Abstract
The characterization of the humoral response in cancer patients is becoming a practical alternative to improve early detection. We prepared phage microarrays containing colorectal cancer cDNA libraries to identify phage-expressed peptides recognized by tumor-specific autoantibodies from patient sera. From a total of 1536 printed phages, 128 gave statistically significant values to discriminate cancer patients from control samples. From this, 43 peptide sequences were unique following DNA sequencing. Six phages containing homologous sequences to STK4/MST1, SULF1, NHSL1, SREBF2, GRN, and GTF2I were selected to build up a predictor panel. A previous study with high-density protein microarrays had identified STK4/MST1 as a candidate biomarker. An independent collection of 153 serum samples (50 colorectal cancer sera and 103 reference samples, including healthy donors and sera from other related pathologies) was used as a validation set to study prediction capability. A combination of four phages and two recombinant proteins, corresponding to MST1 and SULF1, achieved an area under the curve of 0.86 to correctly discriminate cancer from healthy sera. Inclusion of sera from other different neoplasias did not change significantly this value. For early stages (A+B), the corrected area under the curve was 0.786. Moreover, we have demonstrated that MST1 and SULF1 proteins, homologous to phage-peptide sequences, can replace the original phages in the predictor panel, improving their diagnostic accuracy.
Colorectal cancer (CRC)1 is the major cause of cancer-associated mortality in Spain and other developed countries (1). The population over 50 years of age constitutes the major risk segment. They should be screened periodically using some of the available detection methods, such as faecal occult blood testing (FOBT), sigmoidoscopy, colonoscopy, or CT colonography (2). CEA, the only available noninvasive protein marker, is mainly adequate for late stages and recurrence detection (3). Other alternative protein serum markers are needed to cover the entire progression of the disease. There is a need to define new clinically useful markers for accurate diagnosis of colorectal cancer (for a review see (4)).
Humoral response profiling in cancer patients is becoming increasingly used for the discovery of tumor-associated antigens (TAAs) as new biomarkers (5–10). This new area, called “cancer immunomics” or “seromics,” uses autoantibody signatures to classify neoplastic diseases and to find new targets for diagnosis and immunotherapy (11). Two microarray formats are available for TAA detection, recombinant protein microarrays and phage-display microarrays (12, 13). The use of protein microarrays has led to the identification of TAAs with higher prevalences than previously reported (5). Peptide-containing phage microarrays constitute an interesting alternative to commercial protein arrays. They are usually home-made and are more economical to produce than full-length recombinant protein microarrays. They require the construction of phage libraries, usually from T7 phages (8, 14), consisting of cDNA fragments representative of genes expressed in cancer tissues. Peptides encoded by these cDNA fragments are exposed on the surface of the phage fused to the C-terminal end of the capsid 10B protein of the phage. Then, phage libraries are selected through biopanning procedures involving normal and patient's serum (8). Once constructed, the libraries are confronted to a panel of positive and reference serum to identify phages reactive with patient's autoantibodies. Some initial reports made use of nitrocellulose lifts for plaque screening of phages (14), in a process not amenable to high-throughput screening procedures. Combination of phage display with microarray technologies considerably improved the objective evaluation and throughput of the assays, allowing the testing of thousands of phages with only a few microliters of serum (6, 8, 15). This strategy, however, presents some limitations, such as the sequence of peptides that are displayed on the surface of the phage capsid (16), the presence of mimotopes (6) and the batch to batch reproducibility in microarray production, which is a common problem to other protein microarray formats.
Previously, our group identified PIM1, MAPKAPK3, MST1/STK4, SRC, FGFR4, and ACVR2B as autoantibody targets in colorectal cancer using high-density protein microarrays (5). Here, we decided to test CRC cDNA libraries displayed in T7 phages in microarray format for autoantibody screening in colorectal cancer patients' sera. The combination of both proteomic strategies should increase the number of candidate biomarkers and the diagnostic accuracy. Although screening of colorectal cancer sera with phage display libraries grown in Petri dishes was reported by Ran et al. (17), that screening was based on visual interpretation of antibody binding to nitrocellulose lifts of phage plaques using pooled sera, making objective quantification quite difficult.
In this report, we have used a T7 phage display system in combination with a microarray format to survey the humoral response in colorectal cancer patients. We have discovered and validated a new set of TAAs. One of the TAA candidates, MST1/STK4, was previously identified with commercial high-density full-length protein microarrays, indicating a significant concordance between both assays. By ELISA, we tested either phages or the recombinant homologous proteins with cancer and reference sera, including controls and different types of cancer, to validate the diagnostic assays in CRC patients. The final TAA candidates showed a significant accuracy for CRC diagnosis.
EXPERIMENTAL PROCEDURES
CRC and Reference Control Serum
The Institutional Ethical Review Boards of the Centro de Investigaciones Biológicas (CIB) and the Spanish National Research Council (CSIC) approved this study on biomarker discovery in colorectal cancer. Written informed consent was obtained from all patients. Serum samples for microarray and validation, were obtained from patients in the Bellvitge University Hospital, the Institut Catalá d′Oncología, Barcelona, Hospital Puerta de Hierro (Madrid), and the Hospital of Cabueñes (Gijón), Spain. Sample collection was approved by the Ethical Review Boards of these institutions. For selection of CRC-specific T7 phage libraries, three serum samples from CRC patients with Duke's stage B, 3 from stage C, and six from stage D (three with metastasis to liver and three with metastasis to lung) were used. For microarray analysis, serum samples from 15 patients having CRC in different stages were selected. The median age for the CRC patients was 66.3 years (range 54–82). Fifteen serum samples were obtained from control subjects and were selected to match the median age and the same gender proportion that the CRC cohort. For validation, an independent cohort of 50 CRC serum samples, representative of the different Dukes stages (A–D), 46 control samples, 10 asymptomatic patients with familiar antecedents, 2 hyperplasic polyps, 2 ulcerative colitis, and 43 sera from other types of cancer (bladder, breast, lung, pancreas, and stomach) were used (5). A scheme of the training and validation analysis is shown in Fig. 1. Clinical data from all patients are shown in Table I. Samples were handled anonymously according to ethical and legal guidelines at the Spanish National Research Council (CSIC).
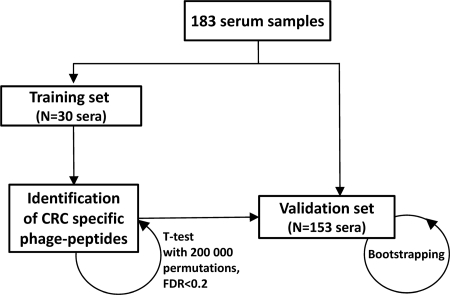
Fig. 1.
Overview of the process followed for the identification and validation of potential biomarkers to diagnose colorectal cancer using phage microarrays.
Table I.
Clinical and pathological information of serum samples used for training and validation assays. B, Bellvitge University Hospital, Institut Catalá d́Oncología (Barcelona, Spain); PH, Puerta de Hierro Hospital (Madrid, Spain), C, Cabueñes Hospital (Gijón, Spain)
| Gender |
Dukes stage |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number (n) | Hospital | Age average (years) | Age range (years) | Male | Female | A | B | C | D | ||
| Total | CRC | 65 | 69.7 | 41–91 | 67.7% | 33.3% | 19% | 22% | 29% | 30% | |
| Controls | 118 | 68.4 | 26–89 | 61.5% | 38.5% | – | – | – | – | ||
| Microarray screening | CRC | 15 | B | 66.2 | 54–82 | 73% | 27% | 40% | 20% | 20% | 20% |
| Healthy | 15 | B | 63.5 | 39–89 | 60% | 40% | – | – | – | – | |
| Validation | CRC | 50 | B, PH | 70.8 | 41–91 | 66% | 34% | 12% | 22% | 32% | 34% |
| Controls | 103 | B, PH, C | 59.2 | 26–89 | 51% | 49% | – | – | – | – | |
| Healthy | 46 | B, PH, C | 59.6 | 34–89 | 63% | 37% | – | – | – | – | |
| CRC familiar antecedents | 10 | C | 49.4 | 26–73 | 25% | 75% | – | – | – | – | |
| Ulcerative colitis | 2 | C | 39 | 28–49 | 50% | 50% | – | – | – | – | |
| Hyperplasic polyp | 2 | C | 67 | 61–73 | 50% | 50% | – | – | – | – | |
| Other tumors | |||||||||||
| Bladder cancer | 11 | B | 67.7 | 58–78 | 64% | 36% | – | – | – | – | |
| Breast cancer | 8 | B | 52 | 30–66 | 0% | 100% | – | – | – | – | |
| Lung cancer | 8 | B | 63 | 55–77 | 75% | 25% | – | – | – | – | |
| Pancreas cancer | 8 | B | 65 | 37–74 | 62% | 38% | – | – | – | – | |
| Stomach cancer | 8 | B | 62 | 37–80 | 25% | 75% | – | – | – | – | |
Serum samples were processed according to an identical protocol in the different hospitals. Blood samples were left at room temperature for a minimum of 30 min (and a maximum of 60 min) to allow clot formation, and then centrifuged at 3000 × g at 4 °C for 10 min. The sera were frozen and stored at −80 °C until use.
T7 Phage Display cDNA Library Synthesis and Biopanning
Construction of phage libraries and biopanning was basically performed as previously described (8). Full methodology is given in supplemental data.
Printing and Use of Phage Microarrays
Following amplification, bacterial lysates were centrifuged and phage-containing supernatants were diluted 1:2 in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBST) and printed in duplicate onto nitrocellulose-coated slides (Whatman/Schleicher and Schuell's) using an OmniGrid Spotter (GeneMachines, San Carlos, CA). Negative controls consisted of BSA (Sigma-Aldrich), buffer alone or empty spots. Human IgG (Sigma-Aldrich), and T7 protein were also spotted as positive controls to verify the array quality.
Serum samples (15 from CRC patients and 15 from healthy individuals) were probed in the phage-peptide microarrays as previously described (6), with minor modifications. Briefly, slides were equilibrated in PBS at room temperature for 5 min and then blocked with 3% skimmed milk in PBS (MPBS) for 1 h at room temperature with agitation. Then, 6.6 μl of human serum (dilution 1:300), 120 μg of E. coli lysate and 0.3 μg of anti T7-Tag monoclonal antibody (Novagen, Madison, WI) in 2 ml of 3% MPBS were incubated for 90 min at room temperature. Slides were washed three times with PBST for 10 min. To detect human antibodies and T7 phages, slides were incubated with Alexa Fluor 647-labeled goat anti-human IgG (Invitrogen, Carlsbad, CA) diluted 1:2000 in 3% MPBS and Alexa Fluor 555-labeled goat anti-mouse IgG (Invitrogen) diluted 1: 40,000 in MPBS, respectively. Arrays were washed three times with PBST, once with PBS and dried by centrifugation at 1200 rpm for 3 min. Finally, slides were read on a ScanArrayTM 5000 (Packard BioChip Technologies). Genepix Pro 7 (Axon Laboratories, Boston, MA) image analysis software was used for spot intensity quantification.
Immunohistochemistry Analysis
All CRC tumor resection specimens (usually hemicolectomies) were fixed in buffered formaldehyde and paraffin-embedded. We selected well-preserved representative areas from the tumor and distant normal mucosa for the immunohistochemical analysis. Immunohistochemistry was performed on 6-μm sections of the blocks following an automated method (Dako autostainer). The primary antibodies for MST1/STK4 (Atlas Antibodies, Stockholm, Sweden) and SULF1 (Sigma) were used at 1:100 and 1:25 dilution, respectively. We counterstained the slides with hematoxylin. Immunoreactivity was graded as 0, absent; 1, mild staining; 2, moderate staining; or 3, intense staining. We classified the cases according to, both, the intensity of the staining and the percentage of areas showing reaction. Because the inflammatory cells showed positivity for MST1 (intense) and SULF1 (mild) antibodies, they were used as internal control. In all cases, an external negative control was included.
ELISA Tests
T7 Phage Capture Plates (Novagen) were blocked for 2 h at 37 °C with 3% MPBS, and then coated overnight with 100 μl of selected phage lysates in 3% MPBS. After washing three times with PBST, plates were blocked with MPBS for 1 h at 37 °C. Then, 100 μl of human serum (dilution 1:50 in 3% MPBS) were incubated for 1 h at 37 °C. After washing, peroxidase-labeled anti-human IgG (1:3000 in 3% MPBS) was added for 2 h at room temperature. Then, the signal was developed with 3,3′,5,5′-tetramethylbenzidine substrate for 10 min (Sigma). The reaction was stopped with 1 m HCl, and the absorbance was measured at 450 nm.
For competition analysis between phage peptides and proteins, T7 Phage Capture Plates were used as above, except that the human sera were pre-incubated overnight with serial dilutions of the proteins MST1, SULF1, or GST. In addition, the preincubated sera were tested in ELISA plates (Maxisorp, Nunc) coated with EBNA1 to verify that the competition between the phage and its respective full-length protein for IgG was specific. EBNA1 was used as a positive control. EBNA1 corresponds to the Epstein-Barr nuclear antigen 1 protein of the Epstein-Barr virus. Over 90% of the human population has been infected with the virus in some moment of their life and presents antibodies to this protein (18). ELISA experiments with full-length proteins MST1, SULF1, and EBNA1 were performed as described before (5). CEA concentration in serum was determined using a specific immunoassay test kit (MP Biomedicals, Santa Ana, CA), following the manufacturer's recommendations.
Statistical Analysis
Microarray data were normalized and processed using the Asterias applications (http://asterias.bioinfo.cnio.es/), a web interface to the limma and marrayNorm Bioconductor packages. After applying a background correction and the global loess normalization (http://dnmad.bioinfo.cnio.es/), data were processed to filter missing values or spots with a too high variance, to merge replicates and then obtain a single value for each phage clone and to transform values in base 2 logarithms (http://prep.bioinfo.cnio.es/). To compare the CRC patients and healthy individuals groups, we performed a t test using pomelo II (http://pomelo2.bioinfo.cnio.es/), where p values were obtained by permutation testing (in our case 200,000). Pomelo II generated a heatmap showing the phages with a FDR-value below 0.15 and an unadjusted p value below 0.05.
For bootstrapping analysis, we fitted a logistic regression model, where we model the probability of being tumoral versus normal as a function of the variables (phages and proteins). We also included in the model the age and sex of the patients, to correct for possible effects of these variables. Models were assessed for adequacy, including the need for nonlinear transformations, using the usual residual plots. To assess predictive ability, we computed the area under the ROC (AUC). However, the AUC computed directly with the original model and the complete data set is too biased toward high values. Thus, we used the bootstrap, with 1000 replicate samples, to obtain a bias-corrected AUC (19). With the bootstrap, we repeatedly sampled with replacement from our original data, and fit the model to that sample, testing the model on the left-out samples. Thus, for each of our 1000 bootstrap samples, we obtained 1000 estimates of AUC from the left-out samples, samples that were not used to fit the model. We refer to this as the bias-corrected AUC. This is, therefore, an estimate of the AUC we would obtain from a future independent validation. All models were fitted using Harrell's Design library (20) with the R statistical computing system (21).
RESULTS
Profiling of Colorectal Cancer Sera with T7 CRC Phage Microarrays
RNA from six patients (three in Dukes' stage A and three in stage C) was used to construct phage cDNA libraries in two vectors (T7Select 415–1 or T7Select 10–3b). Following removal of nonspecific phages and selection of cancer-specific phages, we obtained eight different tumor-specific enriched phage libraries, according to the vector and the serum pool (B, C, Li, and Lu) used during the biopanning procedure (see supplemental data). A total of 1536 individual phages were selected (192 individual phages from each selection) and printed in duplicate onto nitrocellulose slides. The amount of phage printed in the slides was tested by using anti-T7 and anti-human IgG as controls (supplemental Fig. S1A). A homogeneous signal was observed for anti-T7, whereas the anti-human IgG did not give any signal. To determine the intra and inter reproducibility of the arrays, we plotted the intensity of the two spots corresponding to the same phage clone and compared the data from two different microarrays. We verified that intra and inter reproducibility of the arrays were quite good (R2 values were 0.9703 and 0.9091, respectively) (supplemental Fig. S1B). Then, slides were probed with 30 sera (15 from patients at different stages and 15 from healthy controls). Following image quantification and normalization, we compared cancer and normal sera using a t test analysis with 200,000 permutations. One hundred and twenty-eight phage clones showed different reactivity between the two groups, with a FDR < 0.22, 78 phage clones showed increased reactivity, whereas 50 showed a decreased reactivity in CRC sera. A supervised clustering analysis of 50 phage clones with the lowest independent FDR (< 0.15) showed a clear discrimination between CRC patients and healthy individuals (supplemental Fig. S2).
Identification of Phage-inserted Sequences
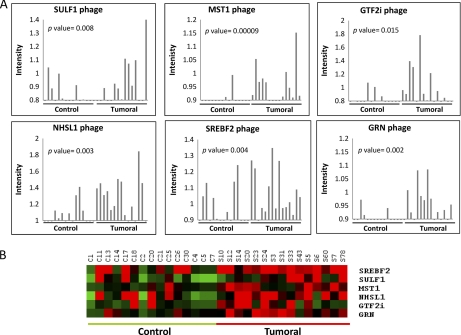
Out of the 78 phages showing an increased reactivity with CRC patients' sera, we obtained 43 unique amino acid sequences as fused to the T7 10B capsid protein (supplemental Table S1). Among these 43 phages, those containing (i) between 8 and 20 residues with the highest possible homology to predicted protein sequences, (ii) highest number of phages with the same sequence, and (iii) lower FDR or p value were selected for further studies. Although most of the inserted sequences corresponded to nonassigned genomic regions, peptides showing homology to proteins MST1/STK4, SULF1, NHSL1, SREBF2, GRN, and GTF2i were identified in the reading frame of the 10B capsid protein. All of them gave a higher microarray signal with tumor sera than control (Fig. 2A). As expected, a significant variation in reactivity was observed between the different patients. Remarkably, MST1/STK4 protein was previously identified as TAA using Protoarrays (5) and SULF1 gene was up-regulated in a CRC transcriptomic analysis (22). Fig. 2B shows a heatmap of the results with the six phage predictor in the training set.
Fig. 2.
Autoantibody response to six CRC-specific phages. A, Microarray signal intensity of cancer and control sera against each phage, following normalization of each serum, in arbitrary units (a.u.). B, Heatmap representation of the microarray signal intensity for the six phages.
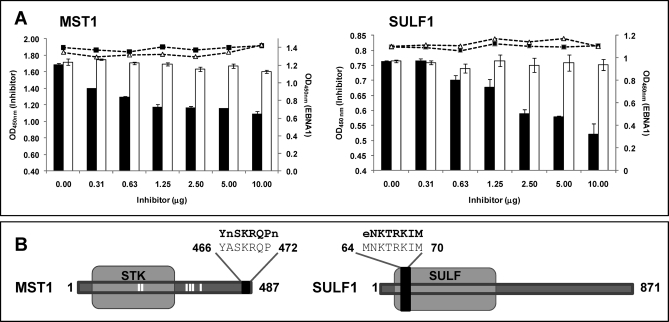
To confirm that peptides expressed in the phages were homologous to MST1 and SULF1 proteins, phages expressing both peptides were subjected to competition analysis with MST1 and SULF1 recombinant proteins. Binding of human cancer sera to both phages was inhibited in a dose-dependent specific manner by MST1 and SULF1 recombinant proteins (Fig. 3A). Antibody binding was almost unaffected when GST was used as a negative control. In contrast, antibody binding to EBNA protein was not affected by incubation with MST1 or SULF1. Phage-inserted sequences were located at the C-terminal region of MST1 and at the N-terminal of SULF1 (Fig. 3B). Collectively, these experiments confirm that the displayed peptides correspond to MST1 and SULF1 proteins.
Fig. 3.
Competition analysis between phage-peptides and homologous proteins. A, A competition ELISA was performed between phages displaying peptides with homology to SULF1 and MST1 and the full-length proteins. GST was used as negative control. Increasing amounts of the recombinant proteins were pre-incubated with the sera and then tested for antibody binding to the phage (vertical bars: black, recombinant protein; white, GST). In the scatter plot, the IgG binding to EBNA1 of the same sera, pre-incubated with increased amounts of recombinant proteins is represented. EBNA 1 was used as a control to demonstrate that the inhibition was protein-specific and no bias was introduced in the experiment (black squares, recombinant protein; white triangles, GST). The Optical Density (OD) at 450 nm of both assays is represented in the figure. Error bars represent standard deviation of three separate experiments. B, Localization of the peptides with homology to SULF1 and MST1 in the full length proteins. Phage-displayed peptide is shown as a black box. White bars correspond to potential phosphorylation sites. Amino acids that were different between the phage-peptide and the wild-type protein are represented in small letter.
Phage-homologous Proteins are Overexpressed in Colorectal Cancer
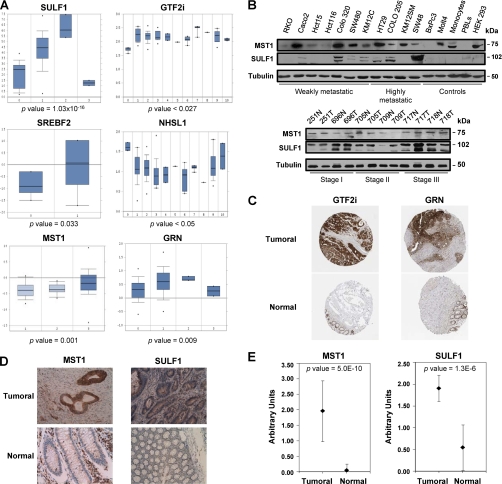
Tumor antigens recognized by autoantibodies are generally overexpressed in tumor cells and cancer tissues (5, 8). A meta-analysis of the mRNA expression levels corresponding to the proteins homologous to the six selected phages was carried out with Oncomine (23), a public open cancer microarray database (Fig. 4A). SULF1 was the most overexpressed gene in different types of colon cancer, followed by GTF2i, MST1, GRN, NHSL1, and SREBF2. In addition, we carried out a Western blot analysis using MST1 and SULF1 antibodies on a panel of 11 colorectal cancer cell lines and CRC tumors representing different progression stages (Fig. 4B). MST1 and SULF1 were expressed in most of the colon cancer cell lines. SULF1 highest expression was mainly observed in metastatic cell lines (SW48, HT29 or COLO205) and at late stage tumor samples. Cellular protein expression patterns of identified proteins were characterized by immunohistochemistry on independent series of CRC tumors contained in custom-made tissue microarrays (MST1/STK4, SULF1) or by meta-analysis according to data retrieved from the Human Protein Atlas in the case of GRN and GTF2i (24) (Fig. 4C). A significant more abundant expression of GRN and GTF2i was reported in neoplastic tissue in comparison to paired normal tissues. For MST1/STK4, most of the tumor tissues showed intense or moderate positivity, whereas the normal mucosa was negative or mildly positive. Tumors were moderately positive for SULF1, whereas normal mucosa displayed a weak staining (Fig. 4D). According to the staining scale (0, low to 3, high) applied for the evaluation of the TMA, we found for MST1 a mean value of 1.96 ± 0.98 and 0.04 ± 0.2 for tumoral and normal tissue, respectively, giving a p value of 5.0E-10, which confirms a statistically significant higher expression of MST1 in tumoral tissue (Fig. 4E). For SULF1, we found a mean value of 1.91 ± 0.30 and 0.55 ± 0.52 (p value 1.2E-6) for tumoral and normal tissue, respectively (Fig. 4E). Collectively, all these data indicate a good correlation between autoantibody targeting, protein abundance and gene expression.
Fig. 4.
Analysis of SULF1, MST1, GTF2i, NHSL1, GRN, and SREBF2 expression in CRC tissues. A, Meta-analysis of gene expression levels corresponding to the proteins homologous to the phage-displayed peptides was assessed by using the Oncomine database. p values are also indicated. Relative gene expression levels were found for NHSL1, SREBF2, GTF2i, SULF1, MST1, and GRN. B, Western blot analysis of SULF1 and MST1 overexpression in tumoral cell lines and paired cancer tissues corresponding to stages A(I), B(II), and C(III). Tubulin was used as a control. C, Tissue microarray data of GTF2i and GRN expression were retrieved from the Human Protein Atlas. D, MST1/STK4 and SULF1 showed intense cytoplasmic staining in well-differentiated enteroid adenocarcinoma of the right colon, whereas normal colonic mucosa far from the tumor was not stained with the antibody. As internal control, we used the positivity of the inflammatory cells in the lamina propria (MST1/STK4 intense staining and SULF1 mild staining). Images were taken at a 200× magnification. E, Immunohistochemistry results for MST1/STK4 and SULF1 in CRC tissue and the normal mucosa of 25 CRC patients were quantified by two pathologists according to the following criteria: 0, no staining; 1, weak staining; 2, normal staining; 3, strong staining. Error bars represent the S.D. of the assay. p values are indicated.
Validation of the Phage-Peptide Detector and Associated Proteins
An independent cohort of 153 samples (50 colorectal cancer, 46 control samples, 10 asymptomatic patients with familiar antecedents, 2 hyperplasic polyps, 2 ulcerative colitis, and 43 sera from other types of cancer (bladder, breast, lung, pancreas, and stomach) (Table I) was used for validation, with 19 samples coming from early colorectal cancer stages (A+B). We tested MST1, SULF1, NHSL1, SREBF2, GRN, and GTF2i-like phage lysates for the ability to discriminate cancer from control sera by using individual ELISA assays. ROC curves were generated for each of these ELISAs. Whereas the sensitivity was relatively low for the individual phages, oscillating between 46 and 58%, the specificity was higher, between 52.2 and 73.9% (Table II). To investigate if different combinations of phages would produce higher accuracy, we fitted the data to a logistic curve, performed logistic regressions and produced different models using different combinations of the phages. When a combination of the six phages was used as a predictor, the area under the curve (AUC) increased up to 0.78, with a sensitivity and specificity of 72 and 73.9%, respectively (Table II). This specificity supported further analysis to assess the clinical relevance of the homologous proteins.
Table II.
Receiver operating-characteristic curves validation of individual and combined phages
| Phage | Specificity (%) | Sensitivity (%) | AUCa |
|---|---|---|---|
| SULF1 | 73.9 | 50 | 0.63 |
| NHSL1 | 52.2 | 52 | 0.52 |
| MST1 | 71.7 | 46 | 0.58 |
| GTF2i | 52.2 | 58 | 0.60 |
| SREBF2 | 60.9 | 48 | 0.53 |
| GRN | 58.7 | 58 | 0.62 |
| Six phages combination | 73.9 | 72 | 0.78 |
a AUC, Area under the curve.
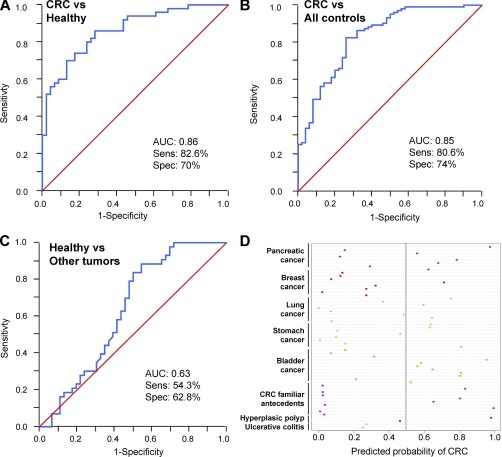
We next tested if the replacement of the phages by the recombinant proteins MST1 and SULF1 could improve the diagnostic accuracy. The results confirmed a significant prediction improvement by using the recombinant proteins, with AUCs of 0.71 and 0.74 for SULF1 and MST1 proteins against 0.63 and 0.58 of the respective phages (supplemental Fig. S3; Table II). By combining the two proteins and four phages, the AUC increased up to 0.86 with a sensitivity of 82.6% and specificity of 70% (Fig. 5A). CEA values were lower (AUC: 0.81) and combined with the rest of the predictor hardly improved the model (AUC: 0.89) (supplemental Fig. S4). Moreover, in the validation step different estimations of AUC were done to compare not only CRC versus healthy but CRC versus all reference sera and healthy versus other tumors (Fig. 5). The most relevant result was the ability of our model to discriminate not only CRC from healthy sera (AUC: 0.86) (Fig. 5A), but also CRC from all the reference sera, which included other related colon pathologies (AUC: 0.85) (Fig. 5B). Remarkably, the panel did not discriminate properly healthy from other tumors (AUC: 0.63) (Fig. 5C). Moreover, the panel seemed to discriminate significantly healthy controls from asymptomatic patients with familiar history of CRC (AUC: 0.78) (data not shown), although the small sample set used will require further verification.
Fig. 5.
Validation of the combination of four phages with MST1 and SULF1 proteins in the diagnosis of colorectal cancer. Performance of the combination of GTF2i, NHSL1, GRN, and SREBF2-like phages and MST1 and SULF1 proteins in the validation set. Receiver-operating-characteristic curves are based on multiplex analyses of the four phages and two proteins from a total of 153 samples (50 samples from CRC patients, 46 healthy controls, 10 samples from controls with CRC familiar antecedents, 2 from ulcerative colitis patients, 2 from patients with hyperplasic polyp, and 43 samples from patients with bladder, breast, lung, pancreatic or stomach cancer). A, Performance of CRC samples versus healthy controls. B, Performance of CRC samples versus all reference sera. C, Performance of healthy sera versus other tumors sera. D, Dotplot showing individual probability of being classified as CRC patient for each of the subjects with different pathologies. The predicted probability is the predicted probability from the final logistic regression model (to differentiate between CRC and reference subjects) following variable selection. Most of the samples were classified below the 0.5 threshold probability (gray line). Therefore, the model did not detect general markers for cancer or inflammatory disease, but particular markers of CRC.
Bootstrapping Analysis and Final Prediction Model
In addition, we performed bootstrapping to obtain a bias-corrected AUC. The initial model included linear terms for all phages and proteins, together with two other variables: gender and age. With this model, the value of the bias-corrected AUC was 0.86. This model was probably more complex than justified. Thus, we carried out variable selection, using backwards selection with Akaike's Information Criterion as stopping rule. The final model retained (GRN phage, MST1, and SULF1 proteins plus the age of the patients (supplemental Table S2). However, to avoid an overestimation of the predictive capacity of the model, we obtained bias-corrected estimates of the AUC, by bootstrapping the complete process of variable selection (i.e. for each bootstrap sample, we started with the complete model with eight variables, and used Akaike's Information Criterion as the stopping rule). The bias-corrected AUC was 0.84. Bootstrapping also provided information on the stability of the selection procedure: among the bootstrapped models, most contained either four, five, six, or seven variables (171, 262, 329, and 172 out of the 1000 bootstrap replicates, respectively). Some of the variables appeared in most of the models: GRN phage in 976, SULF1 protein in 954, age in 952 and MST1 protein in 833.
Moreover, we used this model to predict the probability of being CRC for the set of 57 sera comprising diverse pathologies. We constructed a dotplot representation (Fig. 5D), where we showed the individual probability for each subject. A wide variability in probability is observed within each group, but the median is well below 0.5, indicating a low probability of being CRC.
Then, we tested the value of the prediction according to the stage of the patients for early diagnosis purposes. We started from the model with six markers (4 phages+2 proteins) plus age. The bias-corrected AUC using bootstrapping was 0.786 for stages A+B, 0.857 for stage C and 0.849 for stage D. If we apply the same test with the CEA values, the bias-corrected AUC were 0.742 for A+B, 0.770 for C, and 0.973 for stage D. These results indicate a superiority of our predictor for stages A, B, and C, being CEA only superior for stage D as expected.
DISCUSSION
The use of the microarray format for phage display cancer peptide libraries for autoantibody screening permits an objective identification and quantification not possible by other means. Still, the approach is rather cumbersome and labor-intensive when compared with the use of recombinant protein microarrays. The technique requires considerable effort and resolution. Moreover, the identification of inserted sequences led in most of the cases to mimotopes with no clear protein assignation. All these factors make difficult its widespread use and may explain the relatively low number of reports that have applied this strategy so far. However, protein and phage microarrays have enabled the discovery of relatively large panels of proteins recognized by autoantibodies in colorectal cancer. The number of these TAAs vastly increases the number and prevalence of those antigens previously identified in cancer patients by other approaches (10). Still, we do not know yet how many proteins become autoantibody targets in cancer patients and the molecular basis for this autoimmunity.
As a novelty, this report demonstrates, for the first time, the correspondence between phage-inserted peptides and the corresponding recombinant proteins. Recombinant MST1 and SULF1 proteins were able to compete and displace antibody binding to the phages in ELISA assays. Moreover, they increased the predictive accuracy of the assay. This is an important step to support the reliability of this technology. The classifier using four combined phages and two proteins resulted in high specificity (70%) and sensitivity (82.6%) for CRC sera, improving CEA prediction capacity. Combination with CEA did not improve significantly the diagnostic accuracy of the panel detector (supplemental Fig. S4). The addition of sera from other tumors to the validation step did not change the prediction power of the panel, stressing the value of this approach. Specificity of the test was confirmed by the low AUC obtained after comparing healthy sera from other non-CRC, cancer sera. The significance of this study was to develop a diagnostic assay useful for identification of early adenocarcinomas in CRC, with a bias-corrected AUC of 0.786 for stages A+B. Moreover, preliminary data seem to support that this panel could also discriminate very early stages or asymptomatic patients. Therefore, this panel of biomarkers might be extremely helpful in defining high-risk populations that should go through enhanced screening procedures like colonoscopy as an alternative to FOBT. Although FOBT is relatively inexpensive and non-invasive, it displays high false positive rates (3) and promotes unnecessary colonoscopies (25).
We have observed coincidences between the identified proteins in these phage-arrays and the commercial protein arrays (Protoarrays®). At least two proteins, MST1/STK4 and DNAJ (data not shown), were identified with both types of arrays. DNAJ-specific autoantibodies were previously reported for lung cancer (14). Together with MST1/STK4 and SULF1, other four phages: NHSL1, SREBF2, GRN, and GTF2i, were used for validation of the predictive and diagnostic capacity. The other four sequences will require further verification to prove that they correspond to those hypothetical proteins. It will require the synthesis of the identified peptides or the expression of the full-length recombinant protein. The identification of only small homologous peptides displayed on the phage surface seems to be because of the random cloning of cDNA fragments. Many cDNA inserts correspond to antisense mRNAs, aberrant splicing regions, and other variants. This resulted in the production of phages containing peptide sequences with weak or no homology to known proteins (supplemental Table S1). These peptides have been generally described as mimotopes, epitopes that reflect conformational epitopes and, therefore, have no significant homology to any other known protein. From our results, the concordance between peptides and proteins might be fortuitous and not because of the insertion of cDNA-specific encoding sequences. Thus, the use of random-peptide libraries (26) would be almost equivalent to this approach.
As mentioned before, individual phages offer a lower sensitivity and specificity than the corresponding recombinant proteins, probably because only a single peptide/epitope is involved in the binding. As previously reported (8), we also experienced the necessity of combining multiple phages or phages and proteins (MST1, SULF1) to get a satisfactory diagnostic value.
Expression analysis of MST1 and SULF1 at the tissue level indicated a potential association of SULF1 with late stages of cancer progression and a significant value of these two biomarkers for CRC diagnosis (Fig. 4E). Protein expression data by Western blot were concordant with high mRNA levels of SULF1 in advanced carcinomas according to the meta-analysis of gene expression in tumoral tissues. In agreement with previous results, there was a good correlation between the presence of autoantibodies against a protein and an elevated mRNA and protein expression. Regarding functional activity, SULF1 diminishes HSPG (heparan sulfate proteoglycans) sulfation, inhibits signaling by heparin-dependent growth factors, diminishes proliferation, and facilitates apoptosis in response to exogenous stimulation (27, 28). Messenger RNA down-regulation was observed in ovarian, breast, pancreatic, renal and hepatocellular carcinoma cell lines. However, SULF1 has been reported as up-regulated in CRC tumors (29). This difference in expression between CRC and other tumors might explain the specificity of SULF1 as CRC biomarker.
A previous report of association between MST1 expression and improved survival in colon cancer patients was observed (30). The mechanism underlying this prognostic value might be related to the functional activity of STK4/MST1, which is a stress-activated, pro-apoptotic kinase. MST kinases play important roles in diverse biological processes including cellular responses to oxidative stress and longevity (31).
In summary, we have generated a novel CRC detector based on phages and associated homologous proteins able to generate a diagnostic assay with superior predictive capacity to CEA, especially for early stages, and capable of distinguishing patients with CRC from control subjects or other cancer types. MST1 and SULF1 are candidate biomarkers for CRC diagnosis. The discovery of identical TAAs (MST1) by two different protein array platforms supports the robustness of the application and the significance of autoantibody detection for the early diagnosis of colorectal cancer.
Acknowledgments
RB is recipient of a JAE-DOC Contract of the CSIC. We thank Dr Felix Bonilla (H. Puerta de Hierro) by kindly supplying CRC samples.
Footnotes
* This research was supported by grants from the Spanish Ministry of Education and Science BIO2009-08818, “Proyecto Intramural de Incorporación-CSIC”, Colomics Programme of the regional government of Madrid and grants from the Fundación Médica Mutua Madrileña, Instituto de Salud Carlos III (FIS 05/1006 and 08/1635), the CIBERESP G55, the “Acción transversal del cancer” and the Proteored platform.
 This article contains supplemental material, supplemental Figs. S1 to S4, and supplemental Tables S1 and S2.
This article contains supplemental material, supplemental Figs. S1 to S4, and supplemental Tables S1 and S2.
1 The abbreviations used are:
- AUC
- area under the curve
- CRC
- colorectal cancer
- CEA
- carcinoembryonic antigen
- FDR
- false discovery rate
- FOBT
- faecal occult blood testing
- GRN
- granulin
- GTF2i
- general transcription factor II i
- MST1
- mammalian STE20-like protein kinase 1
- NHSL1
- NHS-like protein 1
- ROC
- receiver operating characteristic
- SREBF2
- sterol regulatory element binding protein 2
- SULF1
- sulfatase 1
- TAA
- tumor-associated autoantigen
- TMA
- tissue microarray.
REFERENCES
- 1. Edwards B. K., Ward E., Kohler B. A., Eheman C., Zauber A. G., Anderson R. N., Jemal A., Schymura M. J., Lansdorp-Vogelaar I., Seeff L. C., van Ballegooijen M., Goede S. L., Ries L. A. (2010) Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer 116, 544–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Winawer S. J. (2007) The multidisciplinary management of gastrointestinal cancer. Colorectal cancer screening. Best Pract. Res. Clin. Gastroenterol. 21, 1031–1048 [DOI] [PubMed] [Google Scholar]
- 3. Duffy M. J., van Dalen A., Haglund C., Hansson L., Holinski-Feder E., Klapdor R., Lamerz R., Peltomaki P., Sturgeon C., Topolcan O. (2007) Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 43, 1348–1360 [DOI] [PubMed] [Google Scholar]
- 4. Barderas R., Babel I., Casal J. I. (2010) Colorectal cancer proteomics, molecular characterization and biomarker discovery. Proteomics Clin. Appl. 4, 159–178 [DOI] [PubMed] [Google Scholar]
- 5. Babel I., Barderas R., Diaz-Uriarte R., Martinez-Torrecuadrada J. L., Sánchez-Carbayo M., Casal J. I. (2009) Identification of tumor-associated autoantigens for the diagnosis of colorectal cancer in serum using high density protein microarrays. Mol. Cell Proteomics 8, 2382–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterjee M., Mohapatra S., Ionan A., Bawa G., Ali-Fehmi R., Wang X., Nowak J., Ye B., Nahhas F. A., Lu K., Witkin S. S., Fishman D., Munkarah A., Morris R., Levin N. K., Shirley N. N., Tromp G., Abrams J., Draghici S., Tainsky M. A. (2006) Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer Res. 66, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sreekumar A., Laxman B., Rhodes D. R., Bhagavathula S., Harwood J., Giacherio D., Ghosh D., Sanda M. G., Rubin M. A., Chinnaiyan A. M. (2004) Humoral immune response to alpha-methylacyl-CoA racemase and prostate cancer. J. Natl. Cancer Inst. 96, 834–843 [DOI] [PubMed] [Google Scholar]
- 8. Wang X., Yu J., Sreekumar A., Varambally S., Shen R., Giacherio D., Mehra R., Montie J. E., Pienta K. J., Sanda M. G., Kantoff P. W., Rubin M. A., Wei J. T., Ghosh D., Chinnaiyan A. M. (2005) Autoantibody signatures in prostate cancer. N. Engl. J. Med. 353, 1224–1235 [DOI] [PubMed] [Google Scholar]
- 9. Scanlan M. J., Chen Y. T., Williamson B., Gure A. O., Stockert E., Gordan J. D., Türeci O., Sahin U., Pfreundschuh M., Old L. J. (1998) Characterization of human colon cancer antigens recognized by autologous antibodies. Int. J. Cancer 76, 652–658 [DOI] [PubMed] [Google Scholar]
- 10. Scanlan M. J., Welt S., Gordon C. M., Chen Y. T., Gure A. O., Stockert E., Jungbluth A. A., Ritter G., Jäger D., Jäger E., Knuth A., Old L. J. (2002) Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 62, 4041–4047 [PubMed] [Google Scholar]
- 11. Lee S. Y., Obata Y., Yoshida M., Stockert E., Williamson B., Jungbluth A. A., Chen Y. T., Old L. J., Scanlan M. J. (2003) Immunomic analysis of human sarcoma. Proc. Natl. Acad. Sci. U.S.A. 100, 2651–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. LaBaer J., Ramachandran N. (2005) Protein microarrays as tools for functional proteomics. Curr. Opin. Chem. Biol. 9, 14–19 [DOI] [PubMed] [Google Scholar]
- 13. Chatterjee M., Ionan A., Draghici S., Tainsky M. A. (2006) Epitomics: global profiling of immune response to disease using protein microarrays. Omics 10, 499–506 [DOI] [PubMed] [Google Scholar]
- 14. Zhong L., Peng X., Hidalgo G. E., Doherty D. E., Stromberg A. J., Hirschowitz E. A. (2004) Identification of circulating antibodies to tumor-associated proteins for combined use as markers of non-small cell lung cancer. Proteomics 4, 1216–1225 [DOI] [PubMed] [Google Scholar]
- 15. Zhong L., Hidalgo G. E., Stromberg A. J., Khattar N. H., Jett J. R., Hirschowitz E. A. (2005) Using protein microarray as a diagnostic assay for non-small cell lung cancer. Am. J. Respir. Crit. Care Med. 172, 1308–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalniņa Z., Siliņa K., Meistere I., Zayakin P., Rivosh A., Abols A., Leja M., Minenkova O., Schadendorf D., Linē A. (2008) Evaluation of T7 and lambda phage display systems for survey of autoantibody profiles in cancer patients. J. Immunol. Methods 334, 37–50 [DOI] [PubMed] [Google Scholar]
- 17. Ran Y., Hu H., Zhou Z., Yu L., Sun L., Pan J., Liu J., Yang Z. (2008) Profiling tumor-associated autoantibodies for the detection of colon cancer. Clin. Cancer Res. 14, 2696–2700 [DOI] [PubMed] [Google Scholar]
- 18. Ascherio A., Munger K. L., Lennette E. T., Spiegelman D., Hernán M. A., Olek M. J., Hankinson S. E., Hunter D. J. (2001) Epstein-Barr virus antibodies and risk of multiple sclerosis: a prospective study. JAMA 286, 3083–3088 [DOI] [PubMed] [Google Scholar]
- 19. Efron B. (1983) Estimating the error rate of a prediction rule: Improvement on cross-validation. J. Am. Stat. Assoc. 78, 316–331 [Google Scholar]
- 20. Harrell F. (2001) Regression Modeling Strategies, Springer, New York [Google Scholar]
- 21. R Core Development Team (2009) R: A Language and Environment for StatisticalComputing. The R Foundation for Statistical Computing; Vienna [Google Scholar]
- 22. Madoz-Gúrpide J., López-Serra P., Martínez-Torrecuadrada J. L., Sánchez L., Lombardía L., Casal J. I. (2006) Proteomics-based validation of genomic data: applications in colorectal cancer diagnosis. Mol. Cell Proteomics 5, 1471–1483 [DOI] [PubMed] [Google Scholar]
- 23. Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C. A., Persson A., Ottosson J., Wernérus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., Hober S., Pontén F., Uhlén M. (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell Proteomics 7, 2019–2027 [DOI] [PubMed] [Google Scholar]
- 25. Juillerat P., Peytremann-Bridevaux I., Vader J. P., Arditi C., Schusselé, Filliettaz S., Dubois R. W., Gonvers J. J., Froehlich F., Burnand B., Pittet V. (2009) Appropriateness of colonoscopy in Europe (EPAGE II). Presentation of methodology, general results, and analysis of complications. Endoscopy 41, 240–246 [DOI] [PubMed] [Google Scholar]
- 26. Mintz P. J., Kim J., Do K. A., Wang X., Zinner R. G., Cristofanilli M., Arap M. A., Hong W. K., Troncoso P., Logothetis C. J., Pasqualini R., Arap W. (2003) Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat. Biotechnol. 21, 57–63 [DOI] [PubMed] [Google Scholar]
- 27. Viviano B. L., Paine-Saunders S., Gasiunas N., Gallagher J., Saunders S. (2004) Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J. Biol. Chem. 279, 5604–5611 [DOI] [PubMed] [Google Scholar]
- 28. Waldow A., Schmidt B., Dierks T., von, Bülow R., von, Figura K. (1999) Amino acid residues forming the active site of arylsulfatase A. Role in catalytic activity and substrate binding. J. Biol. Chem. 274, 12284–12288 [DOI] [PubMed] [Google Scholar]
- 29. Kaiser S., Park Y. K., Franklin J. L., Halberg R. B., Yu M., Jessen W. J., Freudenberg J., Chen X., Haigis K., Jegga A. G., Kong S., Sakthivel B., Xu H., Reichling T., Azhar M., Boivin G. P., Roberts R. B., Bissahoyo A. C., Gonzales F., Bloom G. C., Eschrich S., Carter S. L., Aronow J. E., Kleimeyer J., Kleimeyer M., Ramaswamy V., Settle S. H., Boone B., Levy S., Graff J. M., Doetschman T., Groden J., Dove W. F., Threadgill D. W., Yeatman T. J., Coffey R. J., Jr., Aronow B. J. (2007) Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8, R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams H., Tzankov A., Lugli A., Zlobec I. (2009) New time-dependent approach to analyse the prognostic significance of immunohistochemical biomarkers in colon cancer and diffuse large B-cell lymphoma. J. Clin. Pathol. 62, 986–997 [DOI] [PubMed] [Google Scholar]
- 31. Lehtinen M. K., Yuan Z., Boag P. R., Yang Y., Villén J., Becker E. B., DiBacco S., de, la, Iglesia N., Gygi S., Blackwell T. K., Bonni A. (2006) A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125, 987–1001 [DOI] [PubMed] [Google Scholar]