253(5020):661–665, 1991 [DOI] [PubMed] [Google Scholar]
- 70. Nishisho I, Nakamura Y, Miyoshi Y, et al. : Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 253(5020):665–669, 1991 [DOI] [PubMed] [Google Scholar]
- 71. Korinek V, Barker N, Morin PJ, et al. : Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275(5307):1784–1787, 1997 [DOI] [PubMed] [Google Scholar]
- 72. Morin PJ, Sparks AB, Korinek V, et al. : Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275(5307):1787–1790, 1790 [DOI] [PubMed] [Google Scholar]
- 73. Liang H, Chen Q, Coles AH, et al. : Wnt5a inhibits B cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell 4(5):349–360, 2003 [DOI] [PubMed] [Google Scholar]
- 74. Onichtchouk D, Chen YG, Dosch R, et al. : Silencing of TGF-beta signaling by the pseudoreceptor BAMBI. Nature 401(6752):480–485, 1999 [DOI] [PubMed] [Google Scholar]
- 75. Sekiya T, Adachi S, Kohu K, et al. : Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. J Biol Chem 279(8):6840–6846, 2004 [DOI] [PubMed] [Google Scholar]
- 76. Sekiya T, Oda T, Matsuura K, et al. : Transcriptional regulation of the TGF-beta pseudoreceptor BAMBI by TGF-beta signaling. Biochem Biophys Res Commun 320(3):680–684, 2004 [DOI] [PubMed] [Google Scholar]
- 77. Lin Z, Gao C, Ning Y, et al. : The pseudoreceptor BMP and activin membrane-bound inhibitor positively modulates Wnt/beta-catenin signaling. J Biol Chem 283(48):33053–33058, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Carethers JM: Intersection of transforming growth factor-beta and Wnt signaling pathways in colorectal cancer and metastasis. Gastroenterology 137(1):33–36, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jin L, Hope KJ, Zhai Q, et al. : Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med 12(10):1167–1174, 2006 [DOI] [PubMed] [Google Scholar]
- 80. Huang EH, Wicha MS: Colon cancer stem cells: implications for prevention and therapy. Trends Mol Med 14(11):503–509, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Deng G, Lu Y, Zlotnikov G, et al. : Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science 274(5295):2057–2059, 1996 [DOI] [PubMed] [Google Scholar]
- 82. Li L, Neaves WB: Normal stem cells and cancer stem cells: the niche matters. Cancer Res 66(9):4553–4557, 2006 [DOI] [PubMed] [Google Scholar]
- 83. Todaro M, Alea MP, Di Stefano AB, et al. : Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 1(4):389–402, 2007 [DOI] [PubMed] [Google Scholar]
- 84. Boman BM, Wicha MS, Fields JZ, et al. : Symmetric division of cancer stem cells: a key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther 81(6):893–898, 2007 [DOI] [PubMed] [Google Scholar]
- 85. Whitfield JF: Calcium, calcium-sensing receptor and colon cancer. Cancer Lett 275(1):9–16, 2009 [DOI] [PubMed] [Google Scholar]
- 86. Si-Tayeb K, Noto FK, Nagaoka M, et al. : Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51(1):297–305, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sullivan GJ, Hay DC, Park I, et al. : Generation of functional human hepatic endoderm from human iPS cells. Hepatology 51(1):329–335, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gallicano GI, Mishra L: Hepatocytes from iPS cells: a giant leap forward for hepatology. Hepatology 51(1):20–22, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]