Abstract
Regulated exocytosis of neutrophil intracellular storage granules is necessary for neutrophil participation in the inflammatory response. The signal transduction pathways that participate in neutrophil exocytosis are complex and poorly defined. Several protein kinases, including p38 MAPK and the nonreceptor tyrosine kinases, Hck and Fgr, participate in this response. However, the downstream targets of these kinases that regulate exocytosis are unknown. The present study combined a novel inhibitor of neutrophil exocytosis with proteomic techniques to identify phosphopeptides and phosphoproteins from a population of gelatinase and specific granules isolated from unstimulated and fMLF-stimulated neutrophils. To prevent loss of granule-associated phosphoproteins upon exocytosis, neutrophils were pretreated with a TAT-fusion protein containing a SNARE domain from SNAP-23 (TAT-SNAP-23), which inhibited fMLF-stimulated CD66b-containing granule exocytosis by 100 ± 10%. Following TAT-SNAP-23 pretreatment, neutrophils were stimulated with the chemotactic peptide fMLF for 0 min, 1 min, and 2 min. Granules were isolated by gradient centrifugation and subjected to proteolytic digestion with trypsin or chymotrypsin to obtain peptides from the outer surface of the granule. Phosphopeptides were enriched by gallium or TiO2 affinity chromatography, and phosphopeptides and phosphorylation sites were identified by reversed phase high performance liquid chromatography-electrospray ionization-tandem MS. This resulted in the identification of 243 unique phosphopeptides corresponding to 235 proteins, including known regulators of vesicle trafficking. The analysis identified 79 phosphoproteins from resting neutrophils, 81 following 1 min of fMLF stimulation, and 118 following 2 min of stimulation. Bioinformatic analysis identified a potential Src tyrosine kinase motif from a phosphopeptide corresponding to G protein coupled receptor kinase 5 (GRK5). Phosphorylation of GRK5 by Src was confirmed by an in vitro kinase reaction and by precursor ion scanning for phospho-tyrosine specific immonium ions containing Tyr251 and Tyr253. Immunoprecipitation of phosphorylated GRK5 from intact cells was reduced by a Src inhibitor. In conclusion, targets of signal transduction pathways were identified that are candidates to regulate neutrophil granule exocytosis.
Neutrophils are the primary effector cells of the innate immune system and provide the initial cellular response against microbial infections. Exocytosis of neutrophil storage granules is required for neutrophil participation in, and regulation of, the inflammatory response. Exocytosis provides plasma and/or phagosomal membranes with new membrane components, receptors, signal transduction molecules, adhesion molecules, and components of the NADPH oxidase (1–3). Additionally, molecules necessary for bacterial killing, processing of cytokines and chemokines, induction of increased vascular permeability, and monocyte recruitment are released into phagosomes or extracellularly (3–6). The contents of these granules contribute to tissue damage in autoimmune and inflammatory diseases such as rheumatoid arthritis, ANCA-associated vasculitis, and acute coronary syndromes (7–9). Four classes of neutrophil intracellular storage granules have been proposed based on their protein content, morphology, and separation by density gradient fractionation: the secretory vesicles and gelatinase (tertiary), specific (secondary), and azurophil (primary) granules (10).
Although many of the proteins associated with the different neutrophil granule subsets have been identified using proteomic approaches (11–13), the mechanisms by which neutrophil stimulation leads to exocytosis remain poorly defined. Use of pharmacologic inhibitors and knockout mice has identified several signal transduction pathways that are involved in neutrophil exocytosis. In particular, pathways containing p38 mitogen activated protein kinase (MAPK)1 and the nonreceptor Src tyrosine kinases participate in neutrophil granule exocytosis stimulated by TNF-α, LPS, formylated peptides, and chemokines (2, 14–18). However, the targets of those kinases that are involved in exocytosis are unknown. We postulated that phosphorylation of granule-associated proteins by one or more of those kinases regulates exocytosis. The goal of the present study was to identify granule phosphoproteins and their respective phosphorylation sites as an approach to elucidate the mechanisms of kinase-regulated neutrophil exocytosis. To accomplish this goal, proteomic techniques allowing large-scale identification of phosphopeptides were combined with a novel reagent developed in our laboratory that inhibits granule exocytosis. Granules were enriched from neutrophils treated with N-formyl-methionyl-leucyl-phenylalanine (fMLF) for 0, 1, or 2 min and phosphoproteins on the surface of granules were identified by liquid chromatography-tandem MS (LC-MS/MS) and bioinformatics. The expression and phosphorylation of one of the identified proteins, G protein-coupled receptor kinase 5 (GRK5), was validated.
EXPERIMENTAL PROCEDURES
TAT-SNAP-23 Fusion Protein
The N-terminal 78 residues of synaptosomal-associated protein 23 (SNAP-23), including those amino acids that participate in soluble NSF attachment protein receptor (SNARE) complex formation, were subcloned into the pTAT-HA expression vector (a gift from Dr. Steven Dowdy, UCSD, San Diego, CA). The plasmid was transformed into Escherichia coli arabinose-inducible BL21 cells (Invitrogen Corp., Carlsbad, CA) and protein expression induced with 0.2% arabinose, whereas basal T7 transcription was inhibited with 0.1% glucose, at 37 °C for 4 h. The 6× histidine-tagged fusion protein was purified using the ProBond Ni2+-chelating purification system (Invitrogen) under hybrid conditions according to the manufacturer's instructions. The resulting eluate was dialyzed against 10% glycerol, 0.01% triton X-100 in PBS, pH 7.4 and stored at −80 °C until use.
Human Neutrophils
Neutrophils were isolated from healthy human donors using plasma-Percoll gradients, as previously described (19). Trypan blue staining revealed that at least 97% of cells were neutrophils with >95% viability. Isolated neutrophils were suspended in LPS-free Krebs-Ringer phosphate buffer (115 mm NaCl, 4.7 mm KCl, 2.56 mm CaCl2, 1.2 mm KH2PO4, 1.2 mm MgSO4 × 7 H2O, 20 mm NaHCO3 and 16 mm Hepes, pH 7.2) (Krebs+) at 4 × 107 cells/ml. The Human Studies Committee of the University of Louisville approved the use of human donors.
Granule Enrichment
Human neutrophils (1 × 109 cells) were treated with 10 μm diisopropyl fluorophosphate (Sigma-Aldrich, St. Louis, MO) for 10 min at room temperature to inhibit proteases. To inhibit granule fusion with the plasma membrane, neutrophils were incubated with the fusion protein comprised of the trans-activating transcriptional activator (TAT) cell-penetrating peptide and the N-terminal 78 residues of synaptosomal-associated protein-23 (SNAP-23) (TAT-SNAP-23 fusion protein) (1.0 μg/ml for 10 min at 37 °C), then stimulated with 300 nm fMLF for 0 min, 1 min, or 2 min. Following stimulation, neutrophils were immediately resuspended in 20 ml ice cold disruption buffer (10 mm PIPES, 0.1 m KCl, 3 mm NaCl, 3.5 mm MgCl2, protease inhibitor mixture (Sigma-Aldrich)) and disrupted by nitrogen cavitation at 450 psi at 4 °C. All subsequent enrichment steps were performed on ice or at 4 °C to inhibit protease and phosphatase activity. The cavitate was collected and nuclei and intact cells were removed by centrifugation at 700 × g for 5 min. To isolate granule subsets, the postnuclear supernatant was layered onto a discontinuous Percoll gradient of varying densities (1.050, 1.090, and 1.120 g/ml) and centrifuged at 37,000 × g for 30 min as previously described (20, 21). Four fractions (α, β1, β2, and γ) were recovered from the gradient, as previously described by Kjeldsen et al. (21). Percoll was removed by ultracentrifugation at 100,000 × g for 45 min at 4 °C, and the enriched granule subsets were resuspended in disruption buffer and stored at −20 °C. The β1 fraction, extracted from the interface of the 1.090 and 1.120 g/ml fractions, was used for subsequent proteomic analysis.
Preparation of Granule Membrane Phosphopeptides
The β1 granule fraction (100 μl) was resuspended in 500 μl of reaction buffer (10 mm ammonium bicarbonate, 10 mm CaCl2) and incubated with 14 μg of trypsin or chymotrypsin at 37 °C overnight with rotation. Following proteolysis, samples were centrifuged at 10,000 × g to pellet granules. The supernatant containing peptides derived from proteins on the cytosolic surface of granules was transferred to a 3 kDa cutoff centrifugal concentrator (Pall Corp., Port Washington, NY) and centrifuged at 14,000 × g for 20 min to separate peptides from undigested granules and proteins. The filtrates containing peptides were lyophilized and subjected to methyl esterification by resuspension in 200 μl of methanolic hydrochloride for 2 h at RT. Peptides were reconstituted in either 250 mm acetic acid in 30% acetonitrile for phosphopeptide enrichment by immobilized metal ion affinity chromatography using a Ga3+ chelate silica beads (IMAC) or 5% acetonitrile in 0.05% formic acid (FA) (buffer A) for phosphopeptide enrichment by metal oxide affinity chromatography using a prepacked TiO2 column (MOAC). Peptides intended for MOAC were desalted using a C18 micro trap desalting cartridge (Michrom BioResources, Auburn, CA) equilibrated with buffer A. Peptides were applied to the cartridge and salts were removed by washing with two 100 μl aliquots of buffer A. Peptides were then eluted with 100 μl of 95% acetonitrile and 0.1% FA. Desalted peptides were evaporated to dryness in a speedvac and resuspended in 10 μl of buffer A.
MOAC enrichment was performed using a Dionex Ultimate Plus HPLC system (Dionex Corp. Germering, Germany). Desalted peptides were reconstituted in 50 μl 0.05% FA and loaded onto an Nano Trap Column (TiO2, 5 μm, 100 Å, 200 μm i.d × 1 cm., Dionex). The column was washed with 100 μl 80% ACN with 0.05% FA and eluted with 100 μl of 250 mm NH4HCO3, pH 9.0. All steps were performed at a flow rate of 10 μl/min. IMAC enrichment was performed using a Sigma Phosphoprofile I Phosphopeptide enrichment kit per the manufacturer's instructions. Following phosphopeptide enrichment by IMAC, lyophilized peptides were desalted as described above.
Identification of Phosphopeptides
Phosphopeptides were characterized using a modified version of a previously described LC-MS/MS approach (22). Briefly, IMAC- or MOAC-enriched peptides were loaded onto an analytical microcapillary reversed phase chromatography column (100 × 365 μm fused silica capillary with an integrated, laser pulled emitter tip packed with 10 cm of Jupiter 4 μm RP80A (Phenomenex, Torrance, CA)). Peptides were eluted from the reversed phase column, ionized, and sprayed into a mass spectrometer using a linear reverse phase gradient from 3% to 60% of mobile phase B (80% acetonitrile/0.05% formic acid) over 120 min at a flow rate of 300 nl/min. Spectra were acquired with a LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). During LC-MS/MS analysis, the mass spectrometer performed data-dependent acquisition with a full MS scan between 300 and 2000 mass to charge ratio followed by six MS/MS scans (35% collision energy) on the six most intense ions from the preceding MS scan. Data acquisition was performed using dynamic exclusion with a repeat count of 30 ms and a 3 min exclusion duration window.
Tandem mass spectra were extracted and charge state deconvoluted using the Sequest algorithm (ThermoFinnigan, San Jose, CA; version v.27, rev. 11). Deisotoping was not performed. Sequest searched the human FASTA formatted RefSeq database (from NCBI) assuming either the digestion enzyme trypsin or chymotrypsin with two missed cleavages. The data were searched using Sequest with a fragment ion mass tolerance of 1.200 amu and a precursor ion tolerance of 1.200 amu. Methyl ester of aspartic or glutamic acid and phosphorylation of serine, threonine, and tyrosine were specified in Sequest as variable modifications.
Scaffold (version Scaffold_2_05_01, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. A human RefSeq database dated May 30, 2006 containing 27,916 proteins was searched. Peptide identifications were preliminarily accepted if they could be established at greater than 90% probability as specified by the Peptide Prophet algorithm (23). Protein identifications were preliminarily accepted if they could be established at greater than 90% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (24). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Spectra passing these initial criteria were then manually validated to reduce false positives and confirm peptide assignments (25). Spectra were excluded if the majority of the noise (unassigned peaks) was greater than 25% of the relative intensity. Additionally, spectra were excluded if unassigned peaks comprised the four most intense ions.
Kinase Motif Prediction
The Group-based Prediction System (GPS) 2.1 software (26) was used to predict phosphorylation-specific kinase motifs present in the proteomic data set. Phosphopeptide identifications were submitted to GPS in FASTA format and searched against the database under high stringency settings for Src tyrosine kinases and p38 MAPK motifs. The output files were then manually verified to assure accuracy for the identified phosphorylated residues.
In Vitro Kinase Assay
Phosphorylation of GRK5 was performed by incubating 30 ng of active recombinant Src (Millipore, Billerica, MA) with 200 ng of recombinant GRK5 (Invitrogen) in kinase buffer containing 250 μm ATP, 30 mm Tris HCl, pH 7.2, 8 mm MgCl2, 1.4 mm EDTA, 1 mm EGTA, 4.5 mm NaF, and 0.25 mm Na3VO4. Reactions were incubated at 30 °C for various times. Reactions were terminated by dilution in SDS sample buffer and proteins were separated by SDS-PAGE on 4%–12% Novex Bis-Tris gels (Invitrogen). Proteins were transferred to polyvinylidene fluoride (PVDF) and tyrosine phosphorylation was detected by immunoblot analysis using antiphosphotyrosine antibody (BD Transduction Laboratories, Lexington, KY).
Phosphotyrosine-specific Precursor Ion Scanning
Following the kinase reaction, 100 ng of the reaction mixture was manually loaded onto a Phenomenex Jupiter 4μ Proteo 90A (50 × 1.00 mm) column and washed with >10 bed volumes of buffer A. Peptides were eluted from the column with an Agilent 1100 series pump using a linear gradient from 3% to 60% of mobile phase B (80% acetonitrile/0.05% formic acid) over 15 min at a flow rate of 50 μl/min. Spectra were acquired with a triple stage quadrupole Quantum Discovery Max mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with electrospray ionization. The mass spectrometer performed a full MS scan in Q1 between 600 and 1000 m/z in positive ion mode. A precursor ion scan was then performed in Q3 to analyze product ions yielding 216.043 (± 0.02) m/z following CID fragmentation in Q2. Collision gas (argon) pressure was 2.00 mTorr. Collision energy was calculated relative to the predicted precursor ion mass using the formula: m/z × 0.034 V + 3 V.
Measurement of Exocytosis
Exocytosis of CD66b-expressing granules was determined by measuring plasma membrane expression of CD66b by flow cytometry as previously described (27).
Confocal Microscopy
Neutrophils (6 × 106/ml) were stimulated with 300 nm fMLF for various times (0 min, 1 min, 2 min, and 10 min). The cells were then fixed in 3.7% paraformaldehyde for 15 min at room temperature. Cells were permeabilized and blocked with 5% normal donkey serum in 0.02% saponin in Krebs+ for 1 h at room temperature, washed, and then incubated with GRK5 antibody (1:50, R&D systems, Minneapolis, MN) in blocking buffer overnight. Cells were then incubated with rhodamine-labeled donkey anti-goat IgG (Invitrogen) and FITC-conjugated anti-CD66b (1 μg/ml final concentration). Cells were imaged using an Olympus Fluoview confocal microscope with a PlanApo N 60×/1.42 oil immersion lens employing Olympus Fluoview 1000 (version 2.0) software.
Granule Protein Electrophoresis and Immunoblot Analysis
Granules enriched from neutrophils treated with or without 1.0 μg/ml TAT-SNAP-23 and 300 nm fMLF for 2 min were suspended in SDS sample buffer and heated to 95 °C for 5 min. Five micrograms of granule protein was separated by SDS-PAGE on 4%–12% Novex Bis-Tris gels. Proteins were transferred to PVDF and immunoblot analysis was performed using anti-GRK5 (0.5 μg/ml, R&D Systems) and horseradish peroxidase-linked donkey anti-goat secondary antibody (1:1200, Santa Cruz). Replicate gels were stained with Coomassie. Blots were analyzed on a Typhoon 9400 scanner (GE Healthcare, Piscataway, NJ).
Immunoprecipitation and Immunoblot Analysis
Neutrophils (2 × 107 cells) were pretreated with or without 10 μm PP2 at 37 °C for 10 min, then incubated in the presence or absence of 0.3 μm fMLF for 2 min. Following stimulation, cells were immediately centrifuged and resuspended in 0.6 ml ice-cold lysis buffer (2% Tx-100, 100 mm NaCl, 10 mm EGTA, 1 mm Na2VO3, 50 μm Tris, pH 7.4). Lysates were precleared by adding 100 μl of protein G beads (Santa Cruz) and 2.0 μg of mouse IgG and allowing incubation at 4 °C for 1 h. Beads were pelleted by centrifugation at 800 × g for 5 min at 4 °C. Anti-phosphotyrosine antibody (PY69, BD Biosciences) conjugated to agarose beads was added to the supernatants and incubated at 4 °C overnight. The agarose beads with immunoprecipitated protein were collected by centrifugation at 800 × g for 1 min at 4 °C, washed four times with lysis buffer, and then resuspended in 40 μl of 2× Laemmli buffer and boiled for 5 min. The beads were quickly spun down, and 20 μl of each sample was separated by SDS-PAGE on a 6% gel (Invitrogen). Proteins were transferred to PVDF, blocked in 1% bovine serum albumin/TTBS, and immunoblot analysis was performed using anti-GRK5 (1:300 in 1% bovine serum albumin /TTBS, Santa Cruz) and an HRP-linked rabbit anti-mouse secondary antibody.
RESULTS
Inhibition of Granule Exocytosis by TAT-SNAP-23
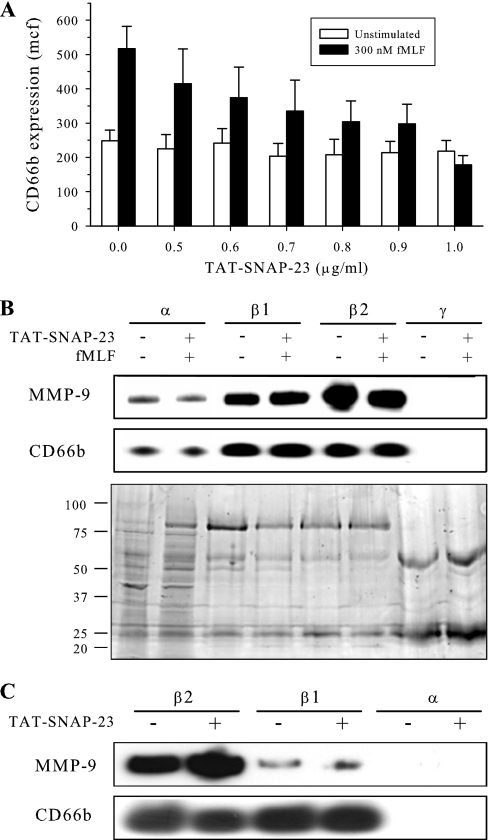
The identification of phosphoproteins on the surface of mobilized granules is complicated by the circumstances of the biology. Upon neutrophil stimulation, granules are mobilized to, and fuse with, the plasma membrane within 5 min (27). The loss of mobilized granules prior to isolation procedures impairs the ability to identify signaling proteins that mediate exocytosis on the granule surface. To capture granules destined for membrane fusion, we generated a fusion protein containing the TAT cell-penetrating peptide sequence and the amino-terminal SNARE domain of SNAP-23 (TAT-SNAP-23). Characterization of that fusion protein will be reported separately (S. M. Uriarte et al., manuscript in preparation). To determine the extent of inhibition of exocytosis, neutrophils were pretreated with TAT-SNAP-23 at various concentrations ranging up to 1.0 μg/ml for 10 min and then stimulated with 300 nm fMLF. Exocytosis was measured by flow cytometric analysis of the plasma membrane expression of a marker for specific granules, CD66b. Fig. 1A demonstrates that TAT-SNAP-23 treatment produced a dose-dependent inhibition of fMLF-stimulated CD66b expression, with maximal inhibition (100 ± 10%) occurring at 1.0 μg/ml. Pretreatment with TAT-SNAP-23 did not reduce cell viability (as measured by trypan blue exclusion) or induce apoptosis (measured as DNA fragmentation using propidium iodide (18)) (data not shown).
Fig. 1.
Enrichment of granules from stimulated neutrophils. A, Human neutrophils (4 × 106/ml) were pretreated with the indicated concentrations of TAT-SNAP-23 for 10 min at 37 °C. Neutrophils were then stimulated with or without 300 nm fMLF for 5 min. Surface expression of CD66b (a marker of specific granules) was measured by flow cytometry. Mean channel fluorescence (mcf) is presented as means ± S.E. for three experiments. The results show a concentration-dependent inhibition of exocytosis that was maximal at 1 μg/ml. B, Neutrophils from a single donor were untreated or pretreated with 1.0 μg/ml TAT-SNAP-23 for 10 min at 37 °C prior to stimulation with 0.3 μm fMLF for 2 min at 37 °C. Granule fractions were isolated by Percoll density centrifugation and then subjected to SDS-PAGE and immunoblot analysis for MMP-9 (gelatinase B) and CD66b. The coomassie-stained replicate gel demonstrates equal protein loading. The immunoblots show that the β1 granule fraction used in the present study contained markers for both specific and gelatinase granules. The distribution of granule markers was similar between fractions unstimulated and fMLF-stimulated cells. C, Neutrophils were treated with or without 1.0 μg/ml TAT-SNAP-23 and granule fractions were enriched and prepared as described above. Immunoblot analysis for MMP-9 and CD66b were performed. TAT-SNAP-23 treatment did not alter the distribution of granule markers in β1 and β2 granule fractions.
Granule enrichment by Percoll density centrifugation resulted in the separation of four granule fractions (α, β1, β2, and γ). To determine whether enrichment of granule subsets was altered by TAT-SNAP-23 treatment, granule fractions isolated from fMLF-stimulated human neutrophils treated with TAT-SNAP-23 and from unstimulated, untreated neutrophils were analyzed by SDS-PAGE followed by Coomassie staining or immunoblot analysis for the specific and gelatinase granule markers CD66b and MMP-9, respectively. Figs. 1B and 1C demonstrate that both β1 and β2 fractions contained markers of specific and gelatinase granules, as has been reported previously (21). Fig. 1B shows that the distribution of granule markers from untreated neutrophils did not differ following fMLF stimulation of neutrophils pretreated with TAT-SNAP-23. Coomassie staining showed that the relative amount and distribution of granule proteins was similar between TAT-SNAP-23 treated and untreated cells. Fig. 1C shows that pretreatment of neutrophils with TAT-SNAP-23 did not alter granule marker distribution upon granule enrichment. Kjeldsen et al. (21) reported that stimulation with fMLF resulted in minimal loss of markers for specific granules, whereas about 25% of gelatinase granule markers were lost. Our data indicate that pretreatment with TAT-SNAP-23 preserved the distribution of markers for gelatinase and specific granules seen in unstimulated cells and prevented loss of granules because of exocytosis. To confirm that increased granule protein phosphorylation could be detected under the described conditions, granules were enriched from unstimulated and fMLF stimulated neutrophils. Twenty micrograms of protein from azurophil granules, specific granules, or neutrophil lysate was subjected to SDS-PAGE and immunoblot analysis performed for phosphotyrosine. Supplemental Fig. 1 demonstrates that tyrosine phosphorylation of granule proteins increases upon fMLF stimulation and that this phosphorylation remains detectable following the granule enrichment procedure.
Identification of Granule Phosphopeptides
Bodenmiller et al. (28) showed that no single method of phosphopeptide enrichment is sufficient to identify an entire phosphoproteome. Thus, the present study used two different proteolytic enzymes to generate peptide fragments of granule proteins and two different metal-based affinity chromatography techniques to enrich for phosphopeptides. Scaffold predicted a false positive rate of 7.7% from the data set. Therefore, manual validation of MS/MS spectra was performed to decrease the assignment error rate. This resulted in the exclusion of 370 protein identifications (this number included loss of identifications because of NCBI database annotations) from the data set. In total, 243 phosphopeptides corresponding to 235 phosphoproteins were identified from the digestion of intact granules. To compare the phosphoprotein identifications from the different enrichment protocols, the presence of proteins identified at greater than 90% probability in any one sample was determined in all other samples regardless of probability (Supplemental Table S1). IMAC enrichment allowed identification of 161 phosphopeptides from 157 proteins, whereas MOAC enrichment identified 82 phosphopeptides from 81 proteins. Twenty-six phosphoproteins (11%) were common to both IMAC- and MOAC-enriched samples. The use of trypsin as a proteolytic enzyme resulted in the identification of 151 phosphopeptides from 150 proteins, whereas 92 phosphopeptides from 89 proteins were identified from chymotrypsin digests. Thirty proteins (13%) were common to both protein digests.
Previous studies suggested that multiply phosphorylated peptides demonstrate a higher affinity for enrichment (29, 30). Thus, we analyzed the distribution of phosphopeptides by the number of phosphorylation sites and the amino acid phosphorylated. 150 singly phosphorylated peptides were identified, whereas 70 were phosphorylated on two sites and 23 were phosphorylated on more than two sites. Of the 414 phosphorylated residues, 228 (62%) were serine, 108 (30%) were threonine, and 30 (8%) were tyrosine.
Changes in the Phosphoprotein Profile upon Stimulation
We showed previously that exocytosis of gelatinase and specific granules was detected as early as 1 min following fMLF stimulation, and was maximal by 5 min (27). To identify phosphoproteins with a high probability of participation in stimulated exocytosis, the current study examined granules obtained following 1 and 2 min of fMLF stimulation, as well as from unstimulated neutrophils. Of the 235 phosphoproteins identified, 79 phosphoproteins were from granules obtained from four separate preparations of unstimulated neutrophils, 81 phosphoproteins were from granules obtained from four separate preparations of cells stimulated with fMLF for 1 min, and 118 phosphoproteins were from five separate preparations of cells stimulated with fMLF for 2 min. Thirty-three phosphoproteins (14%) were identified under different enrichment conditions at multiple time points of stimulation, and 22 (9%) were identified under different enrichment conditions at the same time point. Seven phosphoproteins were present at all three time points; 15 phosphoproteins were present on granules from unstimulated cells and cells stimulated for 1 or 2 min; and 11 phosphoproteins, including protein kinase Cα, integrin αM, and mTOR, were present on granules from cells stimulated for both 1 and 2 min. Supplementary Table S1 lists the identified proteins and their identifying peptide(s) as a function of stimulation time and method of enrichment.
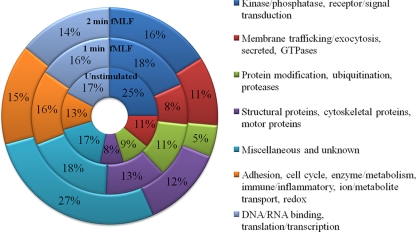
Phosphoproteins Grouped by Cellular Function
The identified phosphoproteins were grouped by cellular function based on Gene Ontology terms derived from the UniProt Knowledge Base (www.uniprot.org). A list of the individual proteins classified by molecular function and the time point of identification and cellular location is presented in Supplemental Tables S1 and S2. Fig. 2 summarizes these functional categorizations by stimulation time. The distribution of phosphoproteins by cellular function was not substantially different among the three time points. Several potential regulators of membrane trafficking and exocytosis were identified in the analysis, including N-ethylmaleimide sensitive factor (NSF); small molecular weight GTPases, GTPase regulators, or GTPase effectors; and proteins known to regulate cytoskeleton rearrangement and stability.
Fig. 2.
Functional classification of phosphoproteins grouped by time point of fMLF stimulation. Cellular protein functions were categorized based on Gene Ontology terms and grouped into broad categories as a function of their temporal identification. The number of identifications per category was compared with the total number of identifications, indicated as a percent (%) of the total. No category demonstrated greater than a 10% change among the three time points of stimulation.
Bioinformatic Prediction of Src and p38 MAPK Phosphorylation Motifs
Although p38 MAPK and Src kinase pathways have been implicated in the regulation of neutrophil exocytosis (2, 16, 18, 31, 32), the substrates of these kinases that mediate exocytosis are unknown. To identify candidate substrates, bioinformatics was used to predict potential p38 MAPK and Src phosphorylation motifs for the phosphopeptides identified in our proteomic analysis. Of the 243 phosphopeptide sequences submitted to Group Prediction System (GPS) 2.1, 16 were predicted to contain a Src family tyrosine kinase motif, 15 contained a p38 MAPK motif, and one protein, barttin, contained both a Src and p38 MAPK motif in the same peptide. The results from this analysis are shown in Table I.
Table I. Bioinformatic Src and p38 MAPK phosphorylation motif predictions. Identified phosphopeptides were submitted to GPS 2.1 to predict potential p38 MAPK and Src Tyr kinase motifs. Proteins were grouped by molecular function and the NCBI accession number, the phosphopeptide identified in the proteomic analysis, and the phosphorylated residue(s) listed. A raised numeral indicates the amino acid-specific phosphorylation of serine, threonine, or tyrosine. Phosphorylated amino acid(s) predicted to comprise a Src family phosphorylation motif are denoted by Y. Those comprising a potential p38 MAPK phosphorylation motif are denoted by S and T.
| Identified Protein | Accession Number | Peptide sequence | Phosphorylated Residue |
|---|---|---|---|
| Adhesion | |||
| Plakophilin 2 isoform 2b | 4758932 | LEIS151PDS154SPER | S151, S154 |
| Cell cycle | |||
| Retinoblastoma-like protein 1 isoform b | 34577077 | EKEAVIT385PVASATQSVS395R | T385 |
| Cytoskeletal regulation | |||
| Drebrin 1 isoform b | 18426913 | S339PS341DSST345AS347T348PVAEQIER | S339, T348 |
| GTPases and GTPase-binding proteins | |||
| ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 | 19923540 | S862PEDGPSAR | S862 |
| Exophilin 5 (Slac2-b) | 21359818 | TSCDSLDLSSAALPDS887SPS890K | S887 |
| Ras protein-specific guanine nucleotide-releasing factor 1 isoform | 24797099 | S232PHADSMR | S232 |
| Immune/inflammatory response | |||
| Lactotransferrin; transferrin | 4505043 | PVAAEVY102GT104ER | Y102 |
| Ion/Metabolite Transport | |||
| Barttin | 17017967 | S91PSPQPPY98VR | S91, Y98 |
| Kinase/Phosphatase | |||
| G protein coupled receptor kinase 5 | 55958649 | GESMALNEKQILEKVNSQFVVNLAY251A-Y253ETK | Y251 |
| Serine/Threonine kinase SMG-1 isoform 2 | 14719394 | SSCT2305T2306PDEWW | T2306 |
| PREDICTED: similar to protein phosphatase 2A 48 kDa regulatory | 51472904 | KT75GPRACSTPLSTSS88PS90PR | S88, S90 |
| Miscellaneous | |||
| Arsenate resistance protein 2 | 33383231 | DEHSS89DPY92 | Y92 |
| T-complex protein 1 subunit theta | 42662316 | IGLS124VSEVIEGY132EIACRK | Y132 |
| Receptor/signal transduction | |||
| Aryl hydrocarbon receptor | 4502003 | SS90PTERNGGQDNC | S90 |
| Chemokine (C-X-C motif) receptor 4 isoform b | 4503175 | T13EEMGSGDY21 | Y21 |
| G-protein coupled receptor 88 | 11545753 | DGAGGSY112 | Y112 |
| Olfactory receptor, family 6, subfamily C, member 76 | 41201775 | PLHY129TTIMS134DR | Y129 |
| Redox | |||
| Cytochrome b5 reductase b5R.2 isoform 1 | 47778923 | AYT70PVSSDDDR | T70 |
| Secreted | |||
| Angiogenin, RNase A family, 5 precursor | 4557313 | GLTS61PCK | S61 |
| Structural proteins | |||
| Fibronectin 1 isoform 3 preproprotein | 16933542 | GDS1527PASS1531K | S1527 |
| Usherin isoform B | 46249406 | S442NFTPY447SRGNVT453F | Y447 |
| Translation/transcription | |||
| Protein CASP | 31652240 | Y493SKAMQEAGSTSMIFSTGPY | Y493 |
| Ubiquitination | |||
| E3 SUMO-protein ligase RanBP2 | 638207 | VT2027GEEDEKVLY2036 | Y2036 |
| Unknown Function | |||
| AlkB, alkylation repair homolog 5 | 40353210 | Y60QEDSDPER | Y60 |
| Hypothetical protein LOC220108 | 31542760 | SSS459DLS462VVS465AY467 | Y467 |
| Hypothetical protein LOC348487 | 32699058 | CFQTTNGY67LSDS71R | Y67 |
| Nucleoporin 214kDa | 33946327 | HGAPS1081PS1083HPISAPQAAAAAALR | S1081 |
| PREDICTED: NHS-like 1 isoform 1 | 51465299 | SRTPPSAASSRY1511SMR | Y1511 |
| Testis expressed sequence 15 | 13775174 | LINVCGDS1842PK | S1842 |
| Tetratricopeptide repeat domain 37 | 7662078 | IKLSDY366DSS369EEAIRT375L | Y366 |
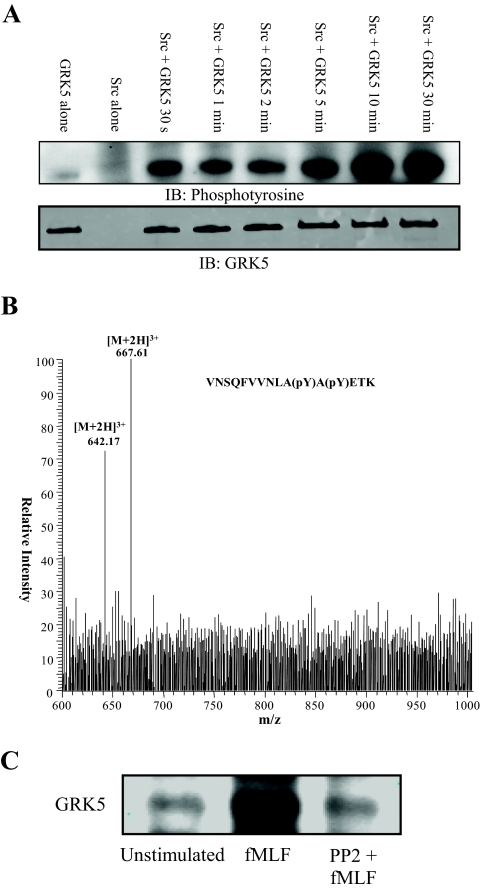
One Src tyrosine kinase motif was predicted for Tyr251 of G protein-coupled receptor kinase 5 (GRK5). This residue comprised one of the two phosphorylation sites from the peptide (GESMALNEKQILEKVNSQFVVNLApYApYETK) identified in the proteomic analysis following 2 min of fMLF stimulation. To confirm that GRK5 is phosphorylated by Src, active recombinant Src was incubated with recombinant GRK5 in the presence of excess ATP for various times up to 30 min. Fig. 3A shows the immunoblot for phosphotyrosine. GRK5 was phosphorylated within 30 s, and maximal phosphorylation occurred by 10 min.
Fig. 3.
Src phosphorylates GRK5. A, Active Src (30 ng) was incubated with or without GRK5 (200 ng) at 30 °C for various times. Proteins were subjected to SDS-PAGE, transferred to PVDF, and analyzed for tyrosine phosphorylation by immunoblot analysis. The blot demonstrates that rapid phosphorylation of GRK5 by Src occurred within 30 s and maximal phosphorylation occurred by 10 min. Parallel blots were probed with anti-GRK5 to confirm similar amounts of GRK5 in each reaction. B, To confirm that Src phosphorylates Tyr 251 and Tyr 253 of GRK5, an in-solution trypsin/Lys-C digest of an in vitro kinase reaction was subjected to mass spectrometry. Precursor ion scanning for product ions yielding the phosphotyrosine-specific immonium ion (216.04 m/z) identified precursor ion masses of 642.17 m/z and 667.61 m/z. These masses correspond to the [M+2H]3+ singly- and doubly phosphorylated GRK5 peptide VNSQFVVNLApYApYETK, where pY represents phosphorylation of tyrosine. C, Neutrophils were pretreated with or without 10 μm PP2 at 37 °C for 10 min. Cells were then treated with or without 0.3 μm fMLF for 2 min after which reactions were stopped by resuspension in ice-cold lysis buffer. Proteins were immunoprecipitated from lysates with anti-phosphotyrosine conjugated to agarose. Proteins were separated by SDS-PAGE and subjected to immunoblot analysis for GRK5. An increased amount of GRK5 was precipitated from cells stimulated with fMLF, whereas pretreatment with the Src inhibitor PP2 returned the amount of precipitated GRK5 to basal levels.
Steen et al. demonstrated the validity of precursor ion scanning in positive ion mode to identify tyrosine phosphorylated peptides (33). To confirm that Src phosphorylated Tyr251and Tyr253 of GRK5, an in-solution trypsin/Lys-C digest of an in vitro kinase reaction was subjected to mass spectrometry. A precursor ion scan for product ions yielding the phosphotyrosine-specific immonium ion (216.04 m/z) was performed. Fig. 3B shows the collective identified precursor ion masses from retention times of 4.08 to 4.78 min. The identified [M+2H]+3 ions of 642.17 m/z and 667.61 m/z correspond, respectively, to the singly and doubly phosphorylated peptide VNSQFVVNLAYAYETK containing Tyr251 and Tyr253. This indicates Src kinase phosphorylated both of these residues on GRK5.
To determine if Src phosphorylates GRK5 in intact cells, cells were pretreated with or without the pharmacological Src kinase family inhibitor PP2 and then treated with or without fMLF for 2 min. As no immunoprecipitating antibody for GRK5 was available, tyrosine phosphorylated proteins were immunoprecipitated from cellular lysates, separated by SDS-PAGE, and transferred proteins were subjected to immunoblot analysis for GRK5. Fig. 3C demonstrates that increased GRK5 was precipitated following fMLF stimulation, and this increased precipitation was inhibited by PP2. These data support the conclusion that GRK5 is phosphorylated by Src in fMLF-stimulated neutrophils.
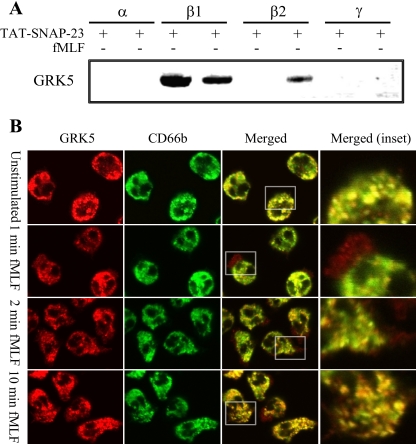
To determine if GRK5 colocalizes with gelatinase and specific granules, immunoblot analysis and confocal microscopy were performed. Granule fractions from unstimulated and fMLF-stimulated neutrophils were subjected to immunoblot analysis. GRK5 was identified in the β2 granule fraction from stimulated neutrophils and in the β1 granule fraction from both unstimulated and stimulated neutrophils, but not in α or γ granule fractions (Fig. 4A). Confocal microscopy of unstimulated and fMLF-stimulated neutrophils showed that GRK5 colocalized with CD66b-containing granules under both conditions (Fig. 4B). In the unstimulated state, a population of CD66b colocalized with GRK5, whereas some CD66b immunofluorescence remained separate. Following fMLF stimulation for 1 and 2 min colocalization of GRK5 with CD66b was observed in the cytosol and GRK5 alone was also present at the plasma membrane and in membrane ruffles. Following 10 min of fMLF, punctate cytosolic staining for GRK5 was again observed to colocalize with CD66b. These data are consistent with the proteomic data and bioinformatic prediction indicating that GRK5 is a novel substrate of Src tyrosine kinase and that GRK5 localizes to neutrophil granules.
Fig. 4.
GRK5 expression on neutrophil granules. A, Granule fractions were enriched from neutrophils treated with or without 1.0 μg/ml TAT-SNAP23 and 300 nm fMLF for 2 mins. Five micrograms of protein from each granule fraction were separated by SDS-PAGE and subjected to immunoblot analysis for GRK5 expression. The results show that GRK5 is expressed on the β2 granule fraction from stimulated neutrophils and on the β1 fraction from both unstimulated and stimulated neutrophils. B, Confocal microscopy demonstrates the cellular localization of GRK5 (red), CD66b (green), and their colocalization (yellow and inset) in neutrophils stimulated with fMLF for 0 min, 1 min, 2 min, or 10 min. In the unstimulated state, a population of CD66b colocalized with GRK5. fMLF stimulation for 1 min and 2 min revealed colocalization of GRK5 with CD66b whereas sharp GRK5 staining was also seen at the plasma membrane and in membrane ruffles. Punctate, cytosolic staining for GRK5 and CD66b was observed following 10 min of fMLF stimulation (inset).
DISCUSSION
Use of pharmacologic inhibitors and knockout mice has identified several signal transduction pathways that are involved in neutrophil granule exocytosis. In particular, pathways containing p38 MAPK and the nonreceptor Src tyrosine kinases regulate neutrophil specific and azurophilic granule exocytosis, and those kinases are activated by several stimuli that induce exocytosis (14–18). However, the targets of those kinases that mediate exocytosis are unknown. The current study identified 243 phosphopeptides corresponding to 235 phosphoproteins from the cytoplasmic face of a subset of neutrophil granules. Bioinformatic analysis identified potential p38 MAPK and/or Src family tyrosine kinase motifs for 30 phosphopeptides identified in the analysis. Phosphorylation of one of these proteins, GRK5, identified as a Src kinase target from our analysis, was confirmed by an in vitro kinase assay and mass spectrometry. Identification of granule proteins phosphorylated upon fMLF stimulation provides several candidates which may participate in regulation of neutrophil granule exocytosis. To our knowledge, this is the first proteomic analysis to identify phosphoproteins associated with human neutrophil granules following cellular stimulation.
The use of cell-penetrating TAT-fusion proteins has provided a viable approach to deliver proteins and peptides into neutrophils (34, 35), as genetic manipulation of neutrophils results in a very low transfection efficiency (36, 37). Cell-penetrating fusion proteins containing aptamers from NSF have previously been used to inhibit release of intracellular granules from vascular endothelial cells (38, 39). To prevent activated granules from fusing with the plasma membrane and allow for their enrichment, the present study used a fusion protein which combined the amino-terminal SNARE domain of SNAP-23 and the cell-penetrating TAT peptide sequence. That fusion protein inhibited exocytosis of secretory vesicles, gelatinase granules, and specific granules (S.M. Uriarte et al., manuscript in preparation). The inhibition of granule fusion by TAT-SNAP-23 did not alter the distribution of granule subsets following density gradient centrifugation. Based on the observed amount of material retrieved from the gradients, TAT-SNAP-23 enhanced recovery of granules following fMLF stimulation. The enriched granule fractions used for our studies expressed markers for both gelatinase and specific granules. This heterogeneity was reported previously to be associated with the enrichment procedure used in this study (21), and TAT-SNAP-23 did not alter the relative distribution of the markers for those granule subsets. Thus, inhibition of exocytosis with TAT-SNAP-23 enhanced the probability that targets of kinases that regulate exocytosis of specific and gelatinase granules were identified.
Bodenmiller et al. showed that optimal phosphopeptide coverage during large-scale proteomic screens required multiple phosphopeptide enrichment techniques (28). Furthermore, identification of transmembrane and membrane-associated proteins remains difficult because of the lack of accessible protease cleavage sites. For those reasons, the current study used two complementary phosphopeptide enrichment procedures and two different proteolytic enzymes to improve protein coverage. The overlap in phosphopeptide identification among these enrichment techniques was low, 13% between trypsin and chymotrypsin and 11% between IMAC and TiO2. Bodenmiller et al. reported a 35% overlap in phosphopeptide identification between IMAC and TiO2 from cytosolic proteins of cultured cells (28). Our data support their conclusion that multiple approaches are required to achieve optimal phosphoprotein recovery in proteomic studies. Despite the use of those multiple approaches, it is unlikely that the complete granular phosphoproteome was identified. Additionally, the transitory nature of exocytosis and the dynamic interaction of kinases and phosphatases allow only a “snapshot” of the phosphoproteins present.
Although the total number of identifications increased with fMLF stimulation, the relative distribution of functional classes was not altered. Thus, broad functional classification did not assist in identifying candidate proteins with a high likelihood of participation in exocytosis. On the other hand, listing proteins by functional class identified several proteins known to regulate membrane trafficking events, including kinases and phosphatases, GTPases, actin- and microtubule-binding proteins, and SNARE-associated proteins. Additionally, the identified proteins were also categorized according to their cellular compartment, as specified by Gene Ontology (Supplemental Table S2). The majority of proteins were either cytoplasmic or membrane-associated. Many of the cytoplasmic proteins, such as motor proteins and GTPases, have been shown to associate with intracellular membranes and transport vesicles. The prevalence of nuclear-, endoplasmic reticulum-, and golgi-associated proteins in the data set also suggests that other intracellular organelles contaminated the granule enrichment procedures. Nonetheless, phosphorylation of these proteins may play a role in other neutrophil functional responses.
The vesicle-fusing ATPase NSF was found to be phosphorylated on Ser343 and Ser356 following 2 min of stimulation. Phosphorylation of NSF on those serine residues has not been reported previously, nor has NSF expression been reported previously in neutrophils. NSF has been implicated in both pre-fusion exocytosis (40) and postfusion recycling of SNAREs (41). Phosphorylation of Ser237 by protein kinase C has been reported to impair NSF binding to SNAP-SNARE complexes (42). Phosphorylation of Tyr83 increased NSF ATPase activity and prevented binding to α-SNAP (43). Seventeen small molecular weight GTPases, GTPase regulators, or GTPase effectors were identified: seven in the basal state, three at 1 min of fMLF stimulation, and eight at 2 min of stimulation (Supplemental Table S1). Phosphorylation of GTPase effectors was reported to be a regulatory step in vesicular motor protein activation (44), GTPase activation (45–47), and GEF cellular localization (48). Rap2a (identified from unstimulated neutrophils, phosphorylated on Ser127) was reported to be associated with specific (49) and gelatinase granules (50) in neutrophils and translocated to the plasma membrane during degranulation via a Src tyrosine kinase-dependent mechanism. Three ADP-ribosylation factors (ARF) were identified including ARFGAP (unstimulated, pSer862), ARFGEF6 (1 min, pThr452), and ARF-like 10c (2 min, pThr61). ARFGAP Git2 was reported to regulate superoxide production in neutrophils via modulation of granule trafficking (51). Rho GTPase nucleotide exchange factors Rho GEF (2 min, pSer999, pSer1000, pSer1008) and Rho GEF3 (2 min, pSer94, pThr96) were identified. Phosphorylation of Rho GEF family members by ERK1/2 and protein kinase A modulated nucleotide exchange activity toward their constituent GTPases (52, 53). Rab27 has been reported to regulated exocytosis of gelatinase, specific, and azurophil neutrophil granules (54, 55). Exophilin5/Slac2b is a Rab27 effector that was found to be phosphorylated on two serine residues at 2 min of fMLF stimulation. Piccolo is one component of the presynaptic active zone that controls the release of synaptic vesicles (56). Our study identified a peptide from piccolo that was phosphorylated on a threonine residue at 2 min of stimulation.
We recently reported that the actin cytoskeleton regulated the rate and extent of neutrophil granule exocytosis (27). The current study identified eight proteins known to regulate cytoskeleton rearrangement and stability. The CD2-associated protein (unstimulated, pThr231) was reported to regulate dynamic actin remodeling, membrane trafficking during endocytosis, and cytokinesis (57). Phosphorylation of CD2-associated protein was shown to control its cellular localization (57). The adaptor-related protein 3 (Arp3) (pSer819, pSer822, 2 min fMLF), in complex with Arp2, regulates fMLF-stimulated actin nucleation in neutrophils (58, 59). The amyloid β-binding protein was phosphorylated on Thr398 at 2 min of stimulation. This protein was shown to bind to syntaxins in a multimeric complex intermediate that regulates synaptic vesicle docking and fusion (60). Hook homolog 1 was phosphorylated on Thr726 at 2 min. Hook proteins comprise the yeast homotypic vesicular sorting complex (HOPS complex) implicated in vesicular trafficking and fusion (61).
It is also notable that only 14% of phosphoproteins were present at more than a single time point. There are several possible explanations for this lack of overlap. Phosphatases activated during neutrophil stimulation mediate dephosphorylation of proteins associated with neutrophil granule exocytosis (62, 63). Supplemental Fig. 1, however, demonstrates that robust granule protein phosphorylation remains detectable even following neutrophil stimulation and granule isolation procedures. Although phosphopeptide enrichment was performed, the transient nature of phosphorylation events limits the stoichiometry of phosphorylation. In addition, ionization efficiency of phosphorylated peptides during electrospray ionization is significantly altered, compared with unmodified peptides, which can hinder identifications as a result of ion suppression (64). Furthermore, as most proteins were identified from one peptide, loss of sample because of enrichment procedures and the sensitivity of the instrumentation may have contributed to the lack of overlapping identifications over multiple time points.
Formyl peptide receptor ligation results in a G protein dependent activation of signal transduction pathways containing p38 MAPK, Src tyrosine kinase, ERK1/2, Akt, protein kinase C, and PI-3K (16, 18, 65, 66). Both p38 MAPK and Src tyrosine kinases were shown to modulate neutrophil exocytosis (2, 15, 16). Bioinformatic analysis predicted potential p38 MAPK and Src tyrosine kinase motifs from the data set. Identification of these potential substrates may help to elucidate some of the downstream effector pathways of formyl peptide receptor signaling mediated by these kinases. Although fMLF is a well-characterized neutrophil chemoattractant, other stimuli, including lipopolysaccharide, tumor necrosis factor-α, phorbol 12-myristate 13-acetate, and interleukin-8, activate these kinases and stimulate exocytosis (67–70). Comparison of the granule phosphoproteome induced by those stimuli with the results of the current study may assist in identifying targets that regulate exocytosis.
Our previous proteomic analyses identified peptides from GRK6 on gelatinase granules and GRK 1 and 2 on secretory vesicles (11, 12). We also identified Src kinase family members on gelatinase and specific granules (11). The application of bioinformatics predicted a Src tyrosine kinase family phosphorylation motif for the residues surrounding Tyr251 of the doubly phosphorylated peptide identified from GRK5. These residues reside within a conserved kinase catalytic domain shared by all GRK proteins. Phosphorylation of GRK2 by c-Src results in increased catalytic activity (71, 72). Src-mediated phosphorylation of GRK5, however, has not been previously demonstrated. The present study confirmed Src-mediated phosphorylation of GRK5 by an in vitro kinase reaction and by phosphotyrosine-specific immonium ion monitoring of the reaction by mass spectrometry. Additionally, immunoprecipitation of tyrosine phosphorylated proteins from fMLF-stimulated neutrophils in the presence and absence of PP2, followed by immunoblot analysis for GRK5, suggests that Src mediates tyrosine phosphorylation of GRK5 in intact neutrophils. The in vitro assays are consistent with the ex vivo finding that a phosphorylated peptide from GRK5 was associated with a neutrophil granule fraction and that GRK5 is a target of Src kinases.
Colocalization of GRK5 with the specific granule marker CD66b by confocal microscopy and validation of GRK5 expression on enriched granule fractions by immunoblot analysis suggests a potential role for GRK5 separate from β-arrestin-mediated endocytosis. Specifically, GRK5 may play an active role in the trafficking of these nonrecycled granules to target membranes. Tyr251 resides in a potential Src homology 2 domain-containing-transforming protein C1 (SHC) phosphotyrosine binding domain consisting of NxxpY that is present only in GRK5 and not other GRKs. SHC is an adaptor protein that mediates the assembly of large signaling complexes during tyrosine kinase signaling (73). The functional role of GRK5 phosphorylation was not established in the current study. However, the ability of fMLF stimulation to induce phosphorylation of GRK5 within a potential SHC binding domain and the association of GRK5 with neutrophil granules indicate that the role of GRK5 in neutrophil exocytosis deserves to be examined.
Acknowledgments
We would like to acknowledge the excellent technical assistance from Angela Kain, Daniel Wilkey, and Terri Manning.
Footnotes
* This work was supported by grants provided by the Department of Veterans Affairs Merit Review Board (KRM), National Institutes of Health Grant HL087924 (SMU), and National Institutes of Health Grant DK176743 (DWP). This study was also supported by funding from an NIEHS award P30ES014443 (DWP and MLM) from the University of Louisville Center for Environmental Genomics and Integrative Biology.
 This article contains supplemental Fig. 1 and Tables 1–2.
This article contains supplemental Fig. 1 and Tables 1–2.
1 The abbreviations used are:
- MAPK
- mitogen-activated protein kinase
- PVDF
- polyvinylidene fluoride
- SNAP-23
- synaptosomal-associated protein-23
- SNARE
- soluble NSF attachment protein receptors
- fMLF
- N-formyl-methionyl-leucyl phenylalanine
- GRK
- G protein-coupled receptor kinase
- SHC
- Src homology 2 domain-containing-transforming protein C1
- TAT
- Trans-activating transcriptional activator
- TAT-SNAP-23
- The fusion protein comprised of the TAT cell-penetrating peptide and the N-terminal 78 residues of SNAP-23
- NSF
- N-ethylmaleimide sensitive factor
- ARF
- ADP ribosylation factor.
REFERENCES
- 1. Nauseef W. M. (2007) How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 219, 88–102 [DOI] [PubMed] [Google Scholar]
- 2. Ward R. A., Nakamura M., McLeish K. R. (2000) Priming of the neutrophil respiratory burst involves p38 mitogen-activated protein kinase-dependent exocytosis of flavocytochrome b558-containing granules. J. Biol. Chem. 275, 36713–36719 [DOI] [PubMed] [Google Scholar]
- 3. Borregaard N., Sørensen O. E., Theilgaard-Mönch K. (2007) Neutrophil granules: a library of innate immunity proteins. Trends Immunol 28, 340–345 [DOI] [PubMed] [Google Scholar]
- 4. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 5. Pham C. T. (2006) Neutrophil serine proteases: specific regulators of inflammation. Nat. Rev. Immunol. 6, 541–550 [DOI] [PubMed] [Google Scholar]
- 6. Soehnlein O., Zernecke A., Eriksson E. E., Rothfuchs A. G., Pham C. T., Herwald H., Bidzhekov K., Rottenberg M. E., Weber C., Lindbom L. (2008) Neutrophil secretion products pave the way for inflammatory monocytes. Blood 112, 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayadas T. N., Tsokos G. C., Tsuboi N. (2009) Mechanisms of immune complex-mediated neutrophil recruitment and tissue injury. Circulation 120, 2012–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loria V., Dato I., Graziani F., Biasucci L. M. (2008) Myeloperoxidase: A New Biomarker of Inflammation in Ischemic Heart Disease and Acute Coronary Syndromes. Mediators Inflamm. 135625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen M., Kallenberg C. G. (2009) New advances in the pathogenesis of ANCA-associated vasculitides. Clin. Exp. Rheumatol. 27 (Suppl. 52), S108–114 [PubMed] [Google Scholar]
- 10. Udby L., Borregaard N. (2007) Subcellular fractionation of human neutrophils and analysis of subcellular markers. Methods Mol. Biol. 412, 35–56 [DOI] [PubMed] [Google Scholar]
- 11. Lominadze G., Powell D. W., Luerman G. C., Link A. J., Ward R. A., McLeish K. R. (2005) Proteomic Analysis of Human Neutrophil Granules. Mol. Cell. Proteomics 4, 1503–1521 [DOI] [PubMed] [Google Scholar]
- 12. Uriarte S. M., Powell D. W., Luerman G. C., Merchant M. L., Cummins T. D., Jog N. R., Ward R. A., McLeish K. R. (2008) Comparison of proteins expressed on secretory vesicle membranes and plasma membranes of human neutrophils. J. Immunol. 180, 5575–5581 [DOI] [PubMed] [Google Scholar]
- 13. Jethwaney D., Islam M. R., Leidal K. G., de Bernabe D. B., Cambell K. P., Nauseef W. M., Gibson B. W. (2007) Proteomic analysis of plasma membrane and secretory vesicles from human neutrophils. Proteome Sci. 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rittner H. L., Labuz D., Richter J. F., Brack A., Schäfer M., Stein C., Mousa S. A. (2007) CXCR1/2 ligands induce p38 MAPK-dependent translocation and release of opioid peptides from primary granules in vitro and in vivo. Brain Behav. Immun. 21, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 15. Fumagalli L., Zhang H., Baruzzi A., Lowell C. A., Berton G. (2007) The Src family kinases Hck and Fgr regulate neutrophil responses to N-formyl-methionyl-leucyl-phenylalanine. J. Immunol. 178, 3874–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mócsai A., Jakus Z., Vántus T., Berton G., Lowell C. A., Ligeti E. (2000) Kinase pathways in chemoattractant-induced degranulation of neutrophils: the role of p38 mitogen-activated protein kinase activated by Src family kinases. J. Immunol. 164, 4321–4331 [DOI] [PubMed] [Google Scholar]
- 17. Hsu M. J., Lee S. S., Lee S. T., Lin W. W. (2003) Signaling mechanisms of enhanced neutrophil phagocytosis and chemotaxis by the polysaccharide purified from Ganoderma lucidum. Br. J. Pharmacol. 139, 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coxon P. Y., Rane M. J., Uriarte S., Powell D. W., Singh S., Butt W., Chen Q., McLeish K. R. (2003) MAPK-activated protein kinase-2 participates in p38 MAPK-dependent and ERK-dependent functions in human neutrophils. Cell Signal 15, 993–1001 [DOI] [PubMed] [Google Scholar]
- 19. Haslett C., Guthrie L. A., Kopaniak M. M., Johnston R. B., Jr., Henson P. M. (1985) Modulation of multiple neutrophil functions by preparative methods or trace concentrations of bacterial lipopolysaccharide. Am. J. Pathol. 119, 101–110 [PMC free article] [PubMed] [Google Scholar]
- 20. Lominadze G., Rane M. J., Merchant M., Cai J., Ward R. A., McLeish K. R. (2005) Myeloid-related Protein-14 is a p38 MAPK Substrate in Human Neutrophils. J. Immunol. 174, 7257–7267 [DOI] [PubMed] [Google Scholar]
- 21. Kjeldsen L., Sengeløv H., Borregaard N. (1999) Subcellular Fractionation of human neutrophils on Percoll density gradients. J. Immunol. Methods. 232, 131–143 [DOI] [PubMed] [Google Scholar]
- 22. Cummins T. D., Barati M. T., Coventry S. C., Salyer S. A., Klein J. B., Powell D. W. (2010) Quantitative mass spectrometry of diabetic kidney tubules identifies GRAP as a novel regulator of TGF-beta signaling. Biochim. Biophys. Acta 1804, 653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 24. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 25. Nichols A. M., White F. M. (2009) Manual validation of peptide sequence and sites of tyrosine phosphorylation from MS/MS spectra. Methods Mol. Biol. 492, 143–160 [DOI] [PubMed] [Google Scholar]
- 26. Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. (2008) GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteom. 7, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jog N. R., Rane M. J., Lominadze G. L., Luerman G. C., Ward R. A., McLeish K. R. (2007) The actin cytoskeleton regulates exocytosis of all neutrophil granule subsets. Am. J. Physiol. Cell Physiol. 292, 1690–1700 [DOI] [PubMed] [Google Scholar]
- 28. Bodenmiller B., Mueller L. N., Mueller M., Domon B., Aebersold R. (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat. Methods 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 29. Aryal U. K., Ross A. R. S. (2010) Enrichment and analysis of phosphopeptides under different experimental conditions using titanium dioxide affinity chromatography and mass spectrometry. Rapid Comm. Mass Spectrometry 24, 219–231 [DOI] [PubMed] [Google Scholar]
- 30. Kweon H. K., Håkansson K. (2006) Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal. Chem. 78, 1743–1749 [DOI] [PubMed] [Google Scholar]
- 31. McLeish K. R., Knall C., Ward R. A., Gerwins P., Coxon P. Y., Klein J. B., Johnson G. L. (1998) Activation of mitogen-activated protein kinase cascades during priming of human neutrophils by TNF-alpha and GM-CSF. J. Leukoc. Biol. 64, 537–545 [PubMed] [Google Scholar]
- 32. Mócsai A., Bánfi B., Kapus A., Farkas G., Geiszt M., Buday L., Faragó A., Ligeti E. (1997) Differential effects of tyrosine kinase inhibitors and an inhibitor of the mitogen-activated protein kinase cascade on degranulation and superoxide production of human neutrophil granulocytes. Biochem. Pharm. 54, 781–789 [DOI] [PubMed] [Google Scholar]
- 33. Steen H., Küster B., Fernandez M., Pandey A., Mann M. (2001) Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Anal. Chem. 73, 1440–1448 [DOI] [PubMed] [Google Scholar]
- 34. Zhao T., Bokoch G. M. (2007) Transduction of proteins into intact neutrophils. Methods Mol Biol. 412, 115–123 [DOI] [PubMed] [Google Scholar]
- 35. Becker-Hapak M., McAllister S. S., Dowdy S. F. (2001) TAT-mediated protein transduction into mammalian cells. Methods 24, 247–256 [DOI] [PubMed] [Google Scholar]
- 36. Gardiner E. M., Pestonjamasp K. N., Bohl B. P., Chamberlain C., Hahn K. M., Bokoch G. M. (2002) Spatial and temporal analysis of Rac activation during live neutrophil chemotaxis. Curr. Biol. 12, 2029–2034 [DOI] [PubMed] [Google Scholar]
- 37. Johnson J. L., Ellis B. A., Munafo D. B., Brzezinska A. A., Catz S. D. (2006) Gene transfer and expression in human neutrophils. The phox homology domain of p47phox translocates to the plasma membrane but not to the membrane of mature phagosomes. BMC Immunol. 7, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eiden L. E. (2005) Fusion polypeptides that inhibit exocytosis: fusing aptamer and cell-penetrating peptide technologies and pharmacologies. Mol Pharmacol. 67, 980–982 [DOI] [PubMed] [Google Scholar]
- 39. Morrell C. N., Matsushita K., Lowenstein C. J. (2005) A novel inhibitor of N-ethylmaleimide-sensitive factor decreases leukocyte trafficking and peritonitis. J. Pharmacol. Exp. Ther. 314, 155–161 [DOI] [PubMed] [Google Scholar]
- 40. Kuner T., Li Y., Gee K. R., Bonewald L. F., Augustine G. J. (2008) Photolysis of a caged peptide reveals rapid action of N-ethylmaleimide sensitive factor before neurotransmitter release. Proc. Natl. Acad. Sci. U.S.A. 105, 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tomes C. N., De Blas G. A., Michaut M. A., Farré E. V., Cherhitin O., Visconti P. E., Mayorga L. S. (2005) alpha-SNAP and NSF are required in a priming step during the human sperm acrosome reaction. Mol. Hum. Reprod. 11, 43–51 [DOI] [PubMed] [Google Scholar]
- 42. Matveeva E. A., Whiteheart S. W., Vanaman T. C., Slevin J. T. (2001) Phosphorylation of the N-ethylmaleimide-sensitive factor is associated with depolarization-dependent neurotransmitter release from synaptosomes. J. Biol. Chem. 276, 12174–12181 [DOI] [PubMed] [Google Scholar]
- 43. Huynh H., Bottini N., Williams S., Cherepanov V., Musumeci L., Saito K., Bruckner S., Vachon E., Wang X., Kruger J., Chow C. W., Pellecchia M., Monosov E., Greer P. A., Trimble W., Downey G. P., Mustelin T. (2004) Control of vesicle fusion by a tyrosine phosphatase. Nat. Cell Biol. 6, 831–839 [DOI] [PubMed] [Google Scholar]
- 44. Kakiashvili E., Speight P., Waheed F., Seth R., Lodyga M., Tanimura S., Kohno M., Rotstein O. D., Kapus A., Szászi K. (2009) GEF-H1 mediates tumor necrosis factor-alpha-induced Rho activation and myosin phosphorylation: role in the regulation of tubular paracellular permeability. J. Biol. Chem. 284, 11454–11466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujishiro S. H., Tanimura S., Mure S., Kashimoto Y., Watanabe K., Kohno M. (2008) ERK1/2 phosphorylate GEF-H1 to enhance its guanine nucleotide exchange activity toward RhoA. Biochem. Biophys. Res. Commun. 368, 162–167 [DOI] [PubMed] [Google Scholar]
- 46. Itoh R. E., Kiyokawa E., Aoki K., Nishioka T., Akiyama T., Matsuda M. (2008) Phosphorylation and activation of the Rac1 and Cdc42 GEF Asef in A431 cells stimulated by EGF. J. Cell Sci. 121, 2635–2642 [DOI] [PubMed] [Google Scholar]
- 47. Birkenfeld J., Nalbant P., Bohl B. P., Pertz O., Hahn K. M., Bokoch G. M. (2007) GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev. Cell 12, 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoshikawa M., Kajiho H., Sakurai K., Minoda T., Nakagawa S., Kontani K., Katada T. (2008) Tyr-phosphorylation signals translocate RIN3, the small GTPase Rab5-GEF, to early endocytic vesicles. Biochem. Biophys. Res. Commun. 372, 168–172 [DOI] [PubMed] [Google Scholar]
- 49. Maridonneau-Parini I., de, Gunzburg J. (1992) Association of rapl and rap2 Proteins with the Specific Granules of Human Neutrophils. J. Biol. Chem. 267, 6396–6402 [PubMed] [Google Scholar]
- 50. Mollinedo F., Perez-Sala D., Gajate C., Jimenez B., Rodriguez P., Lacal J. C. (1993) Localization of rap1 and rap2 proteins in the gelatinase-containing granules of human neutrophils. FEBS Lett. 326, 209–214 [DOI] [PubMed] [Google Scholar]
- 51. Sabe H., Onodera Y., Mazaki Y., Hashimoto S. (2006) ArfGAP family proteins in cell adhesion, migration and tumor invasion. Curr. Opin. Cell Biol. 18, 558–564 [DOI] [PubMed] [Google Scholar]
- 52. Fujishiro S. H., Tanimura S., Mure S., Kashimoto Y., Watanabe K., Kohno M. (2008) ERK1/2 phosphorylate GEF-H1 to enhance its guanine nucleotide exchange activity toward RhoA. Biochem. Biophys. Res. Commun. 368, 162–167 [DOI] [PubMed] [Google Scholar]
- 53. Meiri D., Greeve M. A., Brunet A., Finan D., Wells C. D., LaRose J., Rottapel R. (2009) Modulation of Rho guanine exchange factor Lfc activity by protein kinase A-mediated phosphorylation. Mol. Cell. Biol. 29, 5963–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Herrero-Turrión M. J., Calafat J., Janssen H., Fukuda M., Mollinedo F. (2008) Rab27a regulates exocytosis of teriary and specific granules in human neutrophils. J. Immunol. 181, 3793–3803 [DOI] [PubMed] [Google Scholar]
- 55. Johnson J. L., Brzezinska A. A., Tolmachova T., Munafo D. B., Ellis B. A., Seabra M. C., Hong H., Catz S. D. (2010) Rab27a and Rab27b regulate neutrophil azurophilic granule exocytosis and NADPH oxidase activity by independent mechanisms. Traffic 11, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schoch S., Gundelfinger E. D. (2006) Molecular organization of the presynaptic active zone. Cell Tissue Res. 326, 379–391 [DOI] [PubMed] [Google Scholar]
- 57. Monzo P., Gauthier N. C., Keslair F., Loubat A., Field C. M., Le Marchand-Brustel Y., Cormont M. (2005) Clues to CD2-associated protein involvement in cytokinesis. Mol. Biol. Cell 16, 2891–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Glogauer M., Hartwig J., Stossel T. (2000) Two pathways through Cdc42 couple the N-formyl receptor to actin nucleation in permeabilized human neutrophils. J. Cell Biol. 150, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun C. X., Magalhães M. A., Glogauer M. (2007) Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J. Cell Biol. 179, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okamoto M., Südhof T. C. (1997) Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J. Biol. Chem. 272, 31459–31469 [DOI] [PubMed] [Google Scholar]
- 61. Shen Q., Zheng X., McNutt M. A., Guang L., Sun Y., Wang J., Gong Y., Hou L., Zhang B. (2009) NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp. Cell Res. 315, 1653–1667 [DOI] [PubMed] [Google Scholar]
- 62. Brumell J. H., Chan C. K., Butler J., Borregaard N., Siminovitch K. A., Grinstein S., Downey G. P. (1997) Regulation of Src homology 2-containing tyrosine phosphatase 1 during activation of human neutrophils. Role of protein kinase C. J. Biol. Chem. 272, 875–882 [DOI] [PubMed] [Google Scholar]
- 63. Pereira S., Lowell C. (2003) The Lyn tyrosine kinase negatively regulates neutrophil integrin signaling. J. Immunol. 171, 1319–1327 [DOI] [PubMed] [Google Scholar]
- 64. Gao Y., Wang Y. (2007) A Method to Determine the Ionization Efficiency Change of Peptides Caused by Phosphorylation. J. Am. Soc. Mass Spectrom. 18, 1973–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen Q., Powell D. W., Rane M. J., Singh S., Butt W., Klein J. B., McLeish K. R. (2003) Akt Phosphorylates p47phox and Mediates Respiratory Burst Activity in Human Neutrophils. J. Immunol. 170, 5302–5308 [DOI] [PubMed] [Google Scholar]
- 66. Uriarte S. M., Jog N. R., Luerman G. C., Bhimani S., Ward R. A., McLeish K. R. (2009) Counterregulation of clathrin-mediated endocytosis by the actin and microtubular cytoskeleton in human neutrophils. Am. J. Physiol. Cell Physiol. 296, C857–C867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Güngör N., Pennings J. L., Knaapen A. M., Chiu R. K., Peluso M., Godschalk R. W. L., Van Schooten F. J. (2010) Transcriptional profiling of the acute pulmonary inflammatory response induced by LPS: role of neutrophils. Respir. Res. 11, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chakrabarti S., Zee J. M., Patel K. D. (2006) Regulation of matrix metalloproteinase-9 (MMP-9) in TNF-stimulated neutrophils: novel pathways for tertiary granule release. J. Leukoc. Biol. 79, 214–222 [DOI] [PubMed] [Google Scholar]
- 69. Videm V., Strand E. (2004) Changes in neutrophil surface-receptor expression after stimulation with FMLP, endotoxin, interleukin-8 and activated complement compared to degranulation. Scand. J. Immunol. 59, 25–33 [DOI] [PubMed] [Google Scholar]
- 70. Logan M. R., Odemuyiwa S. O., Moqbel R. (2003) Understanding exocytosis in immune and inflammatory cells: the molecular basis of mediator secretion. J. Allergy Clin. Immunol. 111, 923–932; quiz 933 [PubMed] [Google Scholar]
- 71. Sarnago S., Elorza A., Mayor F., Jr. (1999) Agonist-dependent phosphorylation of the G protein-coupled receptor kinase 2 (GRK2) by Src tyrosine kinase. J. Biol. Chem. 274, 34411–34416 [DOI] [PubMed] [Google Scholar]
- 72. Fan G., Shumay E., Malbon C. C., Wang H. (2001) c-Src tyrosine kinase binds the beta 2-adrenergic receptor via phospho-Tyr-350, phosphorylates G-protein-linked receptor kinase 2, and mediates agonist-induced receptor desensitization. J. Biol. Chem. 276, 13240–13247 [DOI] [PubMed] [Google Scholar]
- 73. Laminet A. A., Apell G., Conroy L., Kavanaugh W. M. (1996) Affinity, Specificity, and Kinetics of the Interaction of the SHC Phosphotyrosine Binding Domain with Asparagine-X-XPhosphotyrosine Motifs of Growth Factor Receptors. J. Biol. Chem. 271, 264–269 [DOI] [PubMed] [Google Scholar]