Abstract
In quantitative proteomics work, the differences in expression of many separate proteins are routinely examined to test for significant differences between treatments. This leads to the multiple hypothesis testing problem: when many separate tests are performed many will be significant by chance and be false positive results. Statistical methods such as the false discovery rate method that deal with this problem have been disseminated for more than one decade. However a survey of proteomics journals shows that such tests are not widely implemented in one commonly used technique, quantitative proteomics using two-dimensional electrophoresis. We outline a selection of multiple hypothesis testing methods, including some that are well known and some lesser known, and present a simple strategy for their use by the experimental scientist in quantitative proteomics work generally. The strategy focuses on the desirability of simultaneous use of several different methods, the choice and emphasis dependent on research priorities and the results in hand. This approach is demonstrated using case scenarios with experimental and simulated model data.
With the advent of high throughput genomics approaches, researchers need appropriate bioinformatic and statistical tools to deal with the large amounts of data generated. In quantitative proteomics work, differences in expression of many individual proteins between treatments or samples might need to be tested. Researchers must then address what has come to be known as the multiple hypothesis testing problem. Suppose 500 features such as protein spots in a two-dimensional electrophoresis (2-DE)1 experiment, or mass spectrum features relating to protein or peptide abundance, are each compared between treatments using a t test. If the conventional a priori significance level of α = 0.05 is used, then 5% or about 25 significant features are expected to occur just by chance even if the null hypothesis of no treatment effect is true for all 500 features. Thus it is easier to make a false positive error when picking out significant results in an experiment with multiple features, than when considering one feature in isolation.
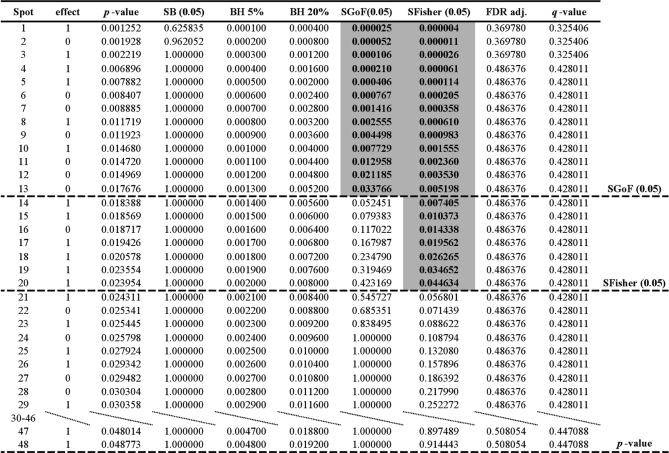
A variety of statistical methods have been devised to deal with the multiple hypothesis testing problem. These are applicable in quantitative proteomics. In this paper we use examples from 2-DE proteomics to demonstrate these methods. In this technique, the intensity of signal from protein spots on 2-DE gels is measured and compared between gels. Use of the word “spot” is obviously not synonymous with use of the word “protein” in that it does not encompass all forms of a given protein such as alternatively spliced variants and post-translational modification variants that might form spots in different positions on the gel. The multiple testing approach is introduced with the following example. Table I shows simulated data for a model of a 2-DE proteomics experiment in which 500 spots have been compared between two treatments using the t test. The third column gives p values significant at α = 0.05 sorted from low to high. A threshold line is shown drawn under spot 70. This has been selected arbitrarily for illustration of some properties of a threshold. The p values for the spots above the threshold are all less than α = 0.05 but we cannot declare them to be significant at the α = 0.05 level because of the multiple hypothesis testing problem. In reality, spots above the threshold will be a mixture of true positives (with treatment effect) and false positives (null hypothesis true); spots below the threshold will be a mixture of true negatives (null hypothesis true) and false negatives (with treatment effect). Multiple hypothesis testing correction methods are used to help position a threshold on the list, different methods placing the threshold in different positions. Spots above the threshold can be declared significant, the null hypothesis is rejected and the alternative hypothesis of treatment effect accepted. Spots below the threshold can be declared nonsignificant and the null hypothesis is accepted. This is in accord with the Neyman-Pearson decision rule method of statistical inference (see 1).
Table I. Simulated results of a model of a proteomics experiment with two treatments and 500 protein spots.
The standard deviation (biological error) within each treatment group was set to 1 for all spots. Treatment effect sizes are, 50 spots effect = 2, 100 spots effect = 1, 350 spots effect = 0 (see text for further explanation). Column headings are Spot, label for spot; effect, defined immediately above; p-value, in t-test between treatments; SB (0.05), probability for sequential Bonferroni at FWER = 0.05; BH 5%, critical value for false discovery rate of 5%; BH 20%, critical value for false discovery rate of 20%; SGoF(0.05), probability for sequential goodness of fit at α = 0.05; SFisher (0.05), probability for sequential combining probabilities test at α = 0.05; FDR adj., FDR adjusted probability; q-value. The table has two breaks incorporating spots 15–51 and 73–86 respectively to save space. See text for further details. Tabulated values are rounded to six decimal places, although spreadsheet calculations were carried out to more than six decimal places.
As the threshold is moved up the table of sorted p values there should be a lower proportion of false positives left above the line. This is useful if our main focus is on being confident that significant results reveal treatment effects, worthy of further investigation. However there will be an increasing proportion of false negatives left below the threshold, we will be failing to recognize treatment effects. Optimal positioning of the threshold for the results in hand is a balancing act, influenced by our perception of whether it is more important to avoid false positive or false negative errors. It has been suggested that there is traditionally too great an emphasis on avoiding false positives (type I errors), and that greater attention should be given to avoiding false negatives (type II errors) (2). False positives can be corrected by further investigation, whereas an experiment with a false negative result might never be repeated, and possible true treatment effects missed. The Fisher view of statistical inference (see 1) could be further applied to spots above the threshold. This is that the lower the p value, the greater the strength of the evidence against the null hypothesis, and the more confident we can be that further investigation will confirm a treatment effect.
There are many statistical problems, relevant to genomics work, that are still under active debate. Examples include the possible arbitrariness of the α = 0.05 critical significance level (e.g. 3–4), the doubt about whether significance testing is even useful as compared with estimation of parameters and confidence intervals (e.g. 4–5), and the interpretation of the concept of probability itself (6). In the particular case of the multiple hypothesis testing problem, many methods and refinements have been proposed (e.g. see 7 for review). The experimental scientist has the problem of evaluating these methods and assessing the debate on their relative merits and validity. In this circumstance we feel that particular methods ought not to be strongly prescribed to the experimental scientist working in proteomics. There should be freedom to apply and choose from the variety of methods available. Our main aim here is to promote the strategy that simultaneous consideration of several multiple hypothesis testing methods is useful, and that particular emphasis on one method rather than another might differ depending on the scientific question and priorities under consideration. We first present a selection of different multiple hypothesis testing approaches that can be applied in proteomics work. We then present a survey analysis which suggests that the use of such methods in the proteomics literature is as yet rather limited. We then demonstrate and discuss the proposed strategy using some example datasets.
Multiple Hypothesis Testing Methods
We describe next some of the most widely used multiple hypothesis testing methods, using as illustration the simulated data of Table I. The data is based on a model with five biological replicates for each of two treatment groups each compared at 500 protein spots. The standard deviation (biological error) within each treatment group was set to one for all spots. Values were chosen at random, assuming normality, using the Excel add-in Poptools (8). For one of the treatment groups, the mean value was set equal to zero for all five biological replicates for all spots. For the second treatment group, for all biological replicates, the spot means were set to 2, 1, and 0 for 50, 100, and 350 spots respectively. Where both treatments have a mean value of zero, the null hypothesis is true, otherwise there is a treatment effect with a mean difference between treatment groups of two (for 50 spots) and one (for 100 spots). The table shows an arbitrarily selected replicate of the model for which the number of spots significant at α = 0.05 is close to the mode for the model, assessed in 100 Monte-Carlo replicates made using Poptools. We emphasize that it is not our intention to investigate the properties of the model in depth, rather to use it to provide some representative data to illustrate multiple hypotheses testing methods.
Most traditional multiple hypothesis testing methods aim to control the number of false positives. An example is the Bonferroni correction (9). This controls the family-wise error rate (FWER) which is the probability of making one or more false positive errors, in a set of tests. For example if FWER = 0.05, the traditional significance level, then each test in a set of m tests needs to have an a priori p value less than or equal to α = 0.05/m to be declared significant a posteriori. Alternatively the p value can be multiplied by m, as in Table I. If the product is less than or equal to 0.05 then the test can be declared significant. If the lowest p value is declared significant, the second lowest p value is assessed using α = 0.05/(m-1) and so on. This is known as the sequential Bonferroni method (SB). It has been criticized as being too conservative. The threshold usually comes near the top of the table, and although the spots above it are unlikely to be false positives, there are many false negatives below the threshold. Thus the method has low power to detect many true treatment differences. In Table I only three spots remain significant after applying the SB method.
An alternative to the SB method was proposed by Benjamini and Hochberg (10). This is the false discovery rate (FDR) method. This aims to determine a threshold such that a proportion or percent of p values above the threshold are false positives, the remainder true positives. FDR is said to be controlled at this percent level. There are different methods to control FDR and we use here the acronym BH to refer to the specific procedure of Benjamini and Hochberg (10). Critical values for control at the BH 5% level are shown in Table I. The critical values for a spot are calculated as α× (i/500) where i is spot number from 1 to 500, after ranking by increasing p value. The BH method is implemented by what is called a step-up procedure. Starting from the bottom of the table, the p values are checked until one value is less than or equal to the critical value in the same row. This p value and all those above it in the table are then declared significant at the BH 5% level and included above this threshold. In Table I, 12 spots are above the threshold and thus declared significant at this level. From the practical viewpoint this means that a proportion 0.05 of these 12 are expected to be false positives, and in FDR terminology these would be false discoveries. The remainder, a proportion 0.95 are expected to be true discoveries, where the null hypothesis is false and the alternative hypothesis true. The BH 5% threshold is lower down the table than the SB threshold. Theoretical studies have indicated that the BH method has greater power to detect true positives than the SB method, assuming of course that there really are some spots with treatment effects. The cost is that shifting the threshold down the table results in some spots that were true negatives now moving above the threshold and being converted into false positives. The BH method can set control at different levels, for example Verhoeven et al. (11) illustrate graphically the different threshold effects of control at 5%, 10%, and 20%. A column for less stringent control, at BH 20% is also given in Table I. At this control level, a proportion 0.2 of spots above the threshold are expected to be false positives. An alternative to controlling FDR at a specific level is to select a region within the list of p values and work out the FDR for it. This is the basis of FDR adjusted probability (12, 13). For example, suppose the chosen region were that cut off above the arbitrary threshold in Table I. The FDR adjusted probability of spot 70, just above this threshold is defined as (p value × 500)/i = 0.212 or 21.2%, where i = 70 in this case. Thus for all spots above the threshold a proportion 0.212 are expected to be false positives. It should be noted that FDR adjusted probability can have the same value for all spots within a set of spots, for example spots 63–70 in Table I. This is imposed for a technical reason that demands that no spot can have a value that is higher than that of a spot further down the table.
The positive false discovery rate (14) is a modified version of the FDR that takes into account that in practice we are only interested in analyzing datasets in which at least one feature has been declared to have a significant p value at the chosen α level. The positive false discovery rate cannot be controlled at specific levels as can FDR, but allows calculation of the q-value (15, 16), which is analogous to FDR adjusted probability. Thus whereas FDR can be used to control at a specific level such as BH 5% or BH 20% (when we use the Benjamini and Hochberg FDR method) both FDR adjusted probability and q-value can be applied to a specific region of p values from the top of the list down to some point in the list. The q-value of spot 70, just above the arbitrary threshold in Table I, is 0.171. This means that the expected proportion of false positives occurring in the set of spots above the arbitrary threshold is 0.171. There is a symmetry between the false positive rate (p) and the false discovery rate (q) (15): p is the probability of getting a significant result at level α given that the null hypothesis is true: q is the probability that the null hypothesis is true given a significant result at level α.
Next we present two approaches that are less widely used than SB and the false discovery method. In the combined probability test of Fisher (17, 18) the aim is to combine the a-priori p values of all the spots into a single p value for the data as a whole, which is then compared with the chosen α value. To do this, the natural logarithm of the k listed p values are summed. This gives a test statistic distributed as a chi-square with 2k degrees of freedom. The null hypothesis for this combined “meta” test statistic is that all the individual spots in the list show no difference between treatments. The probability value for the combined test statistic can be called the meta p value. If the meta p value is significant at level α then it can be concluded that at least one of the spots in the list has a null hypothesis that is false. The best candidate to declare as having a false null hypothesis is the spot with the lowest p value, the one at the top of the list, and the meta p value is thus placed next to it in the same row at the top of the column headed SFisher for sequential combined probability test of Fisher in Table I. The procedure continues by repeating the combined probability test but with the exclusion of the p value at the top of the list. The resulting new meta p value is then placed in the second row in the SFisher column. This sequential procedure continues until the meta p value is no longer significant at level α. At this point it can be concluded that there is no evidence that any of the remaining spots have false null hypotheses, and thus the number of spots with treatment effects is equal to the number of significant meta tests. In Table I the top 59 spots are significant using a meta test α = 0.05. Alternative methods for combining probabilities are available, for example the unweighted Z-test (the standard normal deviate) called Stouffer's test and a weighted version of this test (see 18). Stouffer's test produces similarly positioned thresholds to Fisher's test in the case scenarios considered here and not discussed further. Application of the weighted test is rather complex, depending for example on the kind of test used and effect size (e.g. see 18, 19) and thus cannot be applied to lists of p values without additional information.
The final method is based on the exact binomial test (20, 21), and is explained with reference to the Table I example. If all null hypotheses are true then 500 × 0.05 = 25 spots are expected by chance to have p values significant at α = 0.05. Suppose however that 33 spots, more than expected, are significant at this level. The probability of obtaining a ratio that is 467:33, or more highly skewed toward an excess of spots that are individually significant at α = 0.05, is calculated using the binomial theorem assuming a null expectation of 0.95:0.05. This meta test probability is 0.045, and if we also use the significance level α = 0.05 for the meta test, the meta test is significant at this α level. It can therefore be concluded that at least one of the 500 null hypotheses is false. As in the sequential Fisher test, the best candidate to declare as significant is the spot with the lowest p value at the top of the list. In Table I, 88 spots are individually significant at α = 0.05 and the meta test p value for 412:88 is highly significant. By analogy with the Fisher test, the exact binomial meta test can be applied sequentially, by testing ratios progressively closer to the expected 475:25 until a nonsignificant meta p value is obtained. For each meta test in this sequential procedure, the meta p value is partnered with the spot with lowest individual p value (column 3) at that point. This is the basis of the SGoF (Sequential Goodness of Fit) test (20). The meta p values deriving from this test are given in Table I in the column headed SGoF(0.05) for sequential goodness of fit. Consider the meta p value associated with a particular spot, say 0.034 for spot 53. Because it is less than the meta test α = 0.05, it can be concluded that there is at least one false null hypothesis in the collection of 448 spots incorporated in the meta test at that point. In the next sequential application of the meta test, the meta p value = 0.052 for spot 54, so at this point it can be concluded that there is no evidence to reject the null hypothesis for any of the remaining 447 spots. The sequential procedure stops. In general, the sequential methods SGoF and SFisher have greater power than the SB and BH methods. In the latter, the p value needed for a spot to be declared significant decreases as the number of spots in the dataset increases, and thus the power to detect true effects also decreases (22). In the former it is the opposite because a larger dataset results in more power in the meta test (20).
Survey of Multiple Testing in Proteomics Journals
Multiple hypothesis testing methods such as FDR have been disseminated for more than one decade since the publication of Benjamini and Hochberg (10). We present now the results of a survey undertaken to gauge the current use of multiple hypothesis testing methods in proteomics journals. Issues of the three journals Molecular and Cellular Proteomics, Journal of Proteome Research, and Proteomics were examined for the year 2009 and papers in which authors had presented lists of protein spots comparing different treatments using the quantitative proteomics 2-DE technique were identified and examined. The studies in these papers were thus candidates for the application of multiple hypothesis testing methods. Much smaller samples of papers were taken for 2010, mainly to confirm that there has not been any recent substantial change in behavior in relation to use of multiple hypothesis testing methods. The results of the survey are presented in Table II. A large majority of the papers (89.2%) did not use multiple hypothesis testing methods. This pattern is consistent across journals and years. Those that did use multiple hypothesis testing, mainly used FDR or q-value methods. The low implementation of multiple hypothesis testing methods was not through lack of use of statistical methods generally, for the majority of papers (92.8%) used α = 0.05 or α = 0.01 criteria for declaring spots as having significant expression differences between treatments. In about half of these, fold change, the ratio of the expression between treatments, was used as an associated criterion, although fold change was seldom used alone. It would be difficult to argue from the results of the survey that it is superfluous to further promote the application of multiple hypothesis testing methods on the basis that these methods are currently widely used in proteomics work.
Table II. Numbers of papers published in three proteomics journals assessed for usage of multiple hypothesis testing procedures and other statistical approaches in quantitative proteomics using two dimensional electrophoresis (2-DE).
| JOURNAL |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mol. Cell. Proteomics |
J. Proteome Res. |
Proteomics |
Total |
||||||
| 2009 | 2010 | 2009 | 2010 | 2009 | 2010 | n | % | ||
| Number of 2-DE papers reviewed (n) | n = 21 | n = 4 | n = 50 | n = 10 | n = 71 | n = 10 | 166 | – | |
| Did not use multiple hypothesis test correction? | 17 | 4 | 44 | 10 | 64 | 9 | 148 | 89.2 | |
| Did use multiple test correction? | 4 | – | 6 | – | 7 | 1 | 18 | 10.8 | |
| aBH (FDR)/q-value | 2 | – | 4 | – | 6 | 1 | 13 | 72.2 | |
| bBonferroni (SB) | – | – | 2 | – | 1 | – | 3 | 16.7 | |
| Others | 2 | – | – | – | – | – | 2 | 11.1 | |
| Some statistical analysis carried out | 18 | 4 | 47 | 9 | 66 | 10 | 154 | 92.8 | |
| No statistical analysis carried out | 3 | – | 3 | 1 | 5 | – | 12 | 7.2 | |
| Threshold method for declaring spots significant | cα = 0.05 | 3 | – | 18 | 2 | 44 | 4 | 71 | 42.8 |
| α = 0.01 | – | – | 5 | – | 3 | – | 8 | 4.8 | |
| dFold change (FC) alone | 3 | – | 3 | 1 | 5 | – | 12 | 7.2 | |
| α = 0.05 + FC | 15 | 4 | 24 | 7 | 16 | 5 | 71 | 42.8 | |
| α = 0.01 + FC | – | – | – | – | 3 | 1 | 4 | 2.4 | |
a (BH) Bejamini and Hochberg 1995, (FDR) False Discovery Rate, q-value (Storey and Tibshirani 2003, Storey 2003) or both.
b Bonferroni procedure, including sequential Bonferroni (SB) (Holm 1979).
c Threshold criterion used for assessing p value lists (other α levels might have been used for some other purposes).
d Fold change (FC) defined as the larger treatment mean divided by the smaller treatment mean for a protein spot.
Case Scenarios
To demonstrate how different thresholds might be implemented depending on scientific priorities we consider next some examples of scenarios that might be faced in proteomics studies. Use of model data has the advantage that we can see the true effect sizes though this would not of course be known for experimental data. Suppose that a proteomics experiment is set up to identify a protein marker that can separate cancer and normal cells with high confidence. Investigation of targets might be expensive in resources, which would be wasted investigating false positives. In this situation the threshold could be drawn near the top of the list of ordered p values, and SB might be fine for positioning it. Consider Table I as example. The three spots above the threshold all have the maximum treatment effect size of two, thus the strategy would be effective in this case. If we wished to be a little less conservative, the BH 5% threshold could be chosen, which identifies 12 significant spots. It is reassuring that the SFisher and SGoF methods give low meta p values for these spots. The computed q-value is even more reassuring than the BH 5% with values of 0.036 or less, suggesting that only a proportion 0.036 of the 12 spots are false positives. An intermediate position might be to choose say the top five spots which have even lower q-values of 0.017 or less. For the simulated data of Table I, 9 of the 12 spots above the BH 5% threshold have the largest treatment effect size in the model and only one, spot 11, is a false positive.
A contrasting scenario could be that of an exploratory study to identify biochemical pathways involved in adaptation of molluscs to an environmental stress, comparing with a control environment. Here we might want a liberal threshold placed further down the table and thus declare many more spots as significant targets for further investigation. Subsequent protein identification of only a subset of these might give clues about potential pathways of interest. One possibility would be to set the threshold at BH 20% which includes 62 spots in Table I. Of these, 11 have zero effect in the model and thus are false positives, a number roughly in line with expectation for BH 20%. An alternative could be to use the approach of selecting a threshold and calculating FDR adjusted probability for the region above it. For example, suppose the arbitrary threshold in Table I marked off a convenient region because resources were available for picking and identifying roughly 70 protein spots. As explained above, a proportion 0.212 of false positives are expected in this region above the threshold using FDR adjusted probability. This is potentially useful, for although the level of FDR control has only changed from 20% to 21.2%, eight more spots are included about the threshold. Targeting a few positives that unbeknown to the investigator are false might not be a problem if this minority are irrelevant to an emerging picture of important biochemical pathways.
If we accept α = 0.05 as the appropriate significance level for a single spot then a very liberal position would be to draw the threshold below all spots having a p value of less than 0.05, a total of 88 in Table I. The FDR adjusted probability and q-value at this threshold increase to 0.278 and 0.224 respectively, the difference between the two values arising from the different assumptions of these methods. As these values increase as we move down the table, the increasing number of false positives included above the threshold gives the potential for confusing any emerging pathway picture, though in the simulated data the number of false positives included happens to be only 14 out of the 88. Given that q-values are available to aid decision we could consider being even more liberal. For example, although not shown in Table I, there are 120 spots with a q-value of 0.325 or less, but we might reflect at this point on whether too many false positives are being included above the threshold.
The next scenario considers a model with a smaller average difference between treatment groups. In this model, the standard deviation within treatment groups was set to 1, as before. For one of the treatment groups, the mean value was set to 0 for all biological replicates. For the second treatment group, for all biological replicates, the spot means were set to 1 and 0 for 100 and 400 spots respectively. An example of simulated data from this model is shown in Table III. A total of 48 spots are significant at the level α = 0.05, an excess of 23 over the 25 expected for null data with all spots having effect size = 0. The p values are too high to draw thresholds for SB, BH 5%, and BH 20%. Thus these methods are not useful in providing evidence of a treatment effect. However the SGoF(0.05) and SFisher give 13 and 20 p values, respectively, less than α = 0.05. Thus these spots can be declared significant at this meta test α level for these methods. The q-value in the range 0.325–0.428 suggests that roughly one third are false positives, compared with a proportion of true nulls of 400/500 = 0.8 in the dataset as a whole. Thus picking a selection of spots because they are above these thresholds is certainly better than picking spots at random from the 500. In order to decide which of the meta test results to give priority to, the Fisher approach to statistical inference, which uses the magnitude of the p value as a measure of the strength of the evidence against the null hypothesis (1), could be implemented. Doing this, the 13 p values above the SGoF(0.05) threshold would be favored as candidates for further work. In reality, the proportion of spots with effect size of 1 above the SGoF(0.05) and SFisher(0.05) thresholds in Table III is 6/13 = 0.46 and 12/20 = 0.60, respectively. Thus unfortunately, in these data, applying the Fisher approach and focusing on the 13 spots above the SGoF(0.05) threshold, because they have lower p values, would actually be less fruitful in identifying spots with treatment effects than using the SFisher(0.05) threshold. For the analysis as a whole, we can conclude that treatment effects have been demonstrated that might guide further studies, but progress to advance biochemical or physiological understanding through spot picking and mass spectrometric analysis might be hindered by presence of the false positives.
Table III. Simulated results of a model of a proteomics experiment with two treatments and 500 protein spots.
The standard deviation (biological error) within each treatment group was set to 1 for all spots. Treatment effect sizes are, 100 spots effect = 1, 400 spots effect = 0. Column headings are as in Table 1. The table has a break incorporating spots 30–46 to save space.
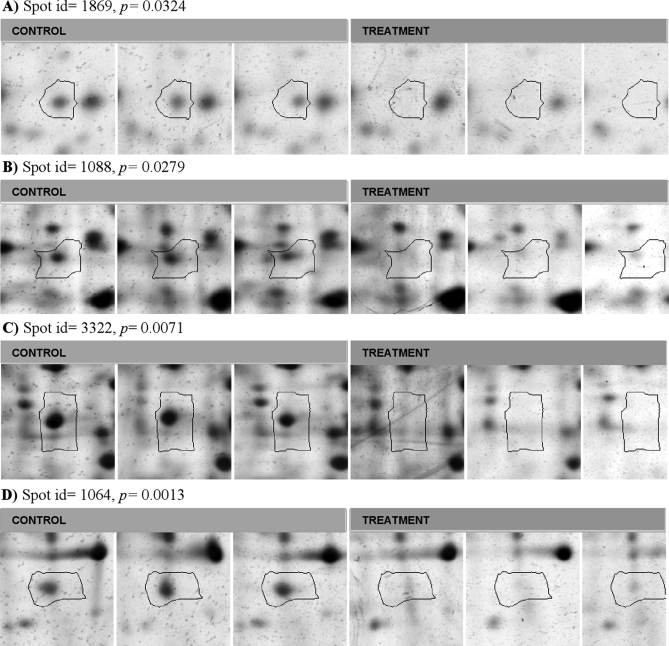
We conclude this section with an example of real experimental data, from a study of a gastropod mollusc exposed to two different environments (treatment and control) with three biological replicates per treatment (Diz A. P. et al. unpublished data). A total of 549 spots were analyzed using the Progenesis SameSpots 2-DE image analysis software from Nonlinear Dynamics Ltd., and of these, 125 were significant at α = 0.05 using the t test, many more than the null expectation of 549 × 0.05 = 27. After applying SB (0.05), BH 5%, and SGoF(0.05) multiple hypothesis testing methods, respectively 1, 1, and 87 spots were declared significant. The first two methods have clearly eliminated many spots for which the null hypothesis is false, and are thus not very useful. SGoF(0.05) is clearly better but eliminates 125−87 = 38 of those spots initially significant at the α = 0.05 level. A montage from Progenesis SameSpots of the six individuals for each of four of these 38 spots is shown in Fig. 1 with p values indicated. Visually all these spots seem to be very different on average between the two treatments, especially when we weigh in our minds the biological variation, which appears relatively small. Psychologically it seems hard to accept that the above three multiple hypothesis testing methods are providing a useful service by excluding these 38 spots. However these spots are found to be significant with SFisher(0.05). Given our prior intention to make use of this multiple hypothesis testing method, we have a justification for including the 38 spots as targets for further work. This decision receives support from consideration of the q-value, which is 0.089 for the 125 spots and 0.075 for the 87 spots, that are significant with SGoF(0.05). Both values could be considered fairly similar in value and satisfactorily low, justifying selection of the larger set of 125 spots for further work.
Fig. 1.
Example of some protein spots (Diz A. P. et al. unpublished data) from a 2-DE analysis from a gastropod mollusc. There were three biological replicates for each of two treatments. Significant p values at α = 0.05 or lower for comparison of treatments with a t test are shown. These spots were not declared significant after applying SB (0.05), BH 5%, and SGoF (0.05). Image analysis and montage preparation were carried out with Progenesis SameSpots versus 4.0 software.
Software for Application of Methods
The multiple hypothesis testing methods described in this paper can be carried out using the SGoF software (20). The QVALUE software (14, 15, 23) provides many alternative options for computing q-values. The Multitest V1.2 software (21) performs the exact binomial test on lists of p values. A useful table that illustrates application of Bonferroni and FDR methods, and that can be adapted in a spreadsheet is given in Fig. 1 of Verhoeven et al. (11). The application of the step-up approach can be seen in Tables I and III by comparing values in the BH 5% and BH 20% columns just above and below the respective thresholds with the p value column. A Supplemental Table in an Excel spreadsheet table, inspired by the Verhoeven et al. (11) table, has been provided which implements methods discussed in this paper. The spreadsheet cells show the formulas and functions needed. The table can be modified and other p value lists pasted in for analysis. We think the table has some didactic value even if in normal practice the methods could be executed by the dedicated software described above. The q-value is excluded from the spreadsheet as the computation is more complicated and this method is best executed using the dedicated software. The Supplemental Table also includes a glossary of some of the terms and methods discussed in this paper.
DISCUSSION AND CONCLUSIONS
This paper presents a selection of multiple hypothesis testing methods and reviews how they might be applied using case scenarios. We hope that this work will help to promote the use of multiple hypothesis testing methods among those researchers who do not currently use them. It has not been our intention to be prescriptive about precisely which methods should be used. Rather we emphasize the strategy of applying several different multiple testing methods to the data in hand. We have suggested appropriate software to apply these methods. A useful strategy might thus be to prepare a table similar to Table I. This could be included as supplementary information to a publication if appropriate. Then, summary tables could be given in the paper with the numbers of features such as proteins spots declared significant by the different methods (e.g. 24, 25). The next stage would be to provide a discussion along the lines of that used here for the case scenarios, giving emphasis to particular methods depending on research priorities and position of thresholds. For example, where some surety is needed that significant features do reflect treatment effects, the SB method might be augmented by a consideration of q-values. Where it is possible to be more liberal, the sequential methods SGoF and SFisher might be used, or FDR control might be set at a level such as BH 5%. In exploratory studies, FDR could be set at a more liberal level such as BH 20%, or FDR adjusted probabilities or q-values could be determined for a specific region such as the threshold marking off all p values significant at α = 0.05. The availability of a table similar to Table I would also allow the readers of the paper to carry out easily their own assessment of the significance of the results.
Other important statistical considerations relating to the use of lists of p values in proteomics research should be mentioned. In the datasets of Table I and Table III, the number of spots above the different thresholds falls far short of the actual number of spots with an effect in the models used. There are many false negatives below the thresholds and the number of true null hypotheses is being over estimated. Several estimation methods are available for experimental data for the proportion of true null hypotheses in a list of p values (e.g. 15, 26, 27). For the 500 spots of the Table I model, the proportion estimated by the Storey and Tibshirani (15) method is 0.79, somewhat greater than the actual 350/500 = 0.7. For the data of the Table III model the estimate is 0.92 also greater than the actual 400/500 = 0.8. An estimated excess of the proportion of true null hypotheses must be because of the use of only five biological replicates per treatment, there is a lack of power. In the design of quantitative proteomics experiments it is important to distinguish between biological and technical replication. Biological replicates would be different individual or pooled organisms allocated among different treatment groups (see 28). Technical replicates occur when the same biological replicate is repeated on different gels. Variation between biological replicates should be tested for significance against variation between technical replicates: variation between treatments should be tested against variation between biological replicates. Thus when testing for treatment differences, priority should be given to maximizing the number of biological replicates to optimize power (29). A further consideration in this context is that a limited number of biological replicates will also affect the precision of p values. This has most severe consequences for the SB where the p values required for FWER = 0.05 for individual spots need to have precision to many decimal places in large datasets (20). If resources permitted, one approach to increasing power would be to repeat the entire experiment, and attempt to confirm or eliminate positive and negative results as true or false.
The use of the q-value is dependent on the assumptions made regarding the distribution of p values. Under the null hypothesis of no treatment effect, a uniform distribution is expected (15, 30). However aspects of experimental design or inappropriate statistics for estimating p values can result in nonuniform statistics that should be taken into consideration (e.g. 7, 31, 32). Many variant tests for combining probabilities or applying the FDR approach have been examined and compared in relation to underlying assumptions and statistical properties (see 7, 18, 33). An important consideration is whether the p values under study are correlated or are independent. Fortunately, many FDR approaches are robust to deviations from the assumption of independence (e.g. see 7).
The results of the survey reported here indicate that use of fold change is a popular criterion for assessing spots. Fold change provides some information on effect size but cannot be used as a criterion for determining significance according to defined α levels. However given that p values confound effect size and precision (1, 4), fold change might provide useful supplementary information. For example, in Table III, spots 4–20 above the SFisher (0.05) threshold have the same q-value. If only a few of these can be selected for further investigation, then fold change might be used as an indicator of effect size as an alternative to using the p value as an indicator of the amount of evidence against the null hypothesis. This approach might be particularly appealing in circumstances where visual evidence strongly suggests a treatment effect as in the results shown in Fig. 1. Finally, further experimental work could also be used to confirm significance of spots above a liberal threshold. This could be done simply by repeating the experiment. However this method of updating the p values simply applies greater power, it does not replace the need for multiple hypothesis testing methods. Another attempt to bypass multiple hypothesis testing would be the in-depth investigation of individual proteins using techniques such as Western blotting (e.g. 34, 35). This would be analogous to confirming candidate transcripts using qRT-PCR in microarray work. But as Pan et al. (36) point out, this might involve a heroic effort if there are many target features, and initial use of multiple hypothesis testing methods would be sensible to narrow down candidates for further experimental work.
This paper has as its main focus the application of a multiple testing strategy to the list of p values obtained in quantitative 2-DE work. However multiple testing is also relevant to gel-free proteomics methods (see 37), and also of course in transcriptomics work where treatment differences in mRNA abundance are considered. Thus the strategy we outline is a generic one. For example, the strategy could be considered for use in any of those bottom-up or top-down quantitative proteomics methods in which p values are obtained for differences in abundance for mass spectrum features determined across samples or treatments. The strategy might also be assessed in protein identification with mass spectrometric methods where each p value in the list corresponds to a different target protein or peptide, and where FDR methods are feasible (see 37). New experimental proteomics methods continue to be developed. Multiple testing strategies could be pertinent to any of these provided that the method generates lists of p values for the features under study.
Acknowledgments
We would like to thank Uña-Álvarez J. and Rolán-Álvarez E. for comments on this manuscript.
Footnotes
* This work was supported in part by Ministerio de Ciencia e Innovación and Fondos Feder (code CGL2009-13278-C02) and Xunta de Galicia (codes INCITE09 310 006PR and 2010/80), Spain. We thank also the Institute of Life Sciences Swansea, College of Medicine, Swansea University (UK) for resources to help meet some costs of this research. APD and AC-R are currently funded by “Isidro Parga Pondal” research fellowships from Xunta de Galicia (Spain).
 This article contains supplemental Table.
This article contains supplemental Table.
1 The abbreviations used are:
- 2-DE
- two-dimensional electrophoresis
- FDR
- false discovery rate
- FWER
- family-wise error rate
- SB
- sequential Bonferroni method
- BH
- Benjamini and Hochberg method
- SFisher
- sequential combined probability test of Fisher
- SGoF
- sequential goodness of fit.
REFERENCES
- 1. Stang A., Poole C., Kuss O. (2010) The ongoing tyranny of statistical significance testing in biomedical research. Eur. J. Epidemiol. 25, 225–230 [DOI] [PubMed] [Google Scholar]
- 2. Lieberman M. D., Cunningham W. A. (2009) Type I and Type II error concerns in fMRI research: re-balancing the scale. Soc. Cogn. Affect. Neurosci. 4, 423–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosnow R. L., Rosenthal R. (1989) Statistical procedures and the justification of knowledge in psychological science. Am. Psychol. 44, 1276–1284 [Google Scholar]
- 4. Rothman K. J. (2010) Curbing type I and type II errors. Eur. J. Epidemiol. 25, 223–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones L. V., Tukey J. W. (2000) A sensible formulation of the significance test. Psychol. Methods 5, 411–414 [DOI] [PubMed] [Google Scholar]
- 6. Gillies D. (2000) Philosophical theories of probability. Routledge. 240 pages [Google Scholar]
- 7. Pounds S. B. (2006) Estimation and control of multiple testing error rates for microarray studies. Brief. Bioinform. 7, 25–36 [DOI] [PubMed] [Google Scholar]
- 8. Hood G. M. (2000) PopTools, Version 3.0.3. Available on the internet [Google Scholar]
- 9. Holm S. (1979) A simple sequentially rejective multiple test procedure. Scand. J. Statist. 6, 65–70 [Google Scholar]
- 10. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 11. Verhoeven K. J. F., Simonsen K. L., McIntyre L. M. (2005) Implementing false discovery rate control: increasing your power. Oikos 108, 643–647 [Google Scholar]
- 12. Yekutieli D., Benjamini Y. (1999) Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J. Stat. Plan. Infer. 82, 171–196 [Google Scholar]
- 13. Reiner A., Yekutieli D., Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 14. Storey J. D. (2002) A direct approach to false discovery rates. J. Roy. Stat. Soc. B 64, 479–498 [Google Scholar]
- 15. Storey J. D., Tibshirani R. (2003) Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Storey J. D. (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 [Google Scholar]
- 17. Fisher R. A. (1932) Statistical Methods for Research Workers. Oliver and Boyd, Edinburgh [Google Scholar]
- 18. Whitlock M. C. (2005) Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 18, 1368–1373 [DOI] [PubMed] [Google Scholar]
- 19. Koziol J. A., Tuckwell H. C. (1994) A weighted nonparametric procedure for the combination of independent events. Biom. J. 36, 1005–1012 [Google Scholar]
- 20. Carvajal-Rodríguez A., de, Uña-Alvarez J., Rolán-Alvarez E. (2009) A new multitest correction (SGoF) that increases its statistical power when increasing the number of tests. BMC Bioinformatics 10, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Meeûs T., Guégan J. F., Teriokhin A. T. (2009) MultiTest V. 1.2, a program to binomially combine independent tests and performance comparison with other related methods on proportional data. BMC Bioinformatics 10, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dudoit S., van der Laan M. J. (2008) Multiple testing procedures with applications to genomics. Springer, New York, pp. 588 [Google Scholar]
- 23. Storey J. D., Taylor J. E., Siegmund D. (2004) Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J. Roy. Stat. Soc. B 66, 187–205 [Google Scholar]
- 24. Walker S. J., Wang Y., Grant K. A., Chan F., Hellmann G. M. (2006) Long versus short oligonucleotide microarrays for the study of gene expression in nonhuman primates. J. Neurosci. Meth. 152, 179–189 [DOI] [PubMed] [Google Scholar]
- 25. Diz A. P., Dudley E., MacDonald B. W., Piña B., Kenchington E. L., Zouros E., Skibinski D. O. F. (2009) Genetic variation underlying protein expression in eggs of the marine mussel Mytilus edulis. Mol. Cell. Proteomics 8, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dalmasso C., Broet P., Moreau T. (2005) A simple procedure for estimating the false discovery rate. Bioinformatics 21, 660–668 [DOI] [PubMed] [Google Scholar]
- 27. Pounds S., Cheng C. (2006) Robust estimation of the false discovery rate. Bioinformatics 22, 1979–1987 [DOI] [PubMed] [Google Scholar]
- 28. Diz A. P., Truebano M., Skibinski D. O. F. (2009) The consequences of samples pooling in proteomics: an empirical study. Electrophoresis 17, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 29. Horgan G.W. (2007) Sample size and replication in 2D gel electrophoresis studies. J. Proteome Res. 6, 2884–2887 [DOI] [PubMed] [Google Scholar]
- 30. Kerr K. F. (2009) Comments on the analysis of unbalanced microarray data. Bioinformatics 25, 2035–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Akey J. M., Biswas S., Leek J. T., Storey J. D. (2007) On the design and analysis of gene expression studies in human populations. Nat. Genet. 39, 807–808 [DOI] [PubMed] [Google Scholar]
- 32. Karp N. A., McCormick P. S., Russell M. R., Lilley K. S. (2007) Experimental and statistical considerations to avoid false conclusions in proteomics studies using differential in-gel electrophoresis. Mol. Cell. Proteomics 6, 1354–1364 [DOI] [PubMed] [Google Scholar]
- 33. Zaykin D. V., Zhivotovsky L. A., Czika W., Shao S., Wolfinger R. D. (2007) Combining p-values in large-scale genomics experiments. Pharm. Stat. 6, 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffrogge R., Beyer S., Hübner R., Mikkat S., Mix E., Scharf C., Schmitz U., Pauleweit S., Berth M., Zubrzycki I. Z., Christoph H., Pahnke J., Wolkenhauer O., Uhrmacher A., Völker U., Rolfs A. (2007) 2-DE profiling of GDNF overexpression-related proteome changes in differentiating ST14A rat progenitor cells. Proteomics 7, 33–46 [DOI] [PubMed] [Google Scholar]
- 35. Roth U., Razawi H., Hommer J., Engelmann K., Schwientek T., Müller S., Baldus S. E., Patsos G., Corfield A. P., Paraskeva C., Hanisch F. G. (2010) Differential expression proteomics of human colorectal cancer based on a syngeneic cellular model for the progression of adenoma to carcinoma. Proteomics 10, 194–202 [DOI] [PubMed] [Google Scholar]
- 36. Pan J. Z., Eckenhoff R. G., Eckenhoff M. F. (2007) Limitations of microarray studies. Anesth. Analg. 104, 1300–1301, Author reply 1301–1302 [DOI] [PubMed] [Google Scholar]
- 37. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]