Abstract
p115-RhoGEF (p115) belongs to the family of RGS-containing guanine nucleotide exchange factors for Rho GTPases (RGS-RhoGEFs) that are activated by G12 class heterotrimeric G protein α subunits. All RGS-RhoGEFs possess tandemly linked Dbl-homology (DH) and plekstrin-homology (PH) domains, which bind and catalyze the exchange of GDP for GTP on RhoA. We have identified that the linker region connecting the N-terminal RGS-homology (RH) domain and the DH domain inhibits the intrinsic guanine nucleotide exchange (GEF) activity of p115, and determined the crystal structures of the DH/PH domains in the presence or absence of the inhibitory linker region. An N-terminal extension of the canonical DH domain (the GEF switch), which is critical to GEF activity, is well folded in the crystal structure of DH/PH alone, but becomes disordered in the presence of the linker region. The linker region is completely disordered in the crystal structure and partially disordered in the molecular envelope calculated from measurements of small angle x-ray scattering (SAXS). It is possible that Gα subunits activate p115 in part by relieving autoinhibition imposed by the linker region.
Keywords: GTP-binding protein alpha subunits, G12, G13, Rho GTPase, guanine nucleotide exchange factors, RGS proteins, DH, PH
Introduction
RGS-containing guanine nucleotide exchange factors for Rho GTPases (RGS-RhoGEFs) are a homologous subfamily of RhoGEFs (guanine nucleotide exchange factors for Rho proteins) that contain RGS (regulator of G protein signaling) domains. The three members of this subfamily, p115-RhoGEF (p115), PDZ-RhoGEF (PRG) and leukemia-associated RhoGEF (LARG), represent potential direct regulatory links between G protein-coupled receptors that activate the G12 class of heterotrimeric G proteins and RhoA-mediated pathways that lead to cytokinesis and transformation.1,2 RGS-RhoGEFs catalyze the exchange of GDP for GTP on RhoA, a small GTPase of the Ras superfamily.3 Activated RhoA bound to GTP can then engage downstream effectors and influence cellular functions. Like all members of the large family of RhoGEFs (about 70 in the human genome), the GEF activity of RGS-RhoGEFs resides in their tandemly linked Dbl homology (DH) and plekstrin-homology (PH) domains.4,5 The RGS-homology (RH or rgRGS) domains are situated N-terminal to the DH/PH domains. RH domains of p115 and LARG function as GAPs (GTPase activating proteins) for Gα13 and Gα12 subunits, and binding of Gα subunits to their respective RhoGEFs stimulates their guanine nucleotide exchange activity toward RhoA.6–9 In addition to the RH domain, PRG and LARG also contain an N-terminal PDZ domain that has been shown to mediate interaction of the RGS-RhoGEFs with regulatory proteins.10–12
Structures of DH/PH domains from two RGS-RhoGEFs have been determined, either in isolation or as complexes with nucleotide-free RhoA.13–15 The cores of these DH domains are elongated bundles composed of six major α helices, similar in structure to those of other DH domains in the greater RhoGEF family. DH domains form extensive contacts with nucleotide-free RhoA and are essential for GEF activity.4 The PH domains of RGS-RhoGEFs also form a small interface with RhoA, suggesting a potential role in catalysis.13 Indeed, both DH and PH domains are required to achieve full GEF activity.13,14,16 It appears that the PH domain increases the exchange activity of the RGS-RhoGEFs, either through stabilization of the DH domain or direct impact on the substrate, RhoA.
The exact mechanism by which Gα13 stimulates the exchange activities of LARG and p115 remains elusive.7–9 It has been proposed that the intrinsic GEF activities of RGS-RhoGEFs, which reside in the DH/PH domains, are auto-inhibited by other regions of the multidomain molecule, and interactions between activated Gα13 and p115 or LARG relieves this autoinhibition. A recent study of PRG suggests that part of the linker region between the RH and DH domains interacts with DH in the basal state and inhibits the exchange activity of the DH/PH domains from PRG.17 However, Gα13 does not stimulate the GEF activity of PRG, even though Gα13 binds its RH domain with high affinity.18
Here, using a combination of molecular cloning, biochemical assays, x-ray crystallography, and small angle x-ray scattering (SAXS), we examined the role of the linker region between the RH and the DH domains of p115, which is a substrate for direct stimulation by Gα13. We show that the linker region interferes with the folding of an N-terminal extension of the DH domain that is crucial for the full intrinsic GEF activity of DH/PH, and, thus, identifies a potential autoinhibitory mechanism. This observation provides new insight into the molecular mechanism by which the exchange activity of RGS-RhoGEFs is regulated.
Results
The linker between RH and DH inhibits the basal GEF activity of p115-RhoGEF
The DH/PH domains of p115-RhoGEF (residues 395–766) and the fragment containing the additional linker region between the RH and the DH/PH domains (L-DH/PH, residues 240–766) were each expressed in bacteria with N-terminal cleavable GST- and C-terminal 6His-tags for purification. Both purified proteins stimulated the exchange of GDP for GTP on RhoA (Fig. 1). However, the observed catalytic rate (kobs) of DH/PH toward RhoA was fourfold greater than that of L-DH/PH [Fig. 1(C)], suggesting that the linker region inhibits the intrinsic GEF activity of the DH/PH domains. The RH domain N-terminal to the linker region has little effect on this autoinhibition, as similar inhibition of GEF activity by the linker region was also observed in the presence of the RH domain of p115 (Supporting Information Fig. 1). The kobs of RH-DH/PH toward RhoA was almost identical to that of DH/PH, with a fourfold increase over the kobs of RH-L-DH/PH or L-DH/PH (Supporting Information Fig. 1).
Figure 1.
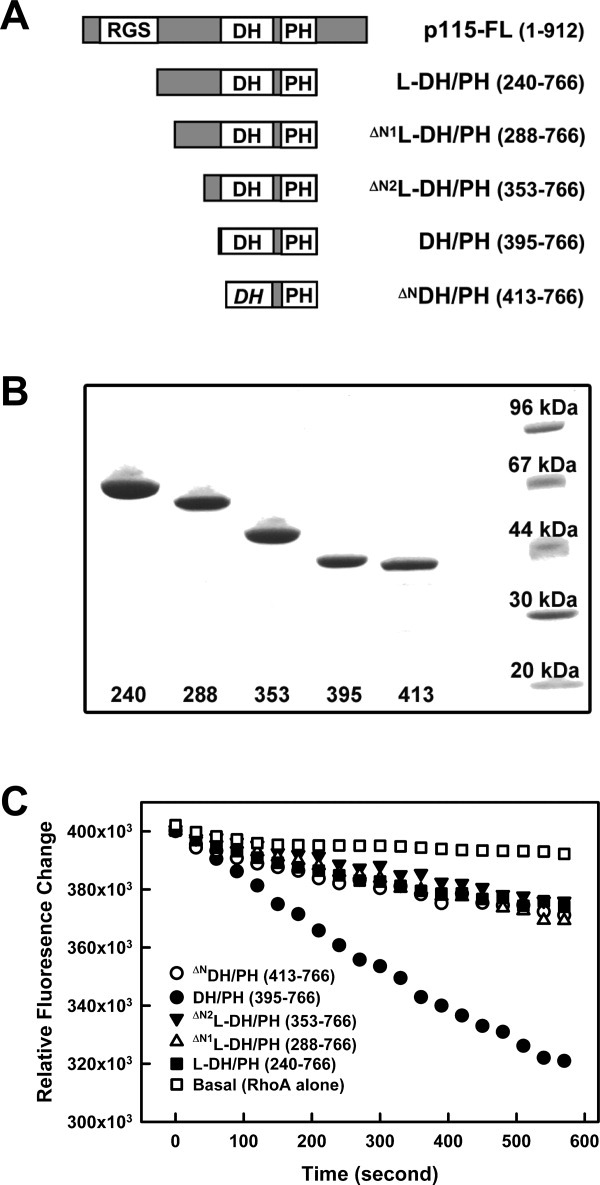
The linker region between the RH and DH domains inhibits GEF activity of DH/PH. (a) Schematic representation of truncated forms of p115 used; included amino acids are listed in parentheses. (b) SDS-PAGE gel showing purified p115 fragments as detailed in panel (a). Numbers underneath represent the first amino acid of the protein. The last lane on the right contains protein standard markers with molecular weight labeled. (c) Nucleotide exchange assays with p115 and RhoA. For each time course, 0.5 μM RhoA loaded with mant-GDP was mixed with 100 μM GTP and the exchange reaction started at room temperature by addition of buffer (Basal, open squares), or 30 nM of p115: L-DH/PH (solid squares), ΔN1L-DH/PH (open triangles), ΔN2L-DH/PH (solid triangles), DH/PH (solid circles), or ΔNDH/PH (open circles). The subsequent decrease in fluorescence (λex = 356 nm, λem = 445 nm) was measured for 10 minutes.
The segment within the linker region that is critical for the autoinhibition of GEF activity was identified by deletion analysis [Fig. 1(A,B)]. Two variations of L-DH/PH, ΔN1L-DH/PH (residues 288–766) and ΔN2L-DH/PH (residues 353–766) were expressed and purified as described in Methods. The results from GEF assays [Fig. 1(C)] reveal that removal of the N-terminal part of the linker region (residues 240–352) does not relieve the autoinhibition of the GEF activity. Thus, the C-terminal part of the linker region (residues 353–394), which is located immediately N-terminal to the DH/PH domains of p115, is sufficient to inhibit the basal exchange activity of DH/PH toward RhoA.
A prior study showed that the very N-terminus of the DH/PH domain in LARG (residues 765–782), which is an extension to the canonical DH fold, is critical for maximal exchange of GDP for GTP on RhoA.13 Deletion of this region or mutations of a conserved tryptophan residue within this region, produced 80% declines in GEF activity of the LARG DH/PH domains toward RhoA. The increased activity was ascribed to direct contacts between residues of this N-terminal extension and the switch regions of RhoA.13 Deletion of the corresponding N-terminal extension in p115 (residues 395–412, ΔNDH/PH) also lowered the GEF activity (Fig. 1). Furthermore, the observed catalytic rate (kobs) of the N-terminally truncated DH/PH was almost identical to those of the L-DH/PH fragments [Fig. 1(C)]. Hereafter, this N-terminal region of the DH domain (residues 395–412) is referred to as “the GEF switch”. It is possible that the linker region inhibits the basal GEF activity of DH/PH by modulating the GEF switch of the DH domain.
Crystal Structure of the DH/PH domains of p115-RhoGEF
The structure of the DH/PH domains of p115 was determined at a resolution of 2.9 Å by molecular replacement using separate search models for the LARG-DH domain and the LARG-PH domain [Fig. 2(A)]. Data collection and refinement statistics for the structure are summarized in Table I. The asymmetric unit of the crystal contains two dyad-related and interacting DH/PH domains [Fig. 2(B)]. The interface between the domains buries about 1,600 Å2 of solvent-accessible surface area, and is stabilized by interactions between a layer of three β-strands near the C-terminus of one PH domain (β5, β6 and β7) and the α4 helix of the dyad related DH domain. However, there is no evidence from size exclusion chromatography that the DH/PH domains are dimeric in solution (data not shown). Members of the RGS-RhoGEF family have been reported to oligomerize, but this interaction involves domains C-terminal to DH/PH.19,20 It is also worth noting that the surface of the PH domain that forms lattice contacts in the structure reported here also mediates protein–protein interactions in crystal structures of LARG-DH/PH13 and PRG-DH/PH.14 We recently found that this surface in PRG interacts with activated, GTP-bound RhoA.21
Figure 2.
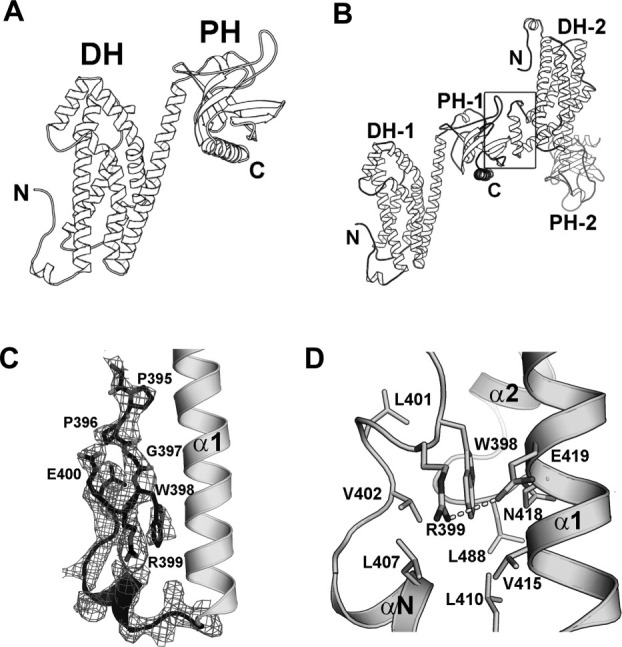
Crystal structure of the p115 DH/PH domains. (a) Ribbon diagrams depicting the tertiary structure of p115 DH/PH. (b) Ribbon diagrams depicting the noncrystallographic dimer of p115 DH/PH (labeled 1 and 2). The dimer interface (marked by a box) involves a layer of β-strands near the C-terminus of one PH domain and the α4 helix from the dyad related DH domain. (c) The GEF switch in the DH domain. Electron density (cages) for the GEF switch from a 2.9 Å σA-weighted 2Fo-Fc total omit map calculated in SFCHECK (CCP4i) is contoured at 1.5 standard deviations above the mean.22 Side chains of residues 395–400 are depicted as stick models. The rest of the DH/PH domain is depicted as ribbon diagrams. (d) Trp-398 from the GEF switch is buried in a hydrophobic pocket between the GEF switch and the canonical DH domain. Side chains of residues involved are depicted as stick models. The putative hydrogen bond between side chains of Arg-399 and Glu-419 is drawn as a dotted line.
Table I.
Data Collection and Refinement Statistics
| DH/PH | ΔN2L-DH/PH | L-DH/PH | R399EDH/PH | |
|---|---|---|---|---|
| Data Collection | ||||
| Source | APS SBC 19ID | APS SBC 19BM | APS SBC 19ID | Rigaku FR-E |
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 | 1.5418 |
| Space group | P41 | P32 | P32 | P32 |
| Unit cell (Å, °) | a = b = 106.0, c = 126.0 | a = b = 111.5, c = 97.8 | a = b = 111.6, c = 97.9 | a = b = 110.8, c = 98.6 |
| α = β = γ = 90 | α = β = 90, γ = 120 | α = β = 90, γ = 120 | α = β = 90, γ = 120 | |
| Dmin (Å) | 2.9 | 3.2 | 3.2 | 2.5 |
| Highest res. Shell (Å) | 2.95–2.90 | 3.26–3.20 | 3.26–3.20 | 2.54–2.50 |
| Unique reflections | 30,747 | 22,503 | 22,476 | 46,979 |
| Redundancy | 4.5 | 4.1 | 2.6 | 3.6 |
| Rsyma | 0.06 (0.75) | 0.06 (0.58) | 0.08 (0.46) | 0.09 (0.58) |
| Completeness (%)a | 99.3 (99.9) | 99.9 (100.0) | 99.5 (99.8) | 99.9 (99.4) |
| <I>/σ<I>a | 31.5 (2.3) | 24.5 (2.1) | 14.8 (2.2) | 22.7 (2.1) |
| Refinement | ||||
| Resolution (Å) | 36.0–2.9 | 48.9–3.2 | 43.7–3.2 | 48.3–2.5 |
| Total reflection used | 29,151 | 21,259 | 21,298 | 44,566 |
| N. nonhydrogen atoms | 6,068 | 5,516 | 5,416 | 5,653 |
| Protein atoms | 6,068 | 5,516 | 5,416 | 5,653 |
| Rwork (%) | 22.1 | 22.5 | 23.1 | 22.6 |
| Rfree (%)b | 27.2 | 27.4 | 28.9 | 26.6 |
| RMS deviations | ||||
| Bond lengths(Å) | 0.018 | 0.020 | 0.017 | 0.023 |
| Bond angles (°) | 1.833 | 1.736 | 1.706 | 2.012 |
| Ramachandran (%) (favored/disallowed) | 90.2/0.4 | 88.5/0.3 | 88.7/0.2 | 91.9/0.0 |
| PDB access code | 3ODO | 3ODX | 3ODW | 3P6A |
Numbers in parentheses correspond to the highest resolution shell.
Rfree is the R-factor obtained for a test set of reflections consisting of a randomly selected 5% of the data. No I/σ cutoff was used in the final calculations of R-factors.
As in previously determined DH/PH structures of RGS-RhoGEFs, the DH domain of p115 is comprised of six major α helices, whereas the PH domain is a six-stranded antiparallel β-barrel, with a C-terminal α-helix packed against the base of the domain. Continuous electron density corresponding to the GEF switch (residues 395–412), composed of an extended segment followed by a short helix, is well defined [Fig. 2(C)]. The interface between the GEF switch and the α1 helix centers on a conserved tryptophan residue (Trp-398) from the GEF switch [Fig. 2(D)]. As observed in the structure of the LARG DH/PH domains, the side chain of this highly conserved tryptophan residue is buried in a hydrophobic pocket formed by residues from both the α1 helix and the GEF switch. A putative hydrogen bond between side chains of Arg-399 from the GEF switch and Glu-419 from the α1 helix further stabilizes this interface. Mutations of the conserved Trp-398 to alanine or Arg-399 to glutamic acid severely decreased the GEF activity of the DH/PH domains (Supporting Information Fig. 2). In the complex of LARG DH/PH with RhoA, part of the GEF switch forms direct contact with switch regions of RhoA. The overall structures of the DH domains of p115 and LARG are very similar [Fig. 3(A)]. Residues involved in the interaction with RhoA are highly conserved [Fig. 3(C)]. Therefore, it is expected that the GEF switch in p115 would form similar contacts with RhoA, and this interaction would be critical for the exchange activity of p115. Indeed, removal of the GEF switch caused a significant decrease in GEF activity of p115 DH/PH domains [Fig. 1(C)].
Figure 3.
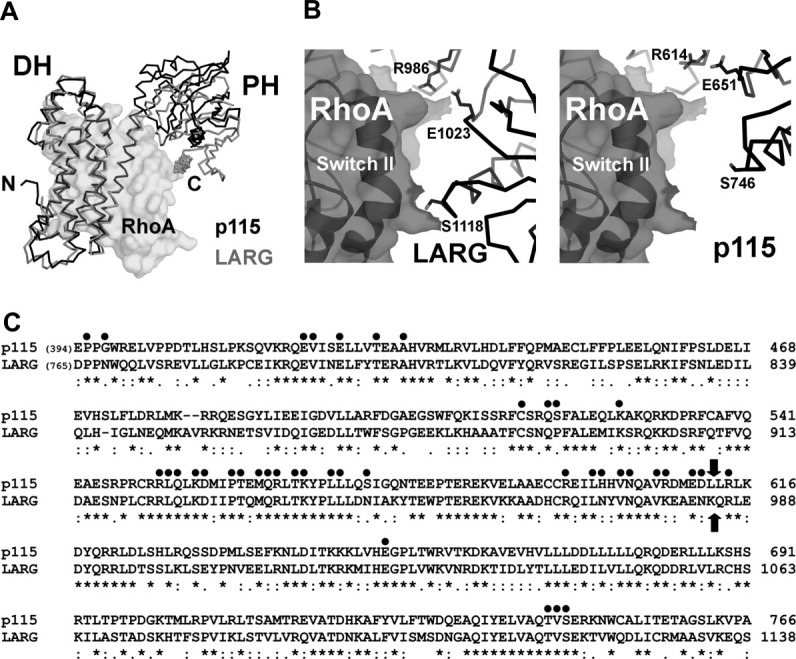
(a) Structural comparison of the DH/PH domains from p115 (black lines) and LARG (gray lines, PDB access code 1X86, with the bound nucleotide-free RhoA depicted as transparent solvent accessible surface). The structural alignment was based on coordinates from the DH domains only. The PH domain in LARG moves closer toward its DH domain upon binding to RhoA. (b) The interface between the PH domain and RhoA. In the crystal structure of the LARG-DH/PH:RhoA complex (left), residues from PH (depicted as sticks) make direct contacts with RhoA (depicted as transparent solvent accessible surface with black ribbons underneath). In the modeled p115-DH/PH:RhoA complex (right), where the two DH domains from p115 and LARG are superimposed, the same set of residues in p115 PH domain can not form direct contacts with RhoA. (c) Sequence alignment of DH/PH domains from p115 and LARG. Residues in LARG that are involved in contacts with the nucleotide-free RhoA are marked with dots on top. Block arrows mark the domain boundary between DH and PH. The sequence alignment is carried out by the program Clustal W.23
The DH/PH domains from p115 and LARG share 50% identity in amino acid sequences [Fig. 3(C)]. Comparison of the structures of p115-DH/PH with LARG-DH/PH reveals only minor differences in the structures of the two domains. The RMS deviation of corresponding Cα atoms is 1.2 Å for the DH domain (residues 407–610 in p115, and 770–982 in LARG), and 0.9 Å for the PH domain (residues 611–759 in p115, and 983–1134 in LARG). However, compared with the LARG-DH/PH structure when bound to RhoA, the DH and PH domains rotate apart in p115 [Fig. 3(A)]. The RMS deviation for all Cα atoms from the DH and the PH domains increases to 4.2 Å. In the case of LARG-DH/PH, several residues in the PH domain make direct contacts with RhoA, including Arg-986 and Ser-1118 [Fig. 3(B)], and these interactions are required for efficient GEF function of the protein.13 As described earlier, interactions between the PH domain and RhoA have been shown to be important for the exchange activity for other RGS-RhoGEFs, including p115. Therefore, in order for its PH domain to make proper contacts with RhoA, significant domain rotation within the DH/PH domains of p115 would be required upon binding to its substrate.
Crystal and solution structures of L-DH/PH
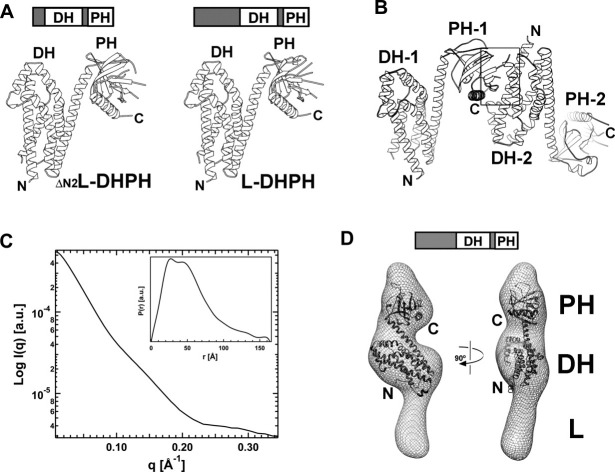
Crystals of L-DH/PH and ΔN2L-DH/PH are nonisomorphous to those of DH/PH, but isomorphous with each other. Structures of both proteins [Fig. 4(A)] were determined at 3.2 Å resolution by molecular replacement (Table I) and are almost identical with an RMS deviation of 0.3 Å for all Cα atoms (residues 411–765). The GEF switch, which is well ordered in the structure of DH/PH (Fig. 2), is disordered in the domains that include the linker segments that precede the GEF switch. Furthermore, there is no electron density for the linker regions themselves, even though the crystal lattice has enough space to accommodate both the DH/PH and the linker region. SDS-PAGE analysis of dissolved crystals showed little degradation of L-DH/PH or ΔN2L-DH/PH. The asymmetric unit of the crystal contains a pair of approximately dyad-related L-DH/PH molecules [Fig. 4(B)], as observed in crystals of DH/PH, but this dimer interface involves a different DH interaction surface [Figs. 2(B) and 4(B)]. The dimer interface buries about 1,900 Å2 of solvent-accessible surface area, and is stabilized by interactions between the layer of three β-strands near the C-terminus of the PH domain (β5, β6, and β7) and the α1 and α2 helices from the dyad-related DH domain, rather than the α4 helix as in the DH/PH dimer. As in the case of the DH/PH domains, there is no evidence from size exclusion chromatography that the L-DH/PH proteins are dimeric in solution (data not shown).
Figure 4.
Crystal and solution structures of the p115 L-DH/PH domains. (a) Ribbon diagrams depicting tertiary structures of ΔN2L-DH/PH (left) and L-DH/PH (right). (b) Ribbon diagrams depicting the noncrystallographic dimer of p115 L-DH/PH (labeled 1 and 2). The dimer interface (marked by a box) involves a layer of β-strands near the C-terminus of one PH domain and the α1-α2 helices from the dyad related DH domain. (c) Solution x-ray scattering profile for p115 L-DH/PH. The distance distribution function, P(r), of L-DH/PH was computed from the x-ray scattering using the program GNOM.36 (d) Solution structure of L-DH/PH of p115. The solution structure (molecular envelope) is depicted as a mesh and overlapped onto the crystal structure of p115 DH/PH domains (ribbon). The crystal structure is relatively well accommodated in the molecular envelope. The large unoccupied region in the molecular envelope beneath the DH domain is likely to be the location for the linker region with limited ordered structure.
The inter-dimer contact of the PH domain with the α1 helix of DH would preclude part of the GEF switch (residues 395–397) from contacts with the canonical DH domain similar to those observed in the DH/PH structure [Fig. 4(B)]. However, there is room for the rest of the GEF switch, including the important tryptophan residue (Trp-398), to fit in between the two L-DH/PH molecules. Removal of the three amino acids preceding Trp-398 led to a twofold decrease of the GEF activity of the DH/PH domains (Supporting Information Fig. 2). However, the decrease in GEF activity caused by the deletion of residues 395–397 is far less severe than the effect of the W398A or R399E mutation, in which the intradomain interactions between the GEF switch and the canonical DH domain are disrupted. The corresponding residues in LARG DH/PH domains [residues 766–768, Fig. 3(C)] interact with RhoA directly.13 The decrease in GEF activity observed here is likely due to the loss of potential contacts between residues 395–397 and the substrate RhoA, rather than from a complete disruption of the GEF switch.
Our hypothesis is that the disruption of the GEF switch – α1 helix interface leads to the formation of the lattice contact observed in the Linker-DH/PH crystal structures. We have crystallized several DH/PH mutants bearing single mutations in the GEF switch that disrupt its interaction with the α1 helix of DH. These included the aforementioned W398A and R399E mutants (Supporting Information Fig. 2). Crystals of the mutant DH/PH (W398A or R399E), grown under the exactly same condition as the wild-type DH/PH, are nonisomorphous to those of DH/PH but isomorphous to those of L-DH/PH and ΔN2L-DH/PH [Supporting Information Fig. 3(A), Table I]. The structure of the R399E mutant of DH/PH [R399EDH/PH, Supporting Information Fig. 3(B)] was determined at 2.5-Å resolution by molecular replacement (Table I). The lattice contact in the R399EDH/PH structure [Supporting Information Fig. 3(C)] is identical to those observed in structures of L-DH/PH and ΔN2L-DH/PH (Fig. 4). The GEF switch is completely disordered in R399EDH/PH, and a dyad-related PH domain packs against the α1 and α2 helices of DH (Supporting Information Fig. 3). Both the wild-type and the R399E mutant of DH/PH crystallize under the same condition; however, this lattice contact between the PH and the α1 and α2 helices of the dyad-related DH can only form in the mutant DH/PH, where the intradomain interface between the GEF switch and the α1 helix has been disrupted. Thus, formation of the lattice contact observed in the L-DH/PH and ΔN2L-DH/PH structures requires disruption of the interface between the GEF switch and the α1 helix of the DH domain.
We also determined the solution structure of L-DH/PH at low (∼20 Å) resolution using SAXS. The molecular envelope of L-DH/PH was obtained by ab initio shape reconstruction from experimental SAXS data. The experimental scattering profile is shown in Figure 4(C). Low-angle scattering intensity calibration with cytochrome c indicated that L-DH/PH exists as a monomer in solution. The distance distribution function, P(r), is characteristic of an elongated molecule [Fig. 4(C), insert]. The crystal structure of p115 DH/PH fits well into the calculated molecular envelope [Fig. 4(D)]. The linker region, which is completely disordered in the crystal structure of L-DH/PH, appears to be at least partially ordered in the solution structure, where it putatively occupies a volume of the molecular envelope below the DH domain. However, this volume is not sufficient to accommodate the additional 156 residues of the linker. Thus, the linker region is likely to be at least partially disordered in solution. Although the crystal structure of p115 DH/PH domains is generally well accommodated in the molecular envelope, the N-terminal GEF switch does not fit well inside the envelope; this strongly suggests that the GEF switch adopts a different conformation in the presence of the linker region preceding it.
Discussion
This study indicates that the linker region in RGS-RhoGEFs, which is located between the RH and the DH domains, inhibits the intrinsic GEF activity of these proteins. This behavior was also observed with the linker region in PRG.17 The disappearance of the GEF switch in the crystal structures of the two L-DH/PH proteins suggests a mechanism for the autoinhibition of the intrinsic GEF activity of these RGS-containing RhoGEFs.
The GEF switch is a critical component for the function of RGS-RhoGEFs. Disruption of its interaction with the canonical DH domain by point mutations or deletion leads to significant reduction of GEF activity (Fig. 1, Supporting Information Fig. 2).13 In the crystal structure of the LARG-DH/PH:RhoA complex, the GEF switch interacts directly with the switch regions of RhoA.13 Given its influence on GEF activity and its conformational flexibility, it was proposed that the GEF switch could be perturbed and/or reorganized by contacts with other domains of RGS-RhoGEFs.13 Our data suggests that the linker region, particularly the segment containing residues 353–394, disrupts the interaction between the GEF switch and the α1 helix of the DH domain that is critical to the intrinsic GEF activity of the protein. If this were the case, either the presence of the autoinhibitory linker or removal of the GEF switch should result in similar exchange activities for the respective DH/PH domains. Indeed, the basal exchange activities of the L-DH/PH proteins are almost identical to that of the ΔNDH/PH where the GEF switch has been deleted (Fig. 1). A similar regulatory mechanism, in which GEF activity can be modulated by modification of a region immediately N-terminal to the DH domain, has been observed in other Dbl family proteins, such as Vav24 and Tim.25 In this case, a tyrosine residue is phosphorylated by Src family kinases to relieve autoinhibition. It is also worth noting that the linker regions between RH and DH domains are not well conserved among RGS-RhoGEFs. Aside from the fact that a seven residue fragment immediately N-terminal to the GEF switch is rich in acidic amino acids, the linkers differ greatly both in length and in amino acid composition. Hence, the autoinhibitory mechanism proposed here for p115 might not be adopted by other members of the RGS-RhoGEF family. An earlier study shows that the linker region from PRG also inhibits the intrinsic GEF activity of the protein; however, inhibition is proposed to be achieved by direct interactions with the canonical DH domain.17
The solution structure of L-DH/PH shows that the linker region is partially ordered, but this is not sufficient to stabilize ordered packing interactions; thus, the linker region appears to be completely disordered in the crystal structures. Secondary structure predication indicates that the linker region contains no regular secondary structure, revealing the flexible nature of this region. We hypothesize that the flexible linker region provides a pivot point that allows the two connected domains (RH and DH/PH) to move relative to each other. The flexibility of the linker region might facilitate proper function of RGS-RhoGEFs by allowing more dynamic ranges of movement to accommodate various regulators and effectors.
One such regulator is Gα13. It interacts directly with the RH domain in RGS-RhoGEFs18,26 and stimulates the intrinsic GEF activity of p115 and LARG. There is evidence that Gα13 also interacts, albeit weakly, with segments of RGS-RhoGEFs outside of the RH domain.9,27 It is plausible that coordinated interaction of Gα13 with the RH domain and the linker, or interaction of a Gα13-RH complex with the linker relieves the autoinhibitory impact of this region. The subsequent alignment of the GEF switch for maximal activity would then account for the increased activity caused by the activated Gα subunits. Activation by perturbation of the linker region might further provide a means for other regulatory inputs to stimulate the exchange activity of RGS-RhoGEFs. Investigation using full-length RGS-RhoGEFs and Gα13 will be required to test and further elucidate this regulatory mechanism.
Materials and Methods
Protein expression and purification
Coding regions of human p115-RhoGEF (p115) were subcloned into a pGEX-KG vector containing the protease recognition site for the Tobacco Etch Virus (pGEX-KG-TEV) for proteolytic cleavage of the expressed domains from Glutathione-S-transferase as described previously.21 A 6His-tag was also inserted at the C-termini of the p115 coding sequences. p115 proteins were expressed in LB medium at 22°C overnight in E. coli strain BL21(DE3) cells with 100 μM isopropyl-β-D-thiogalactopyranoside (IPTG). Frozen cells from 1 L were thawed and suspended with 50 mL of lysis buffer (50 mM NaHEPES, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol and protease inhibitors). Cells were lysed with addition of lysozyme, DNase I and MgCl2 to final concentrations of 2 mg mL−1, 50 ug mL−1, and 5 mM, respectively. GST-tagged fusion proteins were extracted from the soluble fraction of lysates by affinity chromatography with Glutathione Sepharose 4B (Amersham Pharmacia Biotech). Resins with GST-fusion proteins bound were suspended with lysis buffer, then incubated overnight at 4°C in the presence of 1 mg TEV protease to remove the GST-tag. Fragments of p115 released from the resin were further purified by IMAC-Ni2+ affinity chromatography (Bio-Rad). Affinity enriched proteins were subjected to further purification with a Mono Q anion exchange column (Amersham Pharmacia Biotech) that had been pre-equilibrated with Buffer A (25 mM Tris, pH 8.5, 1 mM DTT and 1 mM EDTA). Elution was accomplished with a linear gradient of 0 to 0.5 M NaCl in Buffer A. Mono Q purified p115 fragments were further purified by size-exclusion chromatography using Superdex 200/75 columns (Amersham Pharmacia Biotech) that had been pre-equilibrated with Buffer A and 100-mM NaCl. Fractions that contained the p115 fragments (molecular weight of approximately 40–60 kDa as judged by elution volume) were pooled and concentrated using Amicon-Ultra 4 (10 kDa) concentrators (Millipore) to a final concentration of 15–20 mg mL−1. Aliquots (50 μL) of the concentrated proteins were flash frozen with liquid nitrogen and stored at −80°C. The expression and purification of a C-terminally truncated human RhoA (residues 1–181) was carried out as described previously.21
Nucleotide exchange assay
Fluorescence assays measuring the release of N-methylanthraniloyl-GDP (mant-GDP, Invitrogen) were performed on a Fluorolog-3 spectrofluorometer at room temperature (λex = 356 nm, λem = 445 nm, slits = 1/1 nm), as described previously.21 In each assay, 0.5 μM mant-GDP-loaded RhoA was incubated with 100 μM GTP in reaction buffer (25 mM NaHEPES, pH 8.0, 50 mM NaCl, 1 mM DTT, and 5 mM MgCl2) in a 200 μL cuvette. The exchange reaction was started by the addition of 30 nM p115. Each measurement was repeated at least twice.
Crystallization and data collection
p115 L-DH/PH (residues 240–766) was crystallized by vapor diffusion at 20°C against a solution of 22–26% PEG 3,350, 0.2M ammonium sulfate, and 0.1M NaHEPES, pH 7.2–7.6. Native data with an oscillation range of 165° were measured at 100 K at the Structural Biology Center (Beamline 19ID) at Argonne National Laboratory. p115 ΔN2L-DH/PH (residues 353–766) was crystallized by vapor diffusion at 20°C against a solution of 15–18% PEG 3,350, 0.2 M sodium chloride, and 0.1 M NaHEPES, pH 6.2–6.5. Native data with an oscillation range of 130° were measured at 100 K at the Structural Biology Center (Beamline 19BM) at Argonne National Laboratory. p115 DH/PH (residues 395–766) was crystallized by vapor diffusion at 20°C against a solution of 2.6–2.8M sodium formate. Native data with an oscillation range of 110° were measured at 100 K at the Structural Biology Center (Beamline 19ID) at Argonne National Laboratory. The R399E mutant of p115 DH/PH (R399EDH/PH) was crystallized by vapor diffusion at 20°C against a solution of 2.7–2.9M sodium formate. Native data with an oscillation range of 120° were measured using a Rigaku FR-E copper rotating-anode generator and an R-AXIS IV++ imaging-plate area detector (Rigaku Americas, Houston, Texas, USA). All crystals were cryoprotected with an additional 15% (v/v) ethylene glycol. Diffraction data were reduced using the HKL software package.28
Structure determination and model refinement
Initial phases were generated by molecular replacement using the coordinates of the DH and the PH domains of LARG (PDB access code 1X86) as separate search models, using the program PHASER.29 Model building was performed using the program Coot.30 The model was refined using Refmac5 from the CCP4 software package.31 PROCHECK32 indicates that over 88% of the residues fall in the most favorable regions of φ, ψ conformational space.33 Coordinates and structure factors have been deposited in the Protein Data Bank34 with accession codes 3ODW (L-DH/PH), 3ODX (ΔN2L-DH/PH), 3ODO (DH/PH), and 3P6A (R399EDH/PH). Atomic representations were created using Pymol.35
SAXS data collection
The purified L-DH/PH fragment of p115 was dialyzed overnight in 25 mM NaHEPES, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol, 1 mM EDTA, 10 mM DTT and 5% glycerol to ensure a precise buffer match for background subtraction of the scattering arising from the buffer from that of the protein sample. Triton X-100 (0.5%) was added to the sample before the measurement. The samples for SAXS were at a concentration between 1 and 4 mg mL−1. Measurements were taken at 10°C using the SAXS instrument at the BioCAT beamline of the Advanced Photon Source, Argonne National Laboratory. Data were collected using the Mar 165 CCD area detector at a sample-to-detector distance of 230 cm. Samples were centrifuged at 15,000g for 30 min, and the top 50% of the solution was used for data measurement to eliminate all possible aggregates. Samples were measured in a thermostated quartz capillary flow cell with a diameter of 1.5 mm under a constant flow rate of 10 μL s−1 to prevent potential radiation damage. Ten successive frames with an exposure of 1 s were recorded for each sample and the data averaged for better signal/noise statistics. Each sample measurement was preceded by the measurement of its matching buffer solution. Samples were checked before and after the SAXS measurements for concentration and oligomerization state (by UV absorbance and size-exclusion chromatography). Scattering profiles (intensity I vs. scattering vector Q) were reduced, and the data was merged from measurements at different concentrations using IGOR Pro software (WaveMetrics) with macros written by the BioCAT staff. Structural parameters and the distance distribution function, P(r), were calculated with GNOM36 using data up to a Q of 0.35 Å−1.
Ab Initio modeling
Low-resolution molecular shape reconstructions from the experimental scattering data were performed with DAMMIN.37 The scattering profiles were used up to a Q of 0.35 Å−1 for the reconstruction. Twenty DAMMIN calculations were performed, and the resulted models were averaged by DAMAVER38 to generate the final model, which represents the most probable conformation reconstruction for the protein. The molecular envelope of L-DH/PH was calculated using the program Situs,39 and the crystal structure of DH/PH was then fit into the envelope using the program Chimera.40
Acknowledgments
The authors thank Jana Hadas and Stephen Gutowski for superb technical assistance, and Diana Tomchick, Chad Brautigam and the staff of APS for assistance with data collection. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. Use of the Advanced Photon Source is also supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-ENG-38. BioCAT is a National Institutes of Health-supported Research Center (RR-08630).
Glossary
Abbreviations:
- DH
Dbl-homology
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- GST
glutathione-S-transferase
- mant-GDP
N-methylanthraniloyl-GDP
- mant-GTP
Nmethylanthraniloyl-GTP
- PDZ
post synaptic density protein (PSD95)/Drosophila disc large tumor suppressor (DlgA)/zonula occludens-1 protein (zo-1)
- PH
plekstrin-homology
- RGS
regulators for G protein signaling
- rgRGS
RhoGEF RGS domain
- RH
RGS homology domain
- SAXS
small angle x-ray scattering
- TEV
tobacco etch virus.
References
- 1.Sternweis PC, Carter AM, Chen Z, Danesh SM, Hsiung YF, Singer WD. Regulation of Rho guanine nucleotide exchange factors by G proteins. Adv Protein Chem. 2007;74:189–228. doi: 10.1016/S0065-3233(07)74006-8. [DOI] [PubMed] [Google Scholar]
- 2.Aittaleb M, Boguth CA, Tesmer JJ. Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol Pharmacol. 2010;77:111–125. doi: 10.1124/mol.109.061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 4.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 6.Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. p115 RhoGEF, a GTPase activating protein for Gα12 and Gα13. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 7.Hart MJ, Jiang X, Kosaza T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Gα13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki N, Nakamura S, Mano H, Kozasa T. Galpha 12 activates Rho GTPase through tyrosine-phosphorylated leukemia-associated RhoGEF. Proc Natl Acad Sci USA. 2003;100:733–738. doi: 10.1073/pnas.0234057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki N, Tsumoto K, Hajicek N, Daigo K, Tokita R, Minami S, Kodama T, Hamakubo T, Kozasa T. Activation of leukemia-associated RhoGEF by Galpha13 with significant conformational rearrangements in the interface. J Biol Chem. 2009;284:5000–5009. doi: 10.1074/jbc.M804073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taya S, Inagaki N, Sengiku H, Makino H, Iwamatsu A, Urakawa I, Nagao K, Kataoka S, Kaibuchi K. Direct interaction of insulin-like growth factor-1 receptor with leukemia-associated RhoGEF. J Cell Biol. 2001;155:809–820. doi: 10.1083/jcb.200106139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc Natl Acad Sci USA. 2002;99:12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swiercz JM, Kuner R, Behrens J, Offermanns S. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 2002;35:51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 13.Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J Biol Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 14.Derewenda U, Oleksy A, Stevenson AS, Korczynska J, Dauter Z, Somlyo AP, Otlewski J, Somlyo AV, Derewenda ZS. The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle. Structure. 2004;12:1955–1965. doi: 10.1016/j.str.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Cierpicki T, Bielnicki J, Zheng M, Gruszczyk J, Kasterka M, Petoukhov M, Zhang A, Fernandez EJ, Svergun DI, Derewenda U, Bushweller JH, Derewenda ZS. The solution structure and dynamics of the DH-PH module of PDZRhoGEF in isolation and in complex with nucleotide-free RhoA. Protein Sci. 2009;18:2067–2079. doi: 10.1002/pro.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells CD, Gutowski S, Bollag G, Sternweis PC. Identification of potential mechanisms for regulation of p115 RhoGEF through analysis of endogenous and mutant forms of the exchange factor. J Biol Chem. 2001;276:28897–28905. doi: 10.1074/jbc.M102913200. [DOI] [PubMed] [Google Scholar]
- 17.Zheng M, Cierpicki T, Momotani K, Artamonov MV, Derewenda U, Bushweller JH, Somlyo AV, Derewenda ZS. On the mechanism of autoinhibition of the RhoA-specific nucleotide exchange factor PDZRhoGEF. BMC Struct Biol. 2009;9:36. doi: 10.1186/1472-6807-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Singer WD, Danesh SM, Sternweis PC, Sprang SR. Recognition of the activated states of Galpha13 by the rgRGS domain of PDZRhoGEF. Structure. 2008;16:1532–1543. doi: 10.1016/j.str.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenhaure TM, Francis SA, Willison LD, Coughlin SR, Lerner DJ. The Rho guanine nucleotide exchange factor Lsc homo-oligomerizes and is negatively regulated through domains in its carboxyl terminus that are absent in novel splenic isoforms. J Biol Chem. 2003;278:30975–30984. doi: 10.1074/jbc.M303277200. [DOI] [PubMed] [Google Scholar]
- 20.Chikumi H, Barac A, Behbahani B, Gao Y, Teramoto H, Zheng Y, Gutkind JS. Homo- and hetero-oligomerization of PDZ-RhoGEF, LARG and p115RhoGEF by their C-terminal region regulates their in vivo Rho GEF activity and transforming potential. Oncogene. 2004;23:233–240. doi: 10.1038/sj.onc.1207012. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Medina F, Liu MY, Thomas C, Sprang SR, Sternweis PC. Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation. J Biol Chem. 2010;285:21070–21081. doi: 10.1074/jbc.M110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Cryst A. 1986;42:140–149. [Google Scholar]
- 23.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 25.Yohe ME, Rossman KL, Gardner OS, Karnoub AE, Snyder JT, Gershburg S, Graves LM, Der CJ, Sondek J. Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J Biol Chem. 2007;282:13813–13823. doi: 10.1074/jbc.M700185200. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Singer WD, Sternweis PC, Sprang SR. Structure of the p115RhoGEF rgRGS domain-Galpha13/i1 chimera complex suggests convergent evolution of a GTPase activator. Nat Struct Mol Biol. 2005;12:191–197. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- 27.Wells CD, Liu M-Y, Jackson M, Gutowski S, Sternweis PM, Rothstein JD, Kozasa T, Sternweis PC. Mechanisms for reversible regulation between p115 RhoGEF and GTRAP48 and the G12 class of heterotrimeric G proteins. J Biol Chem. 2002;277:1174. doi: 10.1074/jbc.M105274200. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Collaborative computational project. The CCP4 Suite:programs for protein crystallography. Acta Cryst D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 33.Ramachandran GN, Sassiekharan V. Conformation of polypeptides and proteins. Adv Prot Chem. 1968;28:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 34.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMOL Molecular Graphics System, Version 1.2r3Pre. Schrödinger, LLC: 2002. [Google Scholar]
- 36.Semenyuk AV, Svergun DI. Gnom—a program package for small-angle scattering data-processing. J Appl Cryst. 1991;24:537–540. [Google Scholar]
- 37.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]