Abstract
N-glycosylation is the most common and versatile protein modification. In eukaryotic cells, this modification is catalyzed cotranslationally by the enzyme oligosaccharyltransferase, which targets the β-amide of the asparagine in an Asn-Xaa-Ser/Thr consensus sequon (where Xaa is any amino acid but proline) in nascent proteins as they enter the endoplasmic reticulum. Because modification of the glycosylation acceptor site on membrane proteins occurs in a compartment-specific manner, the presence of glycosylation is used to indicate membrane protein topology. Moreover, glycosylation sites can be added to gain topological information. In this study, we explored the determinants of N-glycosylation with the in vitro transcription/translation of a truncated model protein in the presence of microsomes and surveyed 25,488 glycoproteins, of which 2,533 glycosylation sites had been experimentally validated. We found that glycosylation efficiency was dependent on both the distance to the C-terminus and the nature of the amino acid that preceded the consensus sequon. These findings establish a broadly applicable method for membrane protein tagging in topological studies.
Keywords: C-terminus tagging, glycosylation efficiency, membrane protein topology, oligosaccharyltransferase acceptor site, sequon
Introduction
Membrane proteins represent about a third of the proteins in all living organisms, but structural information is lacking for an understanding of their various functions. Based on the membrane proteins with 3D structures in the membrane protein database maintained in Stephen White's laboratory (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html), at the end of 2009, there were 217 membrane proteins with unique structures. These represented <0.4% of the total protein structures deposited in the Protein Data Bank.1 Compared to soluble proteins, there is a striking paucity of membrane protein structures. Therefore, membrane protein topology (i.e., the number of transmembrane (TM) segments and their orientation in the membrane) provides an important intermediate picture, that is, more informative than the amino acid sequence,2 although less than the fully folded 3D structure.
Our knowledge of the underpinnings of membrane protein structure has grown exponentially in the last few years.3,4 Common membrane protein architectural features are necessary for insertion into the lipid environment of the cell membrane. Hence, the great majority of membrane proteins contain one or more TM α-helices formed by a stretch of ∼20 amino acids with hydrophobic side chains. These hydrophobic TM regions are connected with hydrophilic loops with distinct charge distributions.5 This provides a simple method for predicting the topology of a membrane protein, which is typically confirmed with molecular and biochemical techniques.
In eukaryotic cells, most membrane proteins are integrated into the membrane cotranslationally; that is, at the same time that they are being synthesized by ribosomes. They are incorporated into the endoplasmic reticulum (ER) membrane at sites termed translocons, which comprise a specific set of membrane proteins.6 During this process, the translocon mediates the integration of TM sequences into the nonpolar core of the membrane bilayer and delivers hydrophilic cytoplasmic and luminal domains to the appropriate compartments. Simultaneously, a nascent protein may undergo covalent modifications, like signal sequence cleavage and N-glycosylation. N-glycosylation is performed in the lumen of the ER by the enzyme oligosaccharyltransferase (OST). OST transfers preassembled sugar moieties from a lipid carrier to the β-amino groups of the asparagine residues in the Asn-Xaa-Ser/Thr (NXS/T) consensus sequences.7 Modifications of the glycosylation acceptor sites occur in a compartment-specific manner; thus, the presence of glycosylation can provide valuable topological information.8 This endogenous glycosylation information can be extended experimentally by adding glycosylation tags at the C-terminus of the polypeptide. The aim of this study was to explore the determinants of glycosylation efficiency for added C-terminal tags. Our results showed that a C-terminal tag requires at least six amino acid residues for efficient glycosylation, and that the amino acid preceding the NXS/T sequon is an important determinant of glycosylation efficiency.
Results and Discussion
Glycosylation efficiency increases with distance from the C-terminus
To examine the influence of the distance between the glycosylation site and the C-terminus of the polypeptide, we expressed a truncated protein in an in vitro translation/glycosylation system with or without added dog pancreas microsomes. We utilized the well-characterized Escherichia coli inner membrane protein leader peptidase (Lep) harboring an Asn-Ser-Thr sequon, which is a well-known glycosylation motif. Lep is anchored in the cytoplasmic membrane by two TM segments (H1 and H2) that are connected by a highly positively-charged cytoplasmic domain (P1), which drives membrane topology.9 When Lep was translated/glycosylated in vitro in the presence of dog pancreas microsomes, it inserted into the microsomal membrane with both the N and C termini on the luminal side.10,11 Previous studies have shown that, when an engineered N-glycosylation site was placed downstream of H2 at 11-12 residues distal to the hydrophobic end (Fig. 1, top), it was glycosylated upon correct insertion into the microsomal membrane.10,12 Glycosylation of the molecule resulted in a 2.5 kDa increase in molecular mass relative to that of Lep expressed in the absence of microsomes. To determine whether glycosylation efficiency was affected by the position of the glycosylation acceptor site, we generated truncated proteins that included N-linked glycosylation sites at different distances from the C-terminus. These truncated Lep variants were expressed in a rabbit reticulocyte cell-free translation system supplemented with [35S] Met/Cys and dog pancreas rough microsomes. Translation of each variant yielded two types of protein products: the truncated Lep protein with a single oligosaccharide attached to the tag and the unglycosylated truncated protein. The proportion of glycosylated and unglycosylated proteins directly reflected the efficiency of N-glycosylation by OST. After SDS-PAGE analysis, the proportions of glycosylated and unglycosylated protein were quantified from gel autoradiographs.
Figure 1.
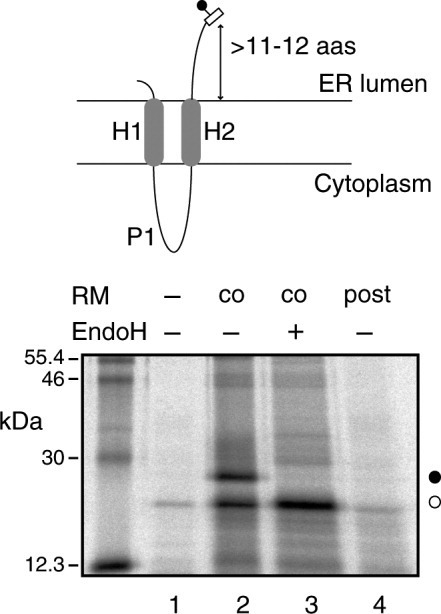
C-terminal-tagged truncated Lep constructs are cotranslationally glycosylated. (Top) Diagram showing the orientation of truncated Lep in the microsomal membrane. (Bottom) C-terminal-tagged (NSTMMS) Lep construct was translated in either the absence (lanes 1 and 4) or presence (lanes 2 and 3) of dog pancreas rough microsomes (RM). After translation, samples were treated with Endo H (lane 3). In lane 4, RMs were added post-translationally; Lep constructs underwent 1 h translation, followed by 10 min cycloheximide treatment, then incubation with RMs was continued for another 1 h. Bands of unglycosylated and glycosylated proteins are indicated with white and black dots, respectively.
We first determined whether truncated Lep proteins that carried a C-terminal glycosylation tag were cotranslationally glycosylated. It has long been reported that the tripeptide sequon Asn-Xaa-Thr is more efficiently glycosylated than Asn-Xaa-Ser;13 in fact, the occurrence rate of the former is about one-third higher than that of the latter (39,161 and 30,579 sequons in our database, respectively), which is in agreement with a recent statistical survey.14 We found that translation of truncated Lep with a six residue glycosylation tag (NSTMMS) in the presence of rough microsomal membranes (RM) was associated with an increase in the molecular mass, indicative of protein glycosylation (Fig. 1, lane 2). This result was further corroborated when the increase in mass was abolished by treatment with endoglycosidase H (Fig. 1, lane 3), a glycan-removing enzyme.15 Notably, when microsomal membranes were included post-translationally, after translation inhibition with cycloheximide, the C-terminal acceptor site was not glycosylated (Fig. 1, lane 4); this suggested that the truncated Lep was integrated into the membrane cotranslationally, via the ER translocon. These results were consistent with an earlier study on truncated Lep, where the glycosylation efficiency was reduced to ∼40% when the glycosylation acceptor site was placed six residues upstream of the stop codon.16 Similarly, it has been shown that the recombinant mammalian concentrative Na+-nucleoside cotransporter rCNT1 could be glycosylated in Xenopus oocytes at an asparagine residue located six residues upstream of the C-terminal end.17
Next, we investigated glycosylation efficiency as a function of the distance between the acceptor Asn residue and the C-terminus of the polypeptide. As shown in Figure 2(A,B), the glycosylation efficiency increased gradually with the distance between the acceptor site and the C-terminus. When the C-terminal glycosylation tag only included the three amino acid residues that formed the acceptor sequon (NST, 3 residues tag), the truncated polypeptide remained unglycosylated [Fig. 2(A), lanes 1 and 2]. Extending the C-terminal tag to four residues (NSTM) slightly increased glycosylation (∼20%, lanes 3 and 4), and a C-terminal tag with five residues (NSTMM) nearly doubled the glycosylation efficiency [Fig. 2(A), lanes 5 and 6]. Further extensions of the tag length rendered similar glycosylation levels [∼50%, see Fig. 2(A), lanes 7-12]. To compare these results with native glycoproteins, we performed a statistical analysis using the sequences and their annotations from the UniProt database (http://www.uniprot.org, release 2010_09).18 After selecting nonredundant N-glycosylated proteins (see Materials and Methods section), the complete set of putative N-glycosylation sites was obtained by selecting only Asn-Xaa-Thr sequons. The final dataset contained 39,161 sequons of which 5,753 were experimentally validated. Native glycosylated sites located at the C-terminal regions were more prominent (13 occurrences) for sequons with the Asn amino acid located six residues upstream from the C-terminus [Fig. 2(C)]. Nevertheless, the total number of glycosylation sites at this position relative to the total sequons (5,753 in native sequences) suggests that protein glycosylation near the C-terminus is a relatively rare event and explains the low glycosylation efficiency (∼50%) in our experiments. Thus, the tag with six amino acid residues (NSTMMS) was selected for further experiments. It should be noted that the presence of a methionine residue following the glycosylation sequon conferred optimal glycosylation efficiency when Thr was present at the hydroxyl (third) position.19
Figure 2.
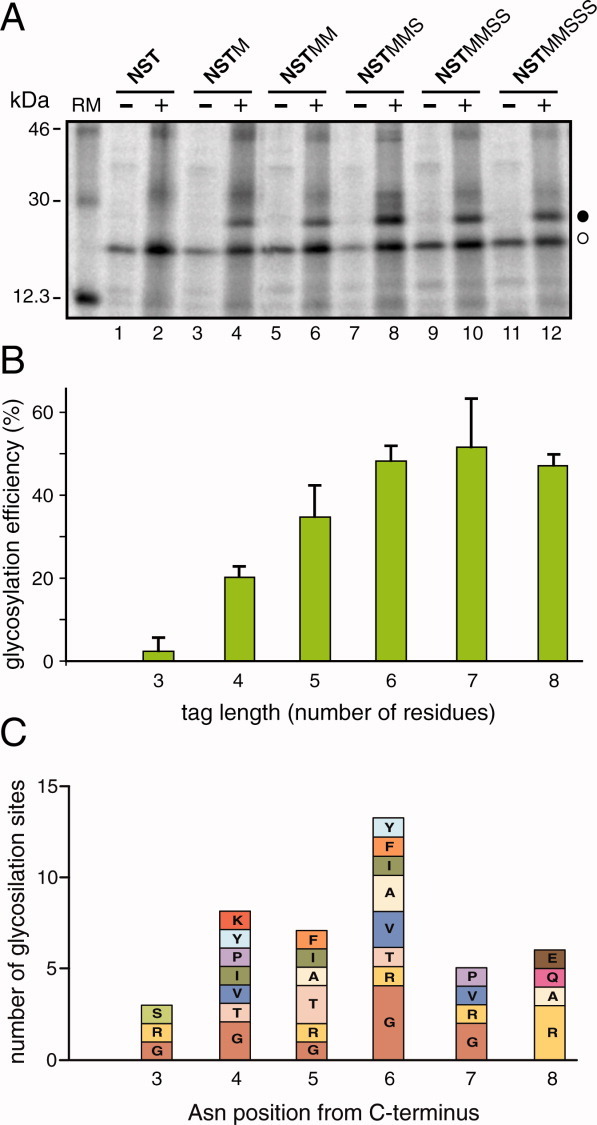
Glycosylation efficiency increases with the distance from the C-terminus. (A) In vitro translation of the truncated proteins with different C-terminal glycosylation tags in the absence (−) and in the presence (+) of RM. As in Figure 1, glycosylated and unglycosylated products are shown with black and white dots, respectively. (B) The glycosylation efficiency is shown as a function of the number of residues between the acceptor Asn and the C-terminus (tag length). To calculate the percent efficiencies, the total glycosylation (100%) was taken as the sum of the signals present in the glycosylated and nonglycosylated forms. Data correspond to averages of at least three independent experiments; error bars show standard deviations. (C) NXT glycosylation sequon distribution at the C-terminal region (positions 3 to 8) in nonredundant experimentally validated glycoproteins. Each bar height is proportional to the number of sequons and displays the distribution of amino acid residues at each position. Nonoccurring amino acid residues at each site are omitted.
Glycosylation of truncated Lep variants
We translated 20 variants of C-terminal-tagged truncated Lep proteins to examine systematically whether the amino acid residue preceding the acceptor Asn affected glycosylation efficiency (Fig. 3). As expected, when a Pro residue preceded the acceptor Asn, glycosylation was significantly inhibited. However, Pro had a stronger inhibitory effect when it was located either at the central Xaa position20 or following the glycosylation sequon.19 It is interesting to note that Pro has never found preceding an experimentally verified glycosylation site in our database when the glycosylated Asn residue in the NXT sequon is located at six residues from the C-terminal end [Fig. 2(C)]. However, this inhibitory effect was not observed when the Pro residue was inserted just before the acceptor Asn in a full-length Lep construct (Fig. 4). In fact, more than 80% of the molecules were glycosylated when this Lep mutant was assayed (Fig. 4, lane 8). This suggested that the residue preceding the glycosylation sequons only impacted glycosylation efficiency when the acceptor Asn residue was close to the end of the polypeptide. Indeed, of the 42 sequons Asn-Xaa-Thr located within the last eight residues, only two were preceded a Pro residue [Fig. 2(C)].
Figure 3.
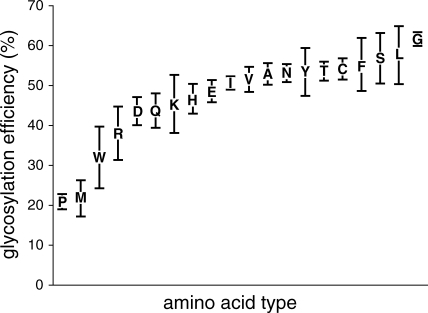
Glycosylation efficiencies of Lep truncates with different amino acid residues preceding the glycosylation sequon. C-terminal-tagged truncated Lep variants contained the indicated amino acid residues in front of the Asn residue of the glycosylation site. Glycosylation levels were determined from gel autoradiographs. Data correspond to averages of at least three independent experiments; error bars show standard deviations.
Figure 4.
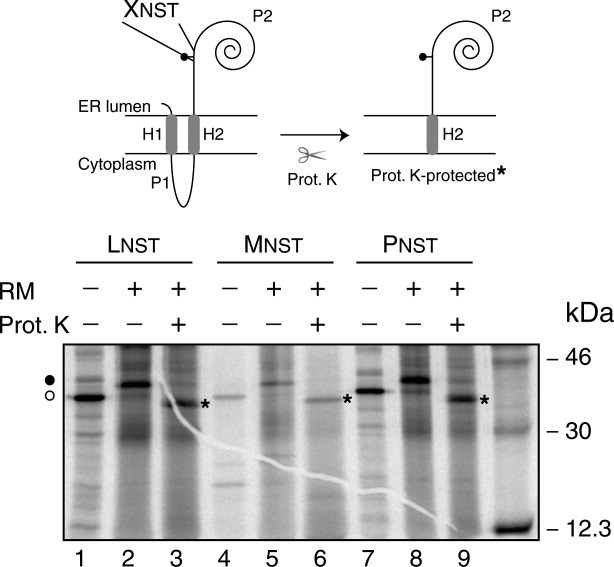
Glycosylation efficiency of full-length Lep mutants. In vitro translation of mRNAs encoding full-length Lep mutants was achieved in the presence (+) and absence (−) of membranes and proteinase K (PK) as indicated. Lep variants contain a single Asn-Ser-Thr sequon (codons 97-99) preceded by Leu (lanes 1-3), Met (lanes 4-6) or Pro (lanes 7-9) in each case. Bands of nonglycosylated protein are indicated by a white dot and glycosylated proteins are indicated by a black dot. The asterisk identifies undigested protein after PK treatment. (Top) Schematic representations of the Lep full-length construct and the proteinase K-protected fragment.
The probability of each amino acid type preceding a verified glycosylation sites has been calculated for the Asn-Xaa-Thr sequons in the nonredundant dataset (Fig. 5). All 20 amino acids can be found preceding the Asn residue of the sequons, although significant differences between their probabilities occur in the experimentally validated glycosylation sites. Experimentally, we found that the glycosylation efficiency of the NST sequon was also significantly lowered when it was preceded by Met, Trp, or Arg residues (Fig. 3), which correlates with the results of our statistical analysis, especially in the case of Met and Trp (Fig. 5). One explanation for this observation might be that the bulky side chains of these residues may block accessibility to the OST active site or the lipid carrier donor; another explanation could be that it may induce an unfavorable local protein conformation. Previous studies revealed that glycosylation was strongly inhibited when Trp was placed at the central Xaa position,20,21 and it was somewhat inhibited when Trp followed the glycosylation sequon.19 Our results also pointed out some average effect caused by the presence of acidic residues immediately before the glycosylation site. It is interesting to note that it has been described a notable reduction in the probability of finding acidic residues preceding the glycosylation site.22 Even more, this is accompanied by an increase probability of finding acidic residues preceding unoccupied glycosylation sites.14 However, both Asp and Glu have been found as average preceding NXT acceptor sites (Fig. 5). The apparent discrepancies between our and the previous studies could arise from the fact that the later surveys included the glycosylation sites with Ser in the third position (NXS/T), whilst our database focussed in NXT glycosylation sites. The other amino acid residues appeared to have only minor effects on glycosylation efficiency; however, the Gly residue consistently induced higher glycosylation levels (Fig. 3), and again, an increased probability of finding Gly preceding glycosylation sites was observed in our analysis (Fig. 5). We assumed that the flexibility that Gly confers on the conformation of the polypeptide chain may provide an advantage for OST catalysis. In fact, the structural conformation of the local region around the glycosylation sequon also influenced its accessibility and, consequently, its site occupancy.23 This supports the hypothesis that unfolding or flexibility is required for protein domains to be efficiently glycosylated. It should be also noted that there is a marked preference for hydrophobic amino acids immediately preceding the glycosylation site in our experimental data, which nicely correlates with previous22 and the present statistical analysis of glycan-protein linkage, especially in the case of Leu that has been also found prevalent in glycosylated sequons (see reference14 and Fig. 5).
Figure 5.
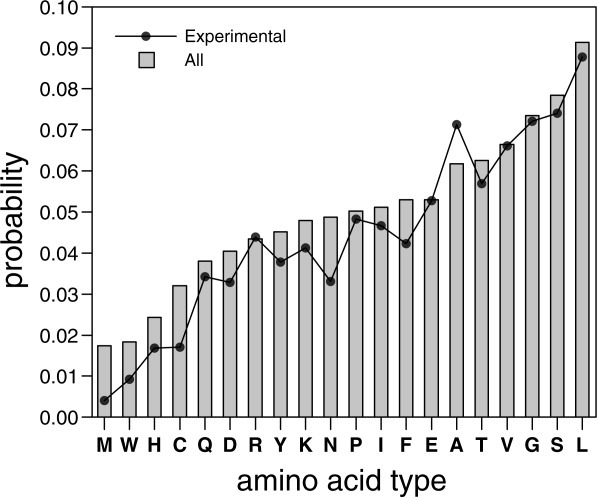
Distribution of amino acid residues preceding NXT glycosylation sequons in all sites (gray bars) and experimentally validated sites (black line).
Interestingly, recent statistical analyses of active bacterial N-glycosylation site consensus sequences showed that Asn, Phe, Ser, and Leu residues frequently precede the acceptor Asn.24 In the present study, we found that these same residues and Gly were the best suited for glycosylation in a eukaryote OST. Based on these results, we propose that bacterial and eukaryotic systems might require similar sequences flanking the acceptor sites to adopt an optimal conformation upon the binding of OST.
Statistical studies have also shown that the glycosylation sequon occurs at the C-terminal end of well-defined glycoproteins at a lower frequency than that expected based on random chance;13,14 and that when N-glycosylation sites are contained within more than one extracytosolic loop, only the first loop is modified.25 Furthermore, those studies found that the glycosylation efficiency for Asn-Xaa-Thr sequons dropped when located close to the C-terminal end of the protein.14 The present work pointed out that this effect was emphasized at the very end of the protein [Fig. 2(C)]. This suggested that it was necessary to use sufficiently large C-terminal glycosylation tags. In fact, we found that at least six residues long C-terminal glycosylation tags were needed to achieve significant glycosylation; this validated their utility in membrane protein topological studies.
To prove our approach, we have fused the N-terminus of bacteriorhodopsin (bR) (from Trp10 to Val101) at the C-terminus of the engineered Lep sequence (see Materials and Methods section). We choose bR because it is a membrane protein with a well-defined topology, in which the N-terminus faces the extracellular side similarly to our chimeric constructs (Fig. 6, top). In vitro transcription/translation of protein truncates using a glycosylable C-terminal tag after bR helix a (the first TM segment) rendered singly-glycosylated forms (Fig. 6, lane 2), indicating the insertion of bR helix a. Truncated polypeptides, which include the first two TM helices of bR, were efficiently doubly-glycosylated (Fig. 6, lane 4), demonstrating translocation of the glycosylation site included as a C-terminal tag, and validating our experimental approach.
Figure 6.
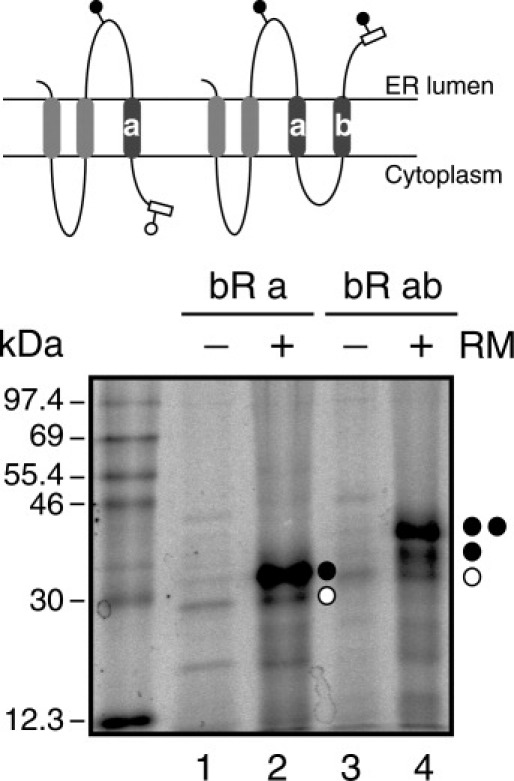
Glycosylation efficiency of Lep/bR truncates. In vitro translation of C-terminal-tagged mRNAs encoding Lep/bR constructs was performed in the presence (+) and absence (−) of membranes as indicated. Bands of nonglycosylated proteins are indicated by a white dot and singly- and doubly-glycosylated proteins are indicated by one and two black dots, respectively. (Top) Schematic representations of the Lep/bR constructs including bR helix a (left) and bR helices a and b (right).
Conclusion
We have investigated N-linked glycosylation efficiency using an in vitro system based on microsomes and a well-characterized model protein. In conclusion, we found that in placing a glycosylation tag on a polypeptide chain, one should consider both the distance from the hydrophobic end of a TM segment and the nature of the amino acid residue preceding the acceptor Asn residue. Taken together, our results provided a rapid and efficient method for the determination of membrane protein topology.
Materials and Methods
Enzymes and chemicals
The pGEM1 plasmid, rabbit reticulocyte lysate, and the TnT coupled transcription/translation system were purchased from Promega (Madison, WI). The ER rough microsomes from dog pancreas and the SP6 RNA polymerase were purchased from tRNA Probes (College Station, TX). The [35S] Met/Cys and 14C-methylated markers were purchased from Perkin-Elmer. The restriction enzymes and endoglycosidase H were purchased from Roche Molecular Biochemicals. The DNA plasmid, RNA clean up, and PCR purification kits were from Qiagen (Hilden, Germany). The PCR mutagenesis kit, QuikChange was from Stratagene (La Jolla, CA). All the oligonucleotides were purchased from Thermo (Ulm, Germany).
DNA manipulations
Full-length Lep DNA was amplified directly from the pGEM1 plasmid, which carried a modified lep gene. In that sequence, the nucleotides that encoded the Asn-Glu-Thr glycosylation acceptor site at position 214-216 in the wild type protein was changed to a nonacceptor sequence Gln-Glu-Thr. In addition, an Asn-Ser-Thr (NST) glycosylation acceptor site was introduced 20 amino acids downstream of H2 at codons 97-99. Alternatively, we prepared templates for in vitro transcription of the truncated wild type lep mRNA with a 3′ glycosylation tag. The truncated lep sequence was prepared by PCR amplification of a fragment of the pGEM1 plasmid that encoded the N-terminal 178 amino acid residues of Lep. The 5′ primer was the same for all PCR reactions and had the sequence 5′-TTCGTCCAACCAAACCGACTC-3′. This primer was situated 210 bases upstream of the lep translational start codon; thus, all amplified fragments contained the SP6 transcriptional promoter from pGEM1. The 3′ primers were designed to have approximately the same annealing temperature as the 5′ primer. They contained a glycosylation tag preceded by one of the 20 natural amino acids, and followed by the tandem translational stop codons, TAG and TAA. PCR amplification comprised a total of 30 cycles with an annealing temperature of 52°C. The amplified DNA products were purified with the Qiagen PCR purification kit, according to the manufacturer's protocol, and verified on a 1% agarose gel. The mutations Leu96 Met and Leu96Pro were performed with the QuikChange mutagenesis kit from Stratagene (La Jolla, CA), according to the manufacturer's protocol.
The N-terminal region from bacteriorhodopsin (residues 10-101) was PCR amplified and cloned into the modified Lep sequence from pGEM plasmid26,27 between SpeI and KpnI sites. The truncated Lep/bR chimeras were prepared by PCR amplification of fragments that encoded up to Lys41 (bR sequence) in the case of Lep/bRa and up to Ile78 for Lep/bRab truncates. All DNA manipulations were confirmed by sequencing the plasmid DNAs.
Expression in vitro
Truncated lep mRNAs with stop codons were transcribed from the SP6 promoter with SP6 RNA polymerase (tRNA probes). Briefly, the transcription mixture was incubated at 37°C for 2 h. The mRNA products were purified with a Qiagen RNeasy clean up kit and verified on a 1% agarose gel.
In vitro translation of in vitro transcribed mRNA was performed in the presence of reticulocyte lysate, [35S] Met/Cys, and dog pancreas microsomes, as described previously.28 For the posttranslational membrane insertion experiments, Lep-derived mRNAs were translated (30°C 1 h) in the absence of RMs. Translation was then inhibited with cycloheximide (10 min, 26°C, 2 mg/mL final concentration), after which RMs were added and incubated for an additional hour at 30°C.29 In all cases, after translation, membranes were collected by ultra-centrifugation and analyzed by sodium-dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Finally, the gels were visualized on a Fuji FLA3000 phosphorimager with ImageGauge software.
For endoglycosidase H (Endo H) treatment, the translation mixture was diluted in four volumes of 70 mM sodium citrate (pH 5.6) and centrifuged (105g for 20 min at 4°C). The pellet was then resuspended in 50 μL of sodium citrate buffer with 0.5% SDS and 1% β-mercaptoethanol, boiled for 5 min, and incubated for 1 h at 37°C with 0.1 mU of Endo H. The samples were analyzed by SDS-PAGE.
Full-length Lep constructs were transcribed and translated in the TnT Quick system (Promega). 1 μg DNA template, 1 μL 35S-Met/Cys (5 μCi) and 1 μL microsomes (tRNA Probes) were added at the start of the reaction, and samples were incubated for 90 min at 30°C. Translation products were analyzed as previously described for the truncated molecules. For the proteinase K protection assay, the translation mixture was supplemented with 1 μL of 50 mM CaCl2 and 1 μL of proteinase K (4 mg/mL), then, digested for 40 min on ice. The reaction was stopped by adding 1 mM phenylmethanesulfonylfluoride before SDS-PAGE analysis.
Statistical analysis of N-glycosilation sites in native proteins
Sequences and their annotations were obtained from the UniProt database (http://www.uniprot.org, release 2010_09).18 Selection of N-glycosylated proteins was done using the UniProt search engine by selecting all sequence annotation (FT field) as glycosylated modified amino acid. Such selection contained both, experimentally validated as well as nonvalidated glycosylation sites. The total number of sequences containing at least one glycosylation site was 25,488, of which 2,533 had been experimentally validated. Next, all the selected sequences were compared to each other using the cd-hit program with default parameters.30 Redundant sequences at the 90% sequence identity cut-off were removed. Finally, only N-glycosylation sites with sequons NXT (being X any of the 20 amino acid types) were maintained. Our final dataset, which contained 39,161 NXT sequons of which 5,753 were experimentally validated, could be considered as an up-to-date set of nonredundant sequences with annotated NXT N-glycosylation sites.
Acknowledgments
The authors thank Prof. Esteve Padrós (Universitat Autònoma de Barcelona) for the sequence of bacteriorhodopsin.
Glossary
Abbreviations:
- bR
bacteriorhodopsin
- ER
endoplasmic reticulum
- Lep
leader peptidase
- RMs
rough microsomal membranes
- SDS-PAGE
sodium dodecylsulfate polyacrylamide-gel electrophoresis
- TM
transmembrane.
References
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 3.White SH. The progress of membrane protein structure determination. Protein Sci. 2004;13:1948–1949. doi: 10.1110/ps.04712004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 5.von Heijne G. Membrane protein structure prediction—hydrophobicity analysis and the positive-Inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 6.Alder NN, Johnson AE. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J Biol Chem. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- 7.Welply JK, Shenbagamurthi P, Lennarz WJ, Naider F. Substrate recognition by oligosaccharyltransferases. Studies on glycosylation of modified asn-x-ser/thr tripeptides. J Biol Chem. 1983;258:11856–11863. [PubMed] [Google Scholar]
- 8.Cheung JC, Reithmeier RA. Scanning N-glycosylation mutagenesis of membrane proteins. Methods. 2007;41:451–459. doi: 10.1016/j.ymeth.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 9.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 10.Nilsson I, von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 11.Martinez-Gil L, Sanchez-Navarro JA, Cruz A, Pallas V, Perez-Gil J, Mingarro I. Plant virus cell-to-cell movement is not dependent on the transmembrane disposition of its movement protein. J Virol. 2009;83:5535–5543. doi: 10.1128/JVI.00393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orzaez M, Salgado J, Gimenez-Giner A, Perez-Paya E, Mingarro I. Influence of proline residues in transmembrane helix packing. J Mol Biol. 2004;335:631–640. doi: 10.1016/j.jmb.2003.10.062. [DOI] [PubMed] [Google Scholar]
- 13.Gavel Y, von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites—implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Dor S, Esterman N, Rubin E, Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. doi: 10.1093/glycob/cwh004. [DOI] [PubMed] [Google Scholar]
- 15.Vilar M, Sauri A, Monne M, Marcos JF, von Heijne G, Perez-Paya E, Mingarro I. Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane. J Biol Chem. 2002;277:23447–23452. doi: 10.1074/jbc.M202935200. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson I, von Heijne G. Glycosylation efficiency of Asn-Xaa-Thr sequons depends both on the distance from the C terminus and on the presence of a downstream transmembrane segment. J Biol Chem. 2000;275:17338–17343. doi: 10.1074/jbc.M002317200. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton SR, Yao SY, Ingram JC, Hadden DA, Ritzel MW, Gallagher MP, Henderson PJ, Cass CE, Young JD, Baldwin SA. Subcellular distribution and membrane topology of the mammalian concentrative Na+-nucleoside cotransporter rCNT1. J Biol Chem. 2001;276:27981–27988. doi: 10.1074/jbc.M100518200. [DOI] [PubMed] [Google Scholar]
- 18.The UniProt Consortium, T.U. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellquist JL, Kasturi L, Spitalnik SL, Shakin-Eshleman SH. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry. 1998;37:6833–6837. doi: 10.1021/bi972217k. [DOI] [PubMed] [Google Scholar]
- 20.Shakin-Eshleman SH, Spitalnik SL, Kasturi L. The amino acid at the X position of an Asn-X-ser sequon is an important determinant of N-linked core-glycosylation efficiency. J Biol Chem. 1996;271:6363–6366. doi: 10.1074/jbc.271.11.6363. [DOI] [PubMed] [Google Scholar]
- 21.Kasturi L, Chen H, Shakin-Eshleman SH. Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochem J. 1997;323:415–419. doi: 10.1042/bj3230415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 23.Jones J, Krag SS, Betenbaugh MJ. Controlling N-linked glycan site occupancy. Biochim Biophys Acta. 2005;1726:121–137. doi: 10.1016/j.bbagen.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF, Wacker M, Aebi M. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landolt-Marticorena C, Reithmeier RAF. Asparagine-linked oligosaccharides are localized to single extracytosolic segments in multi-span membrane glycoproteins. Biochem J. 1994;302:253–260. doi: 10.1042/bj3020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Gil L, Sauri A, Vilar M, Pallas V, Mingarro I. Membrane insertion and topology of the p7B movement protein of Melon Necrotic Spot Virus (MNSV) Virology. 2007;367:348–357. doi: 10.1016/j.virol.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Sauri A, Tamborero S, Martinez-Gil L, Johnson AE, Mingarro I. Viral membrane protein topology is dictated by multiple determinants in its sequence. J Mol Biol. 2009;387:113–128. doi: 10.1016/j.jmb.2009.01.063. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Gil L, Johnson AE, Mingarro I. Membrane insertion and biogenesis of the Turnip crinkle virus p9 movement protein. J Virol. 2010;84:5520–5527. doi: 10.1128/JVI.00125-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]


