Abstract
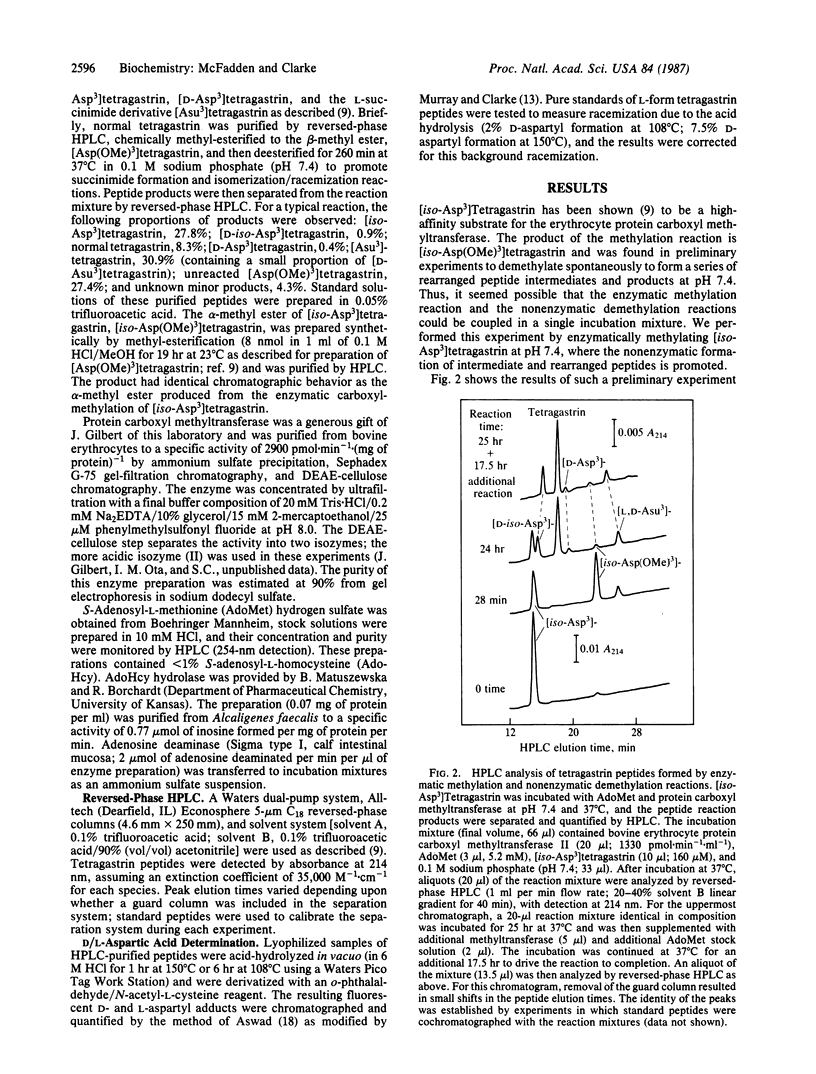
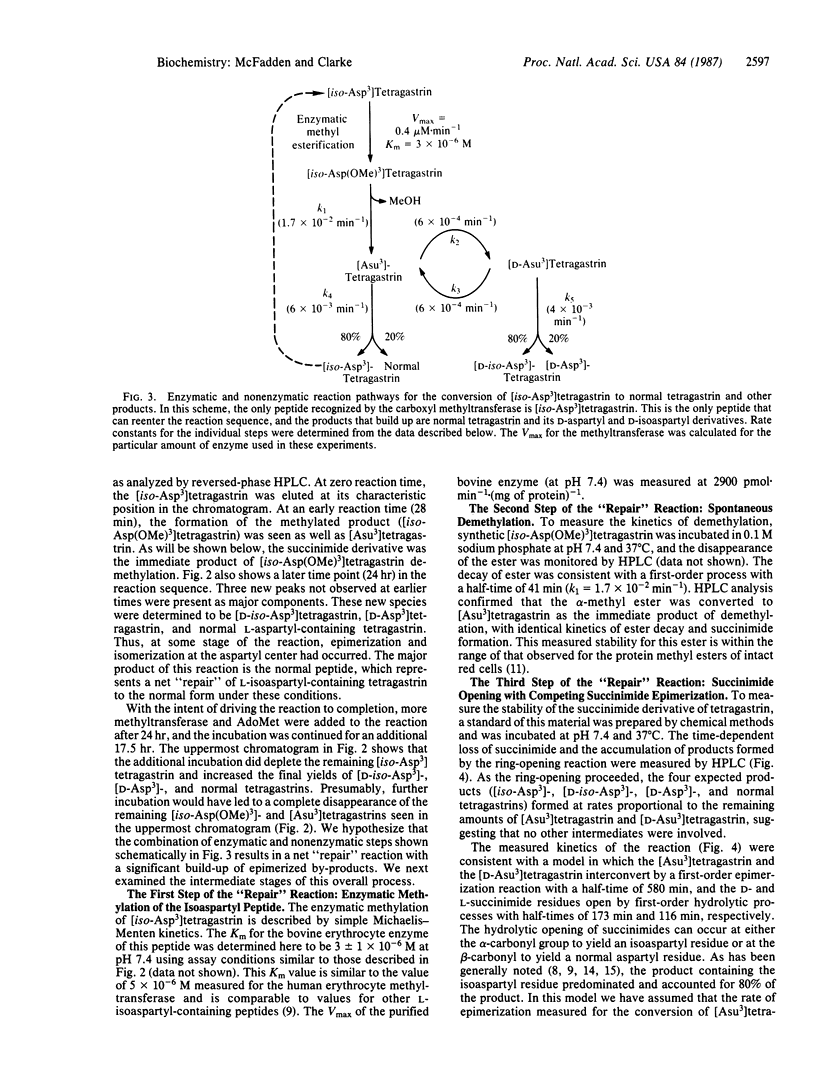
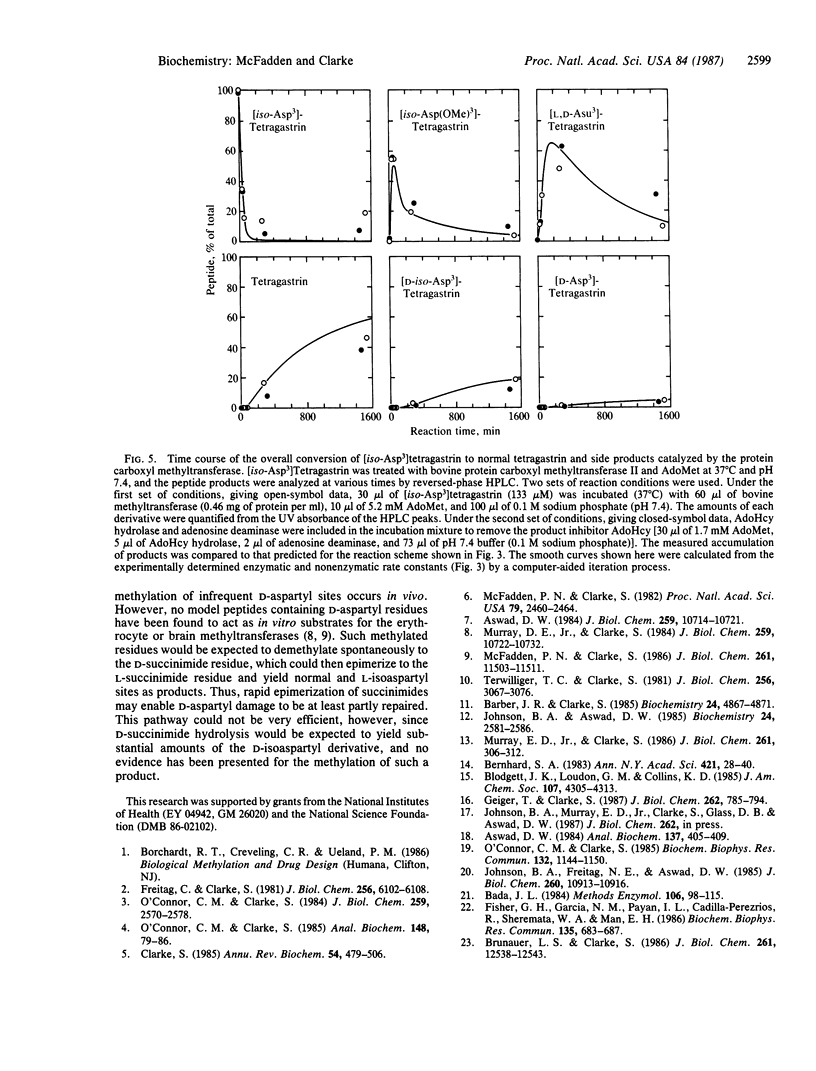
The hypothesis that cellular protein carboxyl-methylation reactions recognize altered aspartyl residues as part of a protein repair pathway has been tested in an in vitro system using tetragastrin (Trp-Met-Asp-Phe-NH2) as a model sequence. The L-isoaspartyl form of tetragastrin, where the phenylalanine residue is linked to the side-chain carboxyl group of the aspartate residue ([iso-Asp3]tetragastrin), is a substrate for the erythrocyte protein carboxyl methyltransferases, while the normal form is not. The enzymatically produced alpha-methyl ester of [iso-Asp3]tetragastrin, [iso-Asp(OMe)3]tetragastrin, is unstable at pH 7.4 and 37 degrees C and spontaneously demethylates with a half-time of 41 min to an intermediate L-succinimide form ([Asu3]tetragastrin) that, in turn, spontaneously hydrolyzes with a half time of 116 min to give a mixture of normal tetragastrin (20%) and [iso-Asp3]tetragastrin (80%). This sequence of enzymatic and nonenzymatic reactions can be coupled in a single reaction mixture; the [iso-Asp3]tetragastrin that is produced upon succinimide hydrolysis can reenter the reaction sequence by enzymatic methylation, and the net result of the process is the conversion of the isomerized peptide to the normal peptide. The efficiency of this "repair" reaction is limited by a side reaction of racemization at the alpha-carbon of the succinimide (half-time = 580 min). In a 24-hr time period, normal L-aspartyl-containing tetragastrin is obtained in about 50% yield from the coupled reaction mixture; other products include [D-iso-Asp3]tetragastrin and [D-Asp3]tetragastrin. The versatile chemistry of succinimide peptides suggests that methylated L-isoaspartyl sites (and possibly methylated D-aspartyl sites) in cellular polypeptides can eventually yield "repaired" normal L-aspartyl sites through succinimide intermediates.
Full text
PDF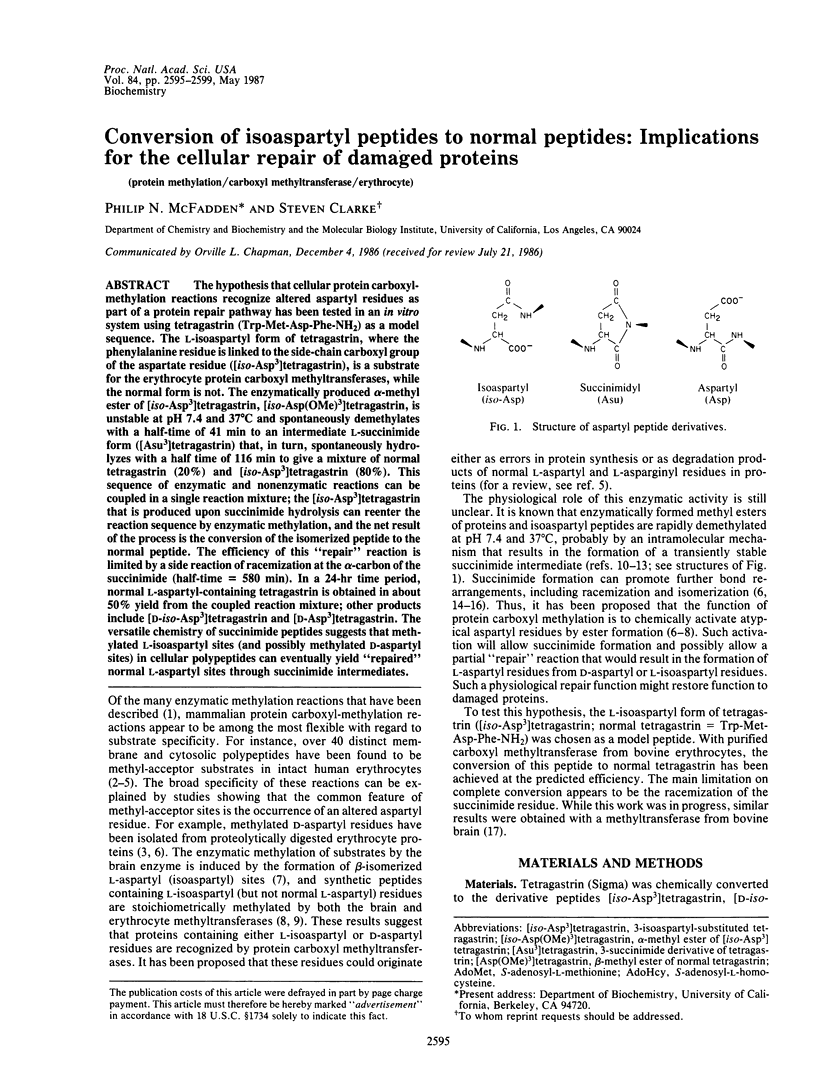

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aswad D. W. Determination of D- and L-aspartate in amino acid mixtures by high-performance liquid chromatography after derivatization with a chiral adduct of o-phthaldialdehyde. Anal Biochem. 1984 Mar;137(2):405–409. doi: 10.1016/0003-2697(84)90106-4. [DOI] [PubMed] [Google Scholar]
- Aswad D. W. Stoichiometric methylation of porcine adrenocorticotropin by protein carboxyl methyltransferase requires deamidation of asparagine 25. Evidence for methylation at the alpha-carboxyl group of atypical L-isoaspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10714–10721. [PubMed] [Google Scholar]
- Bada J. L. In vivo racemization in mammalian proteins. Methods Enzymol. 1984;106:98–115. doi: 10.1016/0076-6879(84)06011-0. [DOI] [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Demethylation of protein carboxyl methyl esters: a nonenzymatic process in human erythrocytes? Biochemistry. 1985 Aug 27;24(18):4867–4871. doi: 10.1021/bi00339a021. [DOI] [PubMed] [Google Scholar]
- Bernhard S. A. Nucleophilic displacement reactions at ester and thiolester bonds. Ann N Y Acad Sci. 1983;421:28–40. doi: 10.1111/j.1749-6632.1983.tb18090.x. [DOI] [PubMed] [Google Scholar]
- Brunauer L. S., Clarke S. Age-dependent accumulation of protein residues which can be hydrolyzed to D-aspartic acid in human erythrocytes. J Biol Chem. 1986 Sep 25;261(27):12538–12543. [PubMed] [Google Scholar]
- Clarke S. Protein carboxyl methyltransferases: two distinct classes of enzymes. Annu Rev Biochem. 1985;54:479–506. doi: 10.1146/annurev.bi.54.070185.002403. [DOI] [PubMed] [Google Scholar]
- Fisher G. H., Garcia N. M., Payan I. L., Cadilla-Perezrios R., Sheremata W. A., Man E. H. D-aspartic acid in purified myelin and myelin basic protein. Biochem Biophys Res Commun. 1986 Mar 13;135(2):683–687. doi: 10.1016/0006-291x(86)90047-1. [DOI] [PubMed] [Google Scholar]
- Freitag C., Clarke S. Reversible methylation of cytoskeletal and membrane proteins in intact human erythrocytes. J Biol Chem. 1981 Jun 25;256(12):6102–6108. [PubMed] [Google Scholar]
- Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem. 1987 Jan 15;262(2):785–794. [PubMed] [Google Scholar]
- Johnson B. A., Aswad D. W. Enzymatic protein carboxyl methylation at physiological pH: cyclic imide formation explains rapid methyl turnover. Biochemistry. 1985 May 7;24(10):2581–2586. doi: 10.1021/bi00331a028. [DOI] [PubMed] [Google Scholar]
- Johnson B. A., Freitag N. E., Aswad D. W. Protein carboxyl methyltransferase selectively modifies an atypical form of calmodulin. Evidence for methylation at deamidated asparagine residues. J Biol Chem. 1985 Sep 15;260(20):10913–10916. [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Chemical conversion of aspartyl peptides to isoaspartyl peptides. A method for generating new methyl-accepting substrates for the erythrocyte D-aspartyl/L-isoaspartyl protein methyltransferase. J Biol Chem. 1986 Sep 5;261(25):11503–11511. [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Methylation at D-aspartyl residues in erythrocytes: possible step in the repair of aged membrane proteins. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2460–2464. doi: 10.1073/pnas.79.8.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E. D., Jr, Clarke S. Metabolism of a synthetic L-isoaspartyl-containing hexapeptide in erythrocyte extracts. Enzymatic methyl esterification is followed by nonenzymatic succinimide formation. J Biol Chem. 1986 Jan 5;261(1):306–312. [PubMed] [Google Scholar]
- Murray E. D., Jr, Clarke S. Synthetic peptide substrates for the erythrocyte protein carboxyl methyltransferase. Detection of a new site of methylation at isomerized L-aspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10722–10732. [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Analysis of erythrocyte protein methyl esters by two-dimensional gel electrophoresis under acidic separating conditions. Anal Biochem. 1985 Jul;148(1):79–86. doi: 10.1016/0003-2697(85)90630-x. [DOI] [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Carboxyl methylation of cytosolic proteins in intact human erythrocytes. Identification of numerous methyl-accepting proteins including hemoglobin and carbonic anhydrase. J Biol Chem. 1984 Feb 25;259(4):2570–2578. [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Specific recognition of altered polypeptides by widely distributed methyltransferases. Biochem Biophys Res Commun. 1985 Nov 15;132(3):1144–1150. doi: 10.1016/0006-291x(85)91926-6. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C., Clarke S. Methylation of membrane proteins in human erythrocytes. Identification and characterization of polypeptides methylated in lysed cells. J Biol Chem. 1981 Mar 25;256(6):3067–3076. [PubMed] [Google Scholar]



