Abstract
Dehydration proteins (Dehydrins) are expressed during dehydration stress in plants and are thought to protect plant proteins and membranes from the loss of water during drought and at cold temperatures. Several different dehydrins have been shown to protect lactate dehydrogenase (LDH) from damage from being frozen and thawed. We show here that a 48 residue K2 dehydrin from Vitis riparia protects LDH more effectively than bovine serum albumin, a protein with known cryoprotective function. Light scattering and 8-anilino-1-naphthalene sulfonate fluorescence experiments show that dehydrins prevent aggregation and unfolding of the enzyme. The cryoprotective effects of LDH are reduced by the addition of salt, suggesting that the positively charged K-segments are attracted to a negatively charged surface but this does not result in binding. Overall K2 is an intrinsically disordered protein; nuclear magnetic resonance relaxation experiments indicate that the two-terminal, Lys-rich K-segments show a weak propensity for α-helicity and are flexible, and that the central, polar rich phi-segment has no secondary structure preference and is highly flexible. We propose that the phi-segments in dehydrins are important for maintaining the disordered structure so that the protein can act as a molecular shield to prevent partially denatured proteins from interacting with one another, whereas the K-segments may help to localize the dehydrin near the enzyme surface.
Keywords: cryoprotection, dehydrin, dynamics, freeze-thaw, intrinsically disordered protein, lactate dehydrogenase
Introduction
The inability of plants to flee an environmental stress has resulted in the evolution of an extensive stress response system. One example of an abiotic stress is dehydration, which can take the form of drought (lack of environmental water), high salinity (high osmolarity), or freezing (lack of liquid water). During periods of dehydration, a wide variety of plants can express dehydration proteins (dehydrins), which are also members of the plant late embryo abundant protein family.1–4 The accumulation of dehydrin transcripts and proteins during dehydration, and a correlation between the level of drought tolerance and the amount of dehydrin present, strongly suggest that they are involved in protecting the plant from the negative effects of dehydration. Numerous in vitro functions have been described and proposed for dehydrins, including cryoprotection of lactate dehydrogenase (LDH),5–8 cryoprotection of purified protoplasts and chloroplasts,9 prevention of water loss,10 binding of excess ions,11 binding of nucleic acids,12 prevention of protein aggregation at elevated temperatures,13 and prevention of ice crystal growth.14,15 Of these functions, the most extensively studied has been the cryoprotection of LDH, where it has been shown that dehydrins are more effective than small molecules such as sucrose or other proteins such as bovine serum albumin (BSA) at protecting LDH activity from freeze-thaw damage.7
The cloning of many dehydrin genes from different plants has revealed a number of features, the most notable being the modular nature of their sequence, which results in a large range of sizes (6–200 kDa). The nomenclature used to describe the dehydrin sequence is the YSK notation, where each letter represents the name of a segment with a conserved sequence.1 By definition, dehydrins contain at least one K-segment (the sequence EKKGIMKIKEKLPG), which is partly conserved among the different dehydrin members.1 The K-segment occurs 1–12 times, with 1 or 2 repeats being the most common. The S-segment contains a tract of Ser residues and is present in one or no copies in a dehydrin. Dehydrins extracted from drought-stressed plants are phosphorylated on these serines.16 The role of phosphorylation is not clear, but may be correlated with translocation of dehydrins to the nucleus,17 or the increased negative charge could enhance the ability of the protein to bind divalent cations such as zinc.18 The Y-segment refers to the sequence (V/T)DEYGNP is similar to the nucleotide binding domain found in bacteria. Typically, 1–3 Y-segments are present at the N-terminus of a dehydrin.1 Although not specifically included in the YSK naming system, dehydrins also contain φ-segments, which are rich in Gly, Thr, and many other polar amino acids. This poorly conserved segment tends to be located between the Y-, S-, and K-segments. Not surprisingly, the overall high-polar residue content of dehydrins causes them to be intrinsically disordered.19 Intrinsically disordered proteins (IDPs) are molecules that, contrary to the structure-function paradigm, do not have a single, well-defined fold (for reviews see, e.g., Refs.20 and21). They are generally highly flexible and have minimal secondary structure, although some IDPs undergo a disorder-to-order transition after binding their target.21 Their many functions include cell cycle control, assembly of protein complexes, and the modification of protein activity.20
Our model system for studying the biochemistry and structure of dehydrins uses the 48 residue K2 protein from Vitis riparia with a segmental architecture of K-φ-K. A previous study has shown that this dehydrin may be a splice variant of YSK2 mRNA, which is transcribed during cold stress.22 The presence of this K2 protein in plants has not yet been demonstrated, but the protein represents a minimal dehydrin construct that can be used to examine the role of K- and φ-segments in its cryoprotective function. Here, we examine the structural propensity and dynamics of K2 in solution, and measure its ability to protect LDH from freeze-thaw damage. Several mechanisms are explored, and it is shown how structural changes correlate with the amount of K2 added.
Results
Segment alignment, structure, and dynamics
An alignment of 20 dehydrin K-φ-K regions is shown in Figure 1, where the conserved and similar residues in the protein are highlighted. As seen with other dehydrins,4 the K-segments are moderately conserved. In the alignment of the first K-segment, the Lys amino acids in the 2nd and 5th positions are completely conserved, whereas the 4th and 8th positions are mostly Lys or a similarly positively charged Arg amino acid. The second K-segment contains more conserved residues, with the Lys present in all sequences in the 3rd, 8th, 10th, and 12th positions. Although the protein is highly hydrophilic, hydrophobic residues are also conserved. In the first K-segment, the 5th position is mostly Ile, whereas the 9th position is mostly Leu. Similarly, in the second K-segment, a hydrophobic Ile is completely conserved in the ninth position, whereas a hydrophobic Leu is completely conserved in the 13th position. In the majority of the alignments, both K-segments end with (hydrophobic)-Pro-Gly. The regions following the second K-segment are 2–8 residues in length, and are rich in His and Gly residues. The φ-segment in the middle of the alignment is highly variable in sequence and length, being rich in Gly, Thr, Ser, and Ala residues (i.e., amino acids with small side chains), with Gln being the most common amino acid with a longer side chain.
Figure 1.
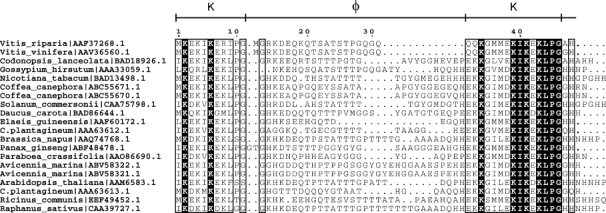
Alignment of K-φ-K sequences. A ClustalW23 generated alignment of 20 K2 regions with the highest sequence identity to Vitis riparia K2. Conserved regions are shown bound by a rectangle, where a white letter on a black background indicates a residue that is conserved in all sequences, and a black letter indicates residues that are partially conserved and/or chemically similar. The sequence numbering for Vitis riparia K2 and the segment names are shown at the top of the alignment. The organism name and protein NCBI accession number are indicated on the left. The figure was generated using ESPript.24
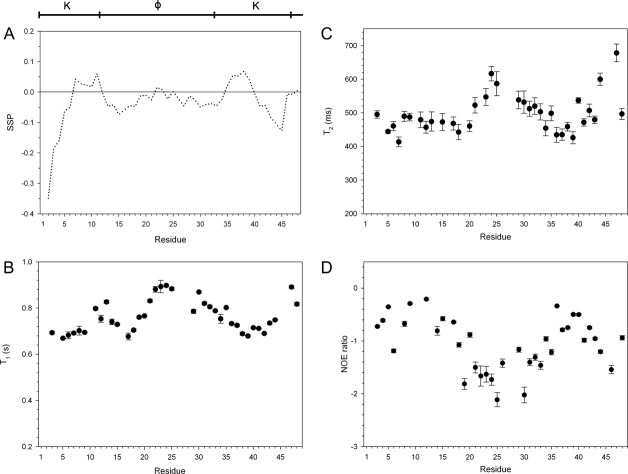
The segmental architecture is also reflected in the structural propensity of V. riparia K2 in solution. The secondary structure propensity (SSP) program can be used to describe the presence of transient secondary structures in IDPs, where positive scores are equated with propensity of the disordered ensemble to be in an α-helical conformation, and negative scores indicate the propensity to be in a β-strand conformation.25 For the K2 studied here, the overall SSP pattern is symmetric about the middle of the sequence except for the first 4 and the last two residues [Fig. 2(A)]. The core of the K-segments has a tendency to form a helix-like structure, whereas the N-terminal end of the first segment and the C-terminal end of the second segment tend to be more strand-like. The final two residues have no secondary structure preference. For the φ-segment, the beginning and end of this region show a propensity toward being extended, while the middle has very little consistent preference for either secondary structure.
Figure 2.
Structure and dynamics of K2 in solution. The arrangement of the K- and φ-segments is shown at the top of the figure. A: The SSP scores of K2 were calculated using Hα, Cα, and Cβ chemical shifts and are plotted on a per residue basis. B: T1, (C) T2, and (D) 15N-NOE relaxation data of K2. Error bars represent the error in fitting the relaxation decay curves as described in CCPNMR.
Like the SSP scores, the heteronuclear 15N relaxation data show that the protein is not highly structured [Fig. 2(B–D)]. For folded, globular proteins, heteronuclear nuclear Overhauser effect (NOE) values are positive and near 1. As can be seen in Figure 2(D), none of the residues have NOEs that are positive, which shows that the K2 protein has high flexibility and is undergoing fast motions. The T1 [Fig. 2(B)] and T2 [Fig. 2(C)] measurements also indicate that the protein is very flexible. All of the relaxation data show the same pattern between the different regions as indicated by the SSP scores, where residues in the K-segments are flexible, and the residues in the φ-segments are even more flexible, with the highest amount of motion seen in the middle of the protein. The last two C-terminal residues show an increase in the amount of flexibility compared with the preceding residues in the K-segment. The lower flexibility in the K-segments and the higher flexibility in the φ-segments support the SSP data, that is, the presence of transient structure in the K-regions and the lack of any structure in the φ-region.
Denaturation and cryoprotection of LDH
One in vitro protective mechanism that has been shown for a number of dehydrins is their ability to protect LDH from losing activity due to being frozen and thawed.5–8 For controls, the assay was also performed in the presence of BSA (a known cryoprotective agent) and lysozyme (a poor cryoprotective agent). As shown in Figure 3, the K2 and YSK2 proteins have cryoprotective abilities that are better than BSA, an observation that has also been made with other dehydrins.7 The fitted curves have a sigmoidal shape when plotted as log concentration versus percent recovery, indicating that below a threshold concentration of ∼0.5 μg/mL K2 there is no ability to protect LDH and, at high concentrations (>4 μg/mL), there is a saturating effect on enzyme protection. YSK2 was able to provide 50% recovery of LDH activity at approximately twofold lower protein concentration, showing that the larger protein provides more efficient protection. LDH protection by lysozyme was weak, with the curve plotted in the figure representing the most effective protection that we measured. On occasion, we observed no protection under identical experimental conditions, but this inconsistency in protection was never observed with BSA, YSK2 or K2. Other researchers have used lysozyme as a negative control in the LDH assay, with one group reporting essentially no protection,8 and the other group reporting weak protection similar to what was observed here.26
Figure 3.
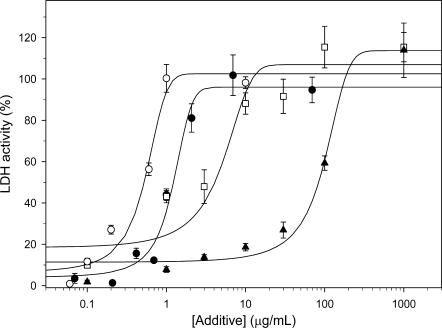
Cryoprotection of LDH. The protection of LDH activity after freezing and thawing of the enzyme was examined in the presence and absence of additives (K2, YSK2, BSA, or lysozyme) over a range of concentrations. The activity of unfrozen LDH is defined as 100% activity. K2, solid circle; YSK2, open circle; BSA, open square; lysozyme, solid triangle. Error bars represent the standard deviation of n = 6 (K2, BSA, lysozyme) or n = 3 (YSK2). The lines represent fits to the sigmoidal equation: % LDH activity =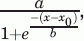 , where x is the concentration of the additive.
, where x is the concentration of the additive.
Several structural properties of LDH before and after freezing were examined to determine how the treatment has damaged the protein. These experiments were repeated in the presence and the absence of K2 at three concentrations, one of which resulted in no recovery of activity, one with ∼50% recovery of activity, and one which recovered 100% of the LDH activity. The use of several concentration of K2 with differing effects on the recovery of LDH allows one to distinguish between structural changes in the enzyme that are correlated with the cryoprotective effects of K2 and those that are not.
The first property examined was the oligomeric state of LDH before and after freezing. Native and fully active LDH is a tetramer, whereas dimers and monomers have no activity.27 Unfrozen and freeze-thawed samples of LDH were loaded onto a Superdex 200 gel filtration column. As shown in Figure 4(A), both treated and untreated proteins eluted at exactly the same position and gave the same peak shape. This shows that the freeze-thaw process used here did not cause the enzyme to irreversibly dissociate.
Figure 4.
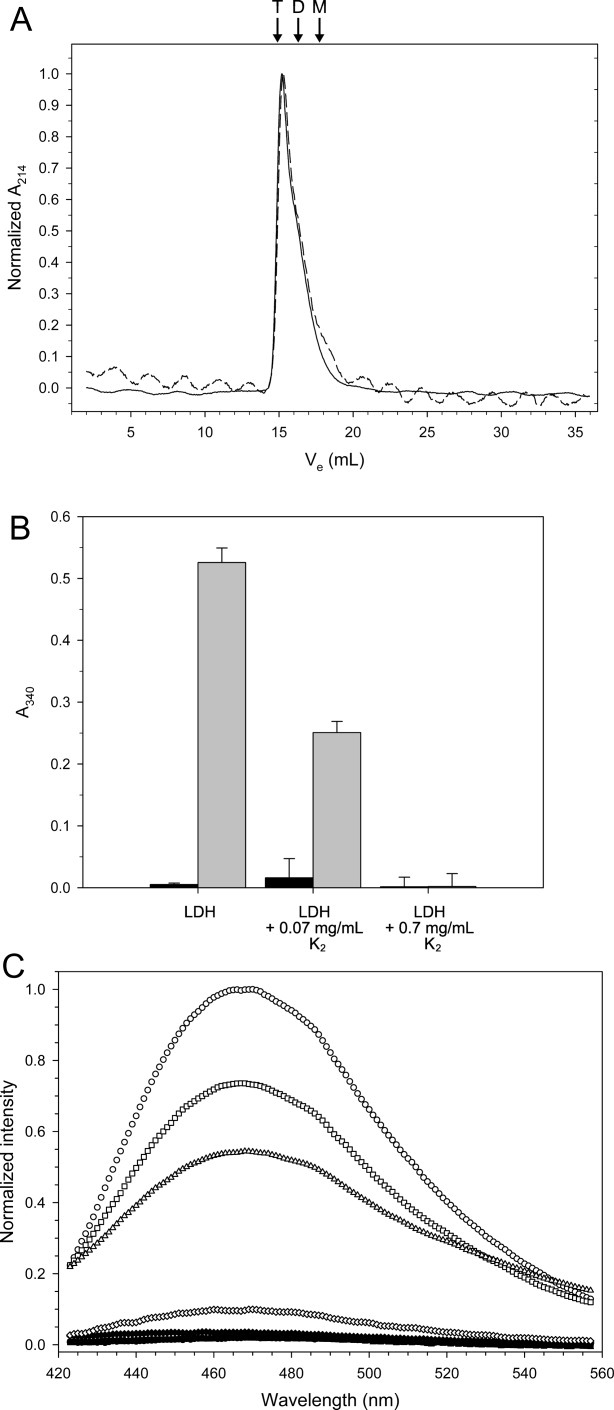
Effect of freezing and freeze-protection on the structure of LDH. A: Gel filtration of 200 μg of LDH before (solid line) and after (dashed line) being frozen and thawed. The arrows and labels indicate the predicted elution volume of the various oligomeric states: T, tetramer; D, dimer; M, monomer. Ve represents the elution volume of the sample. B: Light scatter measured at 340 nm. The bar graph represents data from n = 6 measurements, and the error bars show their standard deviation. The sample conditions are listed below each entry, where black represents unfrozen samples and gray represents frozen and thawed samples. C: Emission scans of ANS fluorescence of 1 μM LDH in the presence and the absence of K2. Samples were scanned before (solid symbols) and after (open symbols) freeze-thaw treatment. Symbols: LDH alone, circle; LDH with 0.028 mg/mL K2, square; 0.28 mg/mL K2, triangle; 0.65 mg/mL K2, diamond.
During gel filtration, a decrease in the amount of eluted LDH was observed after the freeze-thaw cycle, which suggested that the protein was aggregating. To examine the role of aggregation, the scatter at A340 of several enzyme samples before and after freezing were measured [Fig. 4(B)]. The absorbance readings were corrected for scatter caused by buffer and/or K2. For the LDH alone sample, the scatter increased ∼100-fold after the freeze-thaw treatment, indicating that aggregation contributes to the loss of LDH activity. Surprisingly, the addition of K2 at a concentration that produced 50% protection reduced the scatter to background levels, and the addition of K2 that would result in 0% activity increased scatter only 50-fold compared with the 100-fold increase seen for frozen LDH alone [Fig. 4(B)]. This suggests that the prevention of aggregation is not the only factor in the dehydrin cryoprotective mechanism, because it does not account for the loss of activity seen in the presence of lower concentrations of K2.
Therefore, we used the fluorescent dye 8-anilino-1-naphthalene sulfonate (ANS) to explore what further changes may be occurring to the structure of LDH. ANS is used to examine various unfolded states since its fluorescence intensity changes dramatically when the probe binds to solvent exposed hydrophobic patches.28–30 The emission scans of frozen and thawed LDH alone showed that the intensity at 470 nm increased 47-fold compared with the untreated control, demonstrating that hydrophobic residues had been exposed [Fig. 4(C)]. Increasing concentrations of K2 showed decreasing ANS fluorescence to the point where at maximal protection by K2 the fluorescence returned to near background levels. Therefore, the cryoprotective effects of K2 are a combination of preventing aggregation and partial denaturation of LDH.
Cryoprotection by K2
How K2 and LDH may be interacting was subsequently examined using nuclear magnetic resonance (NMR). 15N-heteronuclear single quantum coherence (HSQC) spectra have been extensively used to map interaction sites between a protein and its ligand, because changes in the 1H and 15N chemical shifts can be correlated with the binding of a ligand.31 15N-HSQC spectra of K2 in the presence and the absence of a three molar excess of LDH were overlapped. As can been seen in Figure 5(A), there was absolutely no shift in any of the resonances, demonstrating that there is no direct binding interaction between K2 and LDH (i.e., the interaction has a Kd weaker than the millimolar range).
Figure 5.
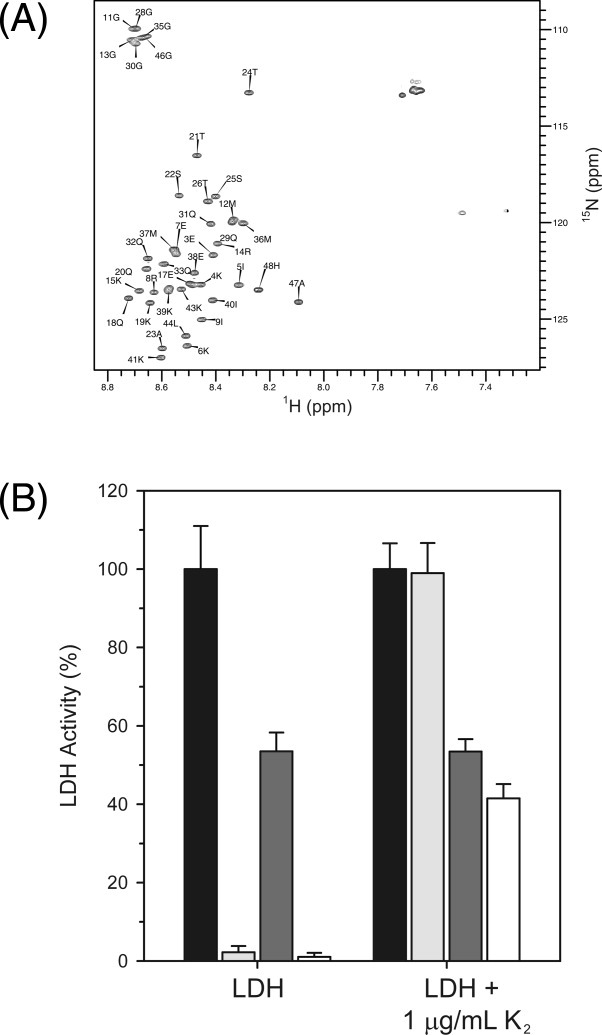
K2 interaction with LDH. (A) 15N-HSQC of 0.1 mM of K2 collected in the absence (black contours) or the presence (gray contours) of 0.3 mM LDH. The peak labels show the residue number and single letter amino acid code of K2. B: Cryoprotection of LDH by K2 is mediated by weak electrostatic interactions. The bar graph shows the percent LDH activity of samples assayed in the presence and the absence of salt with and without 1 μg/mL K2. The data represent n = 6 measurements with the error bar corresponding to the standard deviation. Unfrozen samples with no NaCl, black bars; frozen samples with no NaCl, light gray; unfrozen samples with 1M NaCl, dark gray; frozen samples with 1M NaCl, white.
The lack of a direct interaction suggests that the cryoprotective function of dehydrins may be to act as a shield that prevents denatured LDH from associating with other LDH molecules. The cryoprotection by YSK2 is slightly better than that provided by K2 (see Fig. 3). This is most likely a reflection of the increased size of YSK2 (13.9 kDa vs. 5.4 kDa for K2), making it a larger shield. Further support comes from the cryoprotection studies using the 80 kDa P-80 dehydrin from barley, which is even more effective than the two grape proteins at protecting LDH from freeze-thaw denaturation.32
K2 molecules would need to stay close to an LDH molecule to prevent this aggregation. The conserved K-segments in dehydrins are highly positively charged due to the presence of several Lys residues. To examine whether long-range electrostatics have any role in dehydrin cryoprotection, NaCl was added to the cryoprotective assay, with K2 present at a concentration which should provide ∼100% protection. Figure 5(B) shows the bar graph of changes in LDH activity in the presence and the absence of the salt. All of the measurements are expressed as percent LDH activity, where unfrozen LDH in the absence of salt is defined as 100%. The addition of NaCl reduced the activity of unfrozen LDH in the presence and absence of K2. The presence of salt has reduced the percent activity recovered to 54% for both unfrozen LDH controls. However, the LDH sample with K2 and salt recovered only 41% of its unfrozen activity, suggesting that electrostatic interactions between the positively charged K2 and the negatively charged LDH surface help keep the two proteins in close proximity without binding.
Discussion
Alignment and dynamics of K2
The alignment of the K2 sequences shows the conservation of Lys residues and two hydrophobic residues in the K-segments, and the lack of conservation in the φ-segment (Fig. 1). The conservation patterns are also reflected in the differences in the structural propensity and dynamics data between these two segments. As an IDP, all of K2 is flexible but the K-segments show a weak tendency for forming an α-helix, whereas the φ-segment has no clear secondary structure preference and is even more flexible than the K-regions (Fig. 2). It has been previously proposed that the K-segments form amphipathic helices, but here we show that in solution the protein is very weakly helical. It is only after a dehydrin binds a ligand, such as a micelle, that a notable amount of helical structure is induced.33 We suggest that the high number of small, polar residues in the φ-segment ensures that the protein remains highly flexible and is unable to form even transient structures. The presence of several hydrophobic amino acids are disfavored because they may cause the structure to partly collapse, whereas the presence of only like charged amino acids may cause extended structures to form because of charge–charge repulsion. Therefore, the nature of the amino acids in the φ-region ensures that no structure can form, whereas the K-segments are able to form only transient α-helices.
Denaturation and cryoprotection of LDH
The denaturation of the model enzyme LDH has been studied using a number of chemical and physical stresses. The loss of activity after various treatments has been attributed a loss of its tetrameric state,34 aggregation,5 and/or changes in its three-dimensional structure.35,36 We examined each of these possibilities to see what happens to LDH in our freeze-thaw protocol.
A sample of LDH was applied to a gel filtration column before and after being frozen and thawed. The results show that the oligomeric state was completely unaffected [Fig. 4(A)]. Low temperatures cause LDH to dissociate into monomers,37 but the reversible nature of tetramerization appears to be unaffected by the freeze-thaw cycles. A dramatic loss in absorbance intensity was observed after LDH was treated, suggesting that the protein could be aggregating and thus losing activity. The addition of K2 reduces the scatter of the thawed samples. However, there is a considerable reduction in the amount of scatter observed even at a K2 concentration where all LDH activity is lost, and the addition of K2 that results in a 50% reduction of LDH activity brought the scatter down to background levels [Fig. 4(B)]. This does not suggest that preventing aggregation is unimportant in the cryoprotective function of K2, but instead highlights a difference between in vitro and in vivo dehydrin assays. Performing these experiments with soluble protein extracts from plant cells would result in considerably more aggregation due to the much higher concentration of proteins and other macromolecules compared with in vitro conditions. Such results were seen in the work by Tunnacliffe and coworkers,38 where a disordered protein from a dessicant-tolerant nematode reduced the amount of aggregation of the water soluble proteome.
To explore what other structural changes may be occurring to LDH, the denaturation of the enzyme was followed using ANS as a probe to see if hydrophobic residues were being exposed. The results here show that the freezing and thawing of LDH alone causes a dramatic increase in ANS fluorescence [Fig. 4(C)], indicating that LDH has exposed its hydrophobic residues at some point during the freeze-thaw process. Similar results were observed with a Rhododendron dehydrin.39 Increasing amounts of K2 reduced the ANS fluorescence intensity. These results support the idea that dehydrins protect LDH activity by preventing both the aggregation and partial denaturation of the enzyme.
Cryoprotective mechanism
To determine whether there is a direct interaction between K2 and LDH, the 15N-HSQC spectrum of 15N-K2 alone was compared with that of the dehydrin in the presence of unlabeled LDH [Fig. 5(A)]. The resulting spectra completely overlap, indicating that the two proteins do not bind to each other. The data suggest that dehydrins do not appear to prevent LDH from denaturing by covering the surface of the protein before the freeze process.
We also examined whether there is an extremely weak association between the K-segment of K2 and LDH that was mediated by long-range electrostatic forces. In this model, the dehydrin would not bind LDH, but would stay preferentially localized near the enzyme. The freezing and thawing of LDH in the presence of sodium chloride and K2 at a concentration that should recover all of the pretreatment LDH activity was only ∼75% active relative to the unfrozen sample. This suggests that the K-segments of dehydrins could be important in maintaining a local concentration of K2 around its target enzyme for cryoprotection. Because the “typical” cytoplasmic protein has a negatively charged surface at physiological pH,40 it would make sense that a dehydrin would be positively charged from the Lys residues in the K-segments. It has been shown previously that the removal of the K-segments from several dehydrins lowered the recovery of LDH activity after freezing and thawing the enzyme, whereas the deletion of a φ-like segment had no such effect.39 Interestingly, the same study stated that a K-segment peptide did not show any cryoprotective effect. This would indicate that a flexible φ-region and the presence of two K-segments are required to ensure that the dehydrin is large enough to prevent enzyme denaturation.
Our observations here are similar to those observed for the polyelectrostatic interaction between the intrinsically disordered Sic1 and Cdc4, where Sic1 interacts with its target only after a number of residues have been phosphorylated (i.e., several negative charges are introduced).41 Like K2, Sic1 does not appear to undergo a disorder-to-order transition in the presence of its ligand.
We have demonstrated that a small dehydrin is able to protect LDH from damage due to freezing and thawing. This comes about when the intrinsically disordered K2 acts as a molecular shield to prevent partially denatured LDH from aggregating.
Materials and Methods
Protein purification and preparation
Unlabeled and 15N-labeled K2 were purified as previously described.42 The proteins were desalted by reversed-phase high pressure liquid chromatography (HPLC) using a Biobasic C18 column (Fisher Scientific, Mississauga, ON, Canada) with buffer A (0.1% trifluoroacetic acid (w/v) in water) and buffer B (0.1% trifluoroacetic acid (w/v) in acetonitrile) as the mobile phases. Separation was performed using a linear gradient of 1% buffer B to 100% buffer B over 60 min at a flow rate of 1 mL/min. Samples containing K2 were pooled and lyophilized. Rabbit type II LDH protein (cat. no. L2500, Sigma-Aldrich, Oakville, ON, Canada) was prepared by overnight dialysis against 10 mM sodium phosphate buffer, pH 7.4.
LDH cryoprotection assay and analysis
A modified technique of Lin and Thomashow7 was used to measure LDH activity and examine the cryoprotective effects of the K2 dehydrin and controls. All proteins and reagents were resuspended in 10 mM sodium phosphate, pH 7.4. LDH samples (8 μL of 50 μg/mL) were mixed with protective protein (8 μL of varying concentrations of K2, BSA, or lysozyme) or buffer (8 μL) in a 1.5 mL microcentrifuge tube. Five cycles of freezing (immersion in liquid nitrogen for 30 s) and thawing (immersion in a circulating water bath at 4°C for 5 min) were performed. LDH activity was measured by diluting the enzyme to 0.5 μg/mL into 750 μL of reaction mix (10 mM sodium phosphate, pH 7.4, 2 mM NADH, 10 mM pyruvic acid). Oxidation of NADH was followed by monitoring A340 on a Cary 100 (Varian, Mississauga, ON, Canada) over 2.5 min, during which time the reaction rate was linear. For the cryoprotection assays performed in the presence of salt, the same procedure was followed but 1M NaCl was added to the sample before freeze-thaw treatment.
Gel filtration
LDH (200 μg) was injected onto a Superdex G200 column before and after freeze-thaw treatment as described for the cryoprotection assay. The sample was eluted at a flow rate of 0.5 mL/min using 50 mM sodium phosphate, pH 7.4, 150 mM NaCl as the buffer. The column was calibrated with molecular weight standards using the same buffer and flow rates.
Structure of thawed LDH
For the scatter experiments, 0.15 mg/mL of LDH in 10 mM sodium phosphate, pH 7.4 was frozen and thawed as described in the cryoprotective assay protocol in the presence and the absence of K2 at concentrations of 0.07 and 0.7 μg/mL. The scatter was measured at A340 on a Cary 100 at a temperature of 25°C before and after the samples were frozen and thawed. Absorbance readings were corrected for contribution from buffer and K2.
For ANS fluorescence, 1 μM of LDH and 10 μM of ANS were dissolved in 10 mM sodium phosphate, pH 7.4, in the presence and the absence of K2 at three concentrations (0.028, 0.28, and 0.65 mg/mL). The samples were frozen and thawed as described in the cryoprotection assay protocol. Fluorescence data were acquired on an Alphascan-2 spectrofluorometer (Photon Technology, South Brunswick, NJ), where emission scans were obtained by exciting the sample with λex = 350 nm and scanning from 420 to 560 nm. Scans were corrected for contributions from buffer and K2 alone.
NMR data collection and analysis
Experiments were collected on a Bruker Avance DRX600 spectrometer equipped with a cryogenic triple resonance probe at a temperature of 300 K, and 1H and 15N referencing was performed relative to 2,2′-dimethyl-2-silapentane-5-sulfonate (DSS) as described.43 The T1, T2, and {1H}-15N-NOE relaxation experiments44 were collected using 0.5 mM 15N-K2 in 600 μL of NMR buffer (20 mM sodium phosphate, pH 6.0, 10 mM NaCl, 0.01% sodium azide, 0.1 mM DSS and 5% D2O (v/v)). For all three relaxation experiments, 1024 (1HN) and 128 (15N) complex data points were acquired with a total of eight transients per t1 increment using spectral widths of 8370.5 (1HN) and 1338.0 (15N) Hz. For the T1 experiments, relaxation delays of 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, and 2.0 s were applied. For the T2 experiments, relaxation delays of 16.064, 32.128, 48.192, 64.256, 80.320, 112.450, 144.580, 176.700, 208.830, and 240.960 ms were applied. The {1H}-15N steady-state NOE values represent the intensity ratio from the two HSQC spectra with and without 1H saturation applied prior to the 15N excitation pulse. For the acquisition of 15N-HSQC spectra of 0.1 mM K2 in the presence and the absence of 0.3 mM LDH, 1024 (1HN) and 128 (15N) complex data points were acquired with a total of four transients per t1 increment using spectra widths of 7211.5 (1HN) and 2128.6 (15N) Hz.
Spectra were processed using NMRPipe45 and peak assignments were made using the CCPNMR analysis software version 2.1.46 Chemical shifts of K2 were obtained from Biological Magnetic Resonance Bank (BMRB) accession number 16445.47 The T1 and T2 relaxation times were analyzed by fitting the cross-peak intensity decay to a two parameter, single exponential decay function I(t) = I0e−t/T1,3, where I0 is the intensity at time t = 0 and I(t) is the intensity after a delay time t for the T1 or T2 experiment. The errors were estimated from the uncertainty of the nonlinear fits. For the calculation of the SSP, the program SSP25 was run with the RefDB data set for random coil and secondary structure chemical shift values.48
Acknowledgments
The authors thank the University of Guelph NMR Centre for use of the facility and help with setting up the experiments.
References
- 1.Close TJ. Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiol Plant. 1997;100:291–296. [Google Scholar]
- 2.Rorat T. Plant dehydrins—tissue location, structure and function. Cell Mol Biol Lett. 2006;11:536–556. doi: 10.2478/s11658-006-0044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosova K, Vitamvas P, Prasil IT. The role of dehydrins in plant response to cold. Biol Plant. 2007;51:601–617. [Google Scholar]
- 4.Allagulova CR, Gimalov FR, Shakirova FM, Vakhitov VA. The plant dehydrins: structure and putative functions. Biochemistry. 2003;68:945–951. doi: 10.1023/a:1026077825584. [DOI] [PubMed] [Google Scholar]
- 5.Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momma M, Kaneko S, Haraguchi K, Matsukura U. Peptide mapping and assessment of cryoprotective activity of 26/27-kDa dehydrin from soybean seeds. Biosci Biotechnol Biochem. 2003;67:1832–1835. doi: 10.1271/bbb.67.1832. [DOI] [PubMed] [Google Scholar]
- 7.Lin C, Thomashow MF. A cold-regulated Arabidopsis gene encodes a polypeptide having potent cryoprotective activity. Biochem Biophys Res Commun. 1992;183:1103–1108. doi: 10.1016/s0006-291x(05)80304-3. [DOI] [PubMed] [Google Scholar]
- 8.Kazuoka T, Oeda K. Purification and characterization of COR85-oligomeric complex from cold-acclimated spinach. Plant Cell Physiol. 1994;35:601–611. [Google Scholar]
- 9.Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tompa P, Banki P, Bokor M, Kamasa P, Kovacs D, Lasanda G, Tompa K. Protein-water and protein-buffer interactions in the aqueous solution of an intrinsically unstructured plant dehydrNMR intensity and DSC aspects. Biophys J. 2006;91:2243–2249. doi: 10.1529/biophysj.106.084723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alsheikh MK, Heyen BJ, Randall SK. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J Biol Chem. 2003;278:40882–40889. doi: 10.1074/jbc.M307151200. [DOI] [PubMed] [Google Scholar]
- 12.Hara M, Shinoda Y, Tanaka Y, Kuboi T. DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ. 2009;32:532–541. doi: 10.1111/j.1365-3040.2009.01947.x. [DOI] [PubMed] [Google Scholar]
- 13.Kovacs D, Kalmar E, Torok Z, Tompa P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wisniewski M, Webb R, Balsamo R, Close TJ, Yu XM, Griffith M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: a dehydrin from peach (Prunus persica) Physiol Plant. 1999;105:600–608. [Google Scholar]
- 15.Simpson DJ, Smallwood M, Twigg S, Doucet CJ, Ross J, Bowles DJ. Purification and characterisation of an antifreeze protein from Forsythia suspensa (L.) Cryobiology. 2005;51:230–234. doi: 10.1016/j.cryobiol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Jiang XZ, Wang YS. Beta-elimination coupled with tandem mass spectrometry for the identification of in vivo and in vitro phosphorylation sites in maize dehydrin DHN1 protein. Biochemistry. 2004;43:15567–15576. doi: 10.1021/bi0483965. [DOI] [PubMed] [Google Scholar]
- 17.Godoy JA, Lunar R, Torresschumann S, Moreno J, Rodrigo RM, Pintortoro JA. Expression, tissue distribution and subcellular-localization of dehydrin tas14 in salt-stressed tomato plants. Plant Mol Biol. 1994;26:1921–1934. doi: 10.1007/BF00019503. [DOI] [PubMed] [Google Scholar]
- 18.Goday A, Jensen AB, Culianezmacia FA, Alba MM, Figueras M, Serratosa J, Torrent M, Pages M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear-localization signals. Plant Cell. 1994;6:351–360. doi: 10.1105/tpc.6.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisse T, Bartels D, Kalbitzer HR, Jaenicke R. The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineum displays no defined three-dimensional structure in its native state. Biol Chem. 1996;377:555–561. doi: 10.1515/bchm3.1996.377.9.555. [DOI] [PubMed] [Google Scholar]
- 20.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 22.Xiao H, Nassuth A. Stress- and development-induced expression of spliced and unspliced transcripts from two highly similar dehydrin 1 genes in V. riparia and V. vinifera. Plant Cell Rep. 2006;25:968–977. doi: 10.1007/s00299-006-0151-4. [DOI] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 25.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synucleimplications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Matas MA, Nunez P, Soto A, Allona I, Casado R, Collada C, Guevara MA, Argoncillo C, Gomez L. Protein cryoprotective activity of a cytosolic small heat shock protein that accumulates constitutively in chestnut stems and is up-regulated by low and high temperatures. Plant Physiol. 2004;134:1708–1717. doi: 10.1104/pp.103.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermann R, Jaenicke R, Rudolph R. Analysis of the reconstitution of oligomeric enzymes by cross-linking with glutaraldehyde—kinetics of reassociation of lactic-dehydrogenase. Biochemistry. 1981;20:5195–5201. doi: 10.1021/bi00521a015. [DOI] [PubMed] [Google Scholar]
- 28.Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J Mol Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 29.Daniel E, Weber G. Cooperative effects in binding by bovine serum albumin. I. The binding of 1-anilino-8-naphthalenesulfonate. Fluorimetric titrations. Biochemistry. 1966;5:1893–1900. doi: 10.1021/bi00870a016. [DOI] [PubMed] [Google Scholar]
- 30.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas AF, Gilmanshin RI. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 31.Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Prog Nucl Magn Reson Spectrosc. 2007;51:219–242. [Google Scholar]
- 32.Bravo LA, Gallardo J, Navarrete A, Olave N, Martinez J, Alberdi M, Close TJ, Corcuera LJ. Cryoprotective activity of a cold-induced dehydrin purified from barley. Physiol Plant. 2003;118:262–269. [Google Scholar]
- 33.Koag MC, Wilkens S, Fenton RD, Resnik J, Vo E, Close TJ. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009;150:1503–1514. doi: 10.1104/pp.109.136697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng YB, Meng FG, Chen BY, Wang XC. Inactivation and conformational changes of lactate dehydrogenase from porcine heart in sodium dodecyl sulfate solutions. Int J Biol Macromol. 2002;31:97–102. doi: 10.1016/s0141-8130(02)00063-6. [DOI] [PubMed] [Google Scholar]
- 35.Bai JH, Wang HJ, Zhou HM. Alkaline-induced unfolding and salt-induced folding of pig heart lactate dehydrogenase under high pH conditions. Int J Biol Macromol. 1998;23:127–133. doi: 10.1016/s0141-8130(98)00033-6. [DOI] [PubMed] [Google Scholar]
- 36.Gabellieri E, Strambini GB. ANS fluorescence detects widespread perturbations of protein tertiary structure in ice. Biophys J. 2006;90:3239–3245. doi: 10.1529/biophysj.105.074948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King L, Weber G. Conformational drift and cryoinactivation of lactate dehydrogenase. Biochemistry. 1986;25:3637–3640. doi: 10.1021/bi00360a024. [DOI] [PubMed] [Google Scholar]
- 38.Chakrabortee S, Boschetti C, Walton LJ, Sarkar S, Rubinsztein DC, Tunnacliffe A. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc Natl Acad Sci USA. 2007;104:18073–18078. doi: 10.1073/pnas.0706964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes JL, Campos F, Wei H, Arora R, Yang YI, Karlson DT, Covarrubias AA. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008;31:1781–1790. doi: 10.1111/j.1365-3040.2008.01879.x. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz R, Ting CS, King J. Whole proteome pl values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11:703–709. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- 41.Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci USA. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livernois AM, Hnatchuk DJ, Findlater EE, Graether SP. Obtaining highly purified intrinsically disordered protein by boiling lysis and single step ion exchange. Anal Biochem. 2009;392:70–76. doi: 10.1016/j.ab.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 43.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1 H, 13 C and 15 N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 44.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR spectroscopy: principles and practice. London: Academic Press; 2007. [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 47.Findlater EE, Graether SP. NMR assignments of the intrinsically disordered K2 and YSK2 dehydrins. Biomol NMR Assign. 2009;3:273–275. doi: 10.1007/s12104-009-9192-2. [DOI] [PubMed] [Google Scholar]
- 48.Zhang HY, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]