Abstract
S-nitrosation (SNO) of mitochondrial protein cysteines can be cardioprotective. Several targets have been implicated, yet the scope and identification of specific residues has not been fully assessed. To address this, a comprehensive assessment of mitochondrial SNO-modifiable cysteines was performed to determine nitric oxide (NO) susceptible pathways and identify novel mechanisms of oxidative cardioprotection. The biotin switch assay and mass spectrometry were used on rat cardiac mitochondrial lysates treated with the nitric oxide donor, S-nitrosoglutathione, and controls (n = 3) to map 83 SNO-modified cysteine residues on 60 proteins. Of these, three sites have been reported, 30 sites are new to 21 proteins previously known to be S-nitrosated but which lacked site-specific information and 50 sites were found on 39 proteins not previously implicated in SNO pathways. The SNO-modifications occurred in only a subset of available cysteines, indicating a specific targeted effect. Functional annotation and site-specificity analysis revealed a twofold greater nitric oxide-susceptibility for proteins involved in transport; including regulators of mitochondrial permeability transition suggesting SNO-regulation and a possible protective mechanism. Additionally, we identified many novel SNO-modified proteins with cardioprotective potential involved in the electron transport chain, tricarboxylic acid cycle, oxidative stress defense, fatty acid and amino acid metabolism. These findings suggest that SNO-modification may represent a novel mechanism for the regulation of oxidative phosphorylation and/or cell death. S-nitrosation of mitochondrial permeability transition-associated proteins represents an intriguing potential link to cardioprotection.
Oxidative protein modifications play an important role in the regulation and dysfunction of the cardiac mitochondria (1, 2). In particular, S-nitrosation (SNO)1, the reversible covalent addition of a nitrosonium (NO+) to a cysteine thiol, is emerging as a key cellular mediator (3, 4). Several recent studies have identified SNO as an important reversible signaling modality in the mitochondria (5–9). Specifically, SNO has been observed on several proteins comprising important mitochondrial pathways including oxidative phosphorylation (5–8, 10) the tricarboxylic cycle (6), and fatty-acid oxidation (6, 10). It is beginning to be understood that modification of such pathways through SNO can result in the alteration of mitochondria function. Two major contexts in which mitochondrial SNO has been studied are the phenomena of preconditioning and ischemia/reperfusion injury (10–13), particularly in the cardiac system. Cardiac preconditioning is a phenomenon by which pretreatment of the heart with either brief episodes of ischemia or pharmacological stimuli can protect against damage during future ischemic episodes (14). This cardioprotective phenomenon has been extensively linked to mitochondria and preservation of mitochondrial function during ischemia (15, 16). Thus, it is possible that SNO of critical mitochondrial proteins serves to protect them from the oxidative damage caused by ischemia, preserving their function upon reperfusion (11).
Although the general effects of SNO on mitochondrial proteins have been studied, only a few modified proteins have been identified and fewer still have had the position of the modified amino acid residue(s) determined. The identification of SNO-proteins and the specific modified cysteine(s) can be challenging because of their labile nature. SNO-modifications are sensitive to UV light, reducing conditions (especially in the presence of heavy metals), and are difficult to observe by mass spectrometry (MS) (17). The biotin switch assay, first described in 2001 (18, 19), has emerged as a viable alternative for detection. In the biotin switch assay, SNO-modifications are replaced with a biotin group by first blocking all remaining free thiols and then reducing the SNO-sites with ascorbate and relabeling with a thiol reactive biotin (usually (N-(6-(Biotinomide)hexyl)-3′-(z′-pyridyldithio)-propionamide)). Using this system, labile SNO-sites are tagged with a stable group that can be easily detected and enriched. This method has been successfully coupled with MS in several large-scale SNO-site identification studies (20, 21). A common strategy has been to investigate modifications induced by the application of exogenous nitric oxide (NO)-donors. Small molecule thiols such as S-nitrosoglutathione (GSNO) or SNO-Cys are often used because of the high thiol content of the cell. The precise mechanism of SNO-modification has not been conclusively determined but a leading theory is transnitrosation, the transfer of an NO-moiety between thiols, from either a small molecule (such as glutathione (GSH)) or other proteins (3, 22). GSNO has been found induce preconditioning like effects and modify mitochondrial proteins (10). Broad-scale mapping of mitochondrial SNO-sites is needed to generate new insights into the mechanism of cardioprotection and the regulation of mitochondrial function.
To that end, rat cardiac mitochondrial enriched lysates were treated with GSNO to identify SNO-modifiable proteins using the biotin switch assay and MS. Analysis of the SNO-site susceptibility and functional pathways indicated proteins involved in transport and energy production as possible targets of SNO-regulation. The site-specific identification of SNO-modified cysteines is critical for a deeper understanding of possible regulatory and cardioprotective roles of SNO.
EXPERIMENTAL PROCEDURES
Isolation of Cardiac Mitochondria
Cardiac mitochondria were enriched from rat ventricles according to the protocol outlined in references (23, 24) optimized for cardiac tissue (25). This preparation has previously been shown to be enriched for mitochondria and depleted of myofilament proteins (25). Two independent preparations of 10 and 50 frozen rat hearts (Pel-Freez Biologicals, www.pelfreez-bio.com) were preformed. Hearts were pooled, weighed, and then pulverized in liquid nitrogen. The pulverized tissue was homogenized in 8× volume of mitochondrial isolation buffer (220 mmol/L mannitol, 70 mmol/L sucrose, 2 mmol/L HEPES, pH 7.4 with KOH). All further steps were carried out at 4 °C. Homogenate was centrifuged at 1100 × g for 5 min to remove nuclei and unbroken cells. The supernatant was saved and the pellet washed three times in 1 mL mitochondrial isolation buffer and centrifuged at 1100 × g for 5 min. All supernatants were combined and then centrifuged at 10,000 × g for 15 min to collect crude mitochondria. The crude mitochondrial pellet was then washed twice in 1 mL of mitochondrial isolation buffer and spun at 20,000 × g for 15 min. The crude mitochondrial pellet was then resuspended in 1 mL mitochondrial isolation buffer and was centrifuged at 3000 × g to remove aggregated and myofilament connected mitochondria. This step was repeated for efficient removal of myofilaments (25). Both supernatants were combined and centrifuged at 20,000 × g for 20 min to collect the final mitochondrial pellet. This pellet was flash frozen in liquid nitrogen and stored at −80 °C until further analysis.
Detection of S-Nitrosated Proteins by Biotin Switch Assay
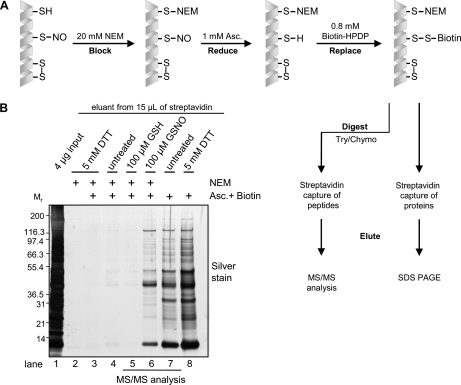
SNO-modifications were detected in GSNO treated mitochondrial lysates using the biotin switch assay described in (21) (discussed in (17, 19, 21, 26)), see Fig. 1A for reaction schema). Mitochondrial enriched preparations were diluted to 0.8 μg/μl in HEN (250 mmol/L HEPES pH 7.7, 1 mmol/L EDTA and 0.1 mmol/L neocuproine) including 0.4% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid (CHAPS) and treated with 100 μmol/L GSNO or three different control treatments (100 μmol/L GSH, 5 mmol/L dithiotreitol (DTT) or untreated vehicle) for 15 min at 37 °C. All steps were performed in the dark or protected from light. Treatment compounds were removed using an HEN equilibrated 5 mL Zeba desalt spin column (Thermo Fisher Scientific, www.thermofisher.com) according to the manufacturer's protocol. The remaining free thiols were blocked with 20 mmol/L NEM in the presence of 2.5% (w/v) SDS and incubated for 20 min at 50 °C. Excess NEM was removed by acetone precipitation. SNO-modified thiols were reduced using 1 mmol/L ascorbate in five volumes of HENS (HEN + 1% (w/v) SDS) and labeled with 0.8 mmol/L Biotin-HPDP (Thermo Fisher Scientific) for 1 hour at room temperature. Excess biotin-HPDP was removed by acetone precipitation (two volumes) and the resultant pellets were carefully washed with an additional volume of acetone. Pellets were resuspended to 5 μg/μL with HENS and either digested directly for MS studies or analyzed by gel electrophoresis. For gel based studies, 250 μg of labeled protein was diluted 20× in neutralization buffer (20 mmol/L HEPES, 150 mmol/L NaCl 1 mmol/L EDTA, 0.5% (v/v) Triton X-100). Biotinylated proteins were captured by incubation with 15 μL of washed, packed ultralink immobilized streptavidin beads (Thermo Fisher Scientific) for 1 hour at room temperature. Beads were washed four times in 50-bead volumes of wash buffer (20 mmol/L HEPES, 600 mmol/L NaCl 1 mmol/L EDTA, 0.5% (v/v) Triton X-100) and twice with elution buffer (20 mmol/L HEPES pH 7.7, 100 mmol/L NaCl, 1 mmol/L EDTA). Captured proteins were eluted with 40 μL of elution buffer containing 100 mmol/L DTT, mixed with 15 μL of 4× LDS sample buffer, boiled, separated by SDS-PAGE (4–12%) and silver stained according to the protocol described in (27).
Fig. 1.
Detection and site mapping of cardiac mitochondrial SNO-modifications. A, Biotin switch assay schema outlining the blocking, reducing and biotin labeling steps as well as capture of intact proteins or digested peptides for LC/MS/MS (19, 21). B, Representative silver stained gel of SNO-modified proteins captured from 250 μg of rat cardiac mitochondria treated with 100 μmol/L GSNO, 100 μmol/L GSH, 5 mmol/L DTT or untreated vehicle with and without NEM blocking and subjected to the biotin switch assay (n = 3). A description of each of the control treatments can be found in the text.
Mass Spectrometry
For MS studies, 1 mg of GSNO-, GSH-treated and untreated positive control biotin switch samples were diluted 50-fold with ddH2O, divided and digested overnight with 10 μg of trypsin (Promega, Fitchburg, WI, www.promega.com) or chymotrypsin (Roche Applied Science, www.roche.com) (n = 3 for each treatment with each enzyme). Digestions were halted by 0.25 mmol/L phenylmethylsulfonyl fluoride and then biotinylated peptides were captured with 30 μL of streptavidin as described above. To ensure MS compatibility, beads were washed an additional ten times with 5 mmol/L ammonium bicarbonate/20% acetonitrile before being eluted in the same solution containing 100 mmol/L DTT. Eluted peptides were dried and then desalted using an Omix C18 tip column following the manufacture's protocol (Varian, www.varianinc.com). For identification of captured peptides, samples were resuspended in 10 μL of 0.5% (v/v) formic acid. Peptide identification by liquid chromatography/tandem mass spectrometry (LCMS/MS) analysis was performed using an LTQ ion trap MS (Thermo Fisher Scientific, www.thermofisher.com) interfaced with a two-dimensional nanoLC system (Eksigent, Dublin, CA, www.eksigent.com). Peptides were desalted on a C18 trap (75 μm × 3 cm, 5–10 μm, 120Å, YMC Gel) at 8 μL/min for 5 min with Buffer A (0.1% formic acid). After desalting, peptides were separated on a C18 column (75 μm × 10 cm, 5 μm, 120Å, YMC ODS-AQ, Waters, Milford, MA, www.waters.com) with an 8 μm emitter tip (New Objective, Woburn, MA, www.newobjective.com) using 5%–60% B (90% acetonitrile in 0.1% formic acid) gradient over 60 min at 300 nL/min. Each MS1 scan was followed by collision induced dissociation of the five most abundant precursor ions with dynamic exclusion for 30 s. MS2 spectra were acquired in normal scan mode using a target setting of 104 ions and an accumulation time of 30 ms.
Data Analysis
MS data was searched against the International Protein Index rat primary sequence database (v3.62, 39924 entries) (28) using Sorcerer 2™-SEQUEST® (version v.27, rev. 11) (Sage-N Research, Milpitas, CA) with postsearch analysis performed using Scaffold 2 (Proteome Software Inc., Portland OR, www.proteomesoftware.com) and the trans-proteome pipeline implementing the PeptideProphet (29) and ProteinProphet (30) algorithms. All raw data peak extraction was performed using the Sorcerer 2™-SEQUEST® default settings. Search parameters included semi-enzyme digest with either trypsin (after Arg or Lys) or chymotrypsin (after Phe, Trp, Try, or Leu) with up to 2 missed cleavages. SEQUEST® was searched with a parent ion tolerance of 1.2 Da and a fragment ion mass tolerance of 1.00 Da with N-ethylmaleimide-modified of cysteine specified as a differential modification. Peptide identifications were accepted if they had an Xcorr score greater than 2.0, a probability greater than 80.0% as specified by the PeptideProphet algorithm (29), and met with visual validation. Protein identifications were accepted if they could be established at greater than 95.0% probability as assigned by the ProteinProphet algorithm (30) and contained at least 1 identified peptide. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. False discovery rates were calculated by the ProteinProphet algorithm (30). The data presented in Supplementary Table S1 contains only those peptides that were present in the GSNO-treated samples containing a cysteine residue and were absent from the GSH-treated samples. By subtracting out any peptides detected in the GSH samples we were able to control for possible reduction of glutathionylation, endogenous disulfide bonds or pre-existing oxidative thiol modification. The position of each modified cysteine was determined by mapping the peptide against the full protein sequence. In some instances where more than one cysteine is present in the identified peptide, the site of SNO-modification could not be unambiguously determined.
Determination of Protein Localization and Function
Protein localizations and function were determined by annotation in the UniProt database (www.uniprot.org). Only proteins annotated as mitochondrial or mitochondrial associated were considered for potential SNO-modifications.
Consensus Sequence Analysis
50 out of 83 SNO-modified sites were randomly chosen and 20 residues on either side of the nitrosated cysteines were selected. After alignment with ClustalW2 (www.ebi.ac.uk/Tools/clustalw2/index.html), the sequences were screened for conserved motifs using the CONSENSUS, TEIRESIAS, and PRATT algorithms (http://coot.embl.de/Alignment/consensus.html; http://cbcsrv.watson.ibm.com/Tspd.html; www.ebi.ac.uk/Tools/pratt/index.html), respectively (31). Frequencies of flanking residues were computed with WebLogo (http://weblogo.berkeley.edu/). For secondary structure and solvent accessibility predictions the NetSurfP server was employed (www.cbs.dtu.dk/services/NetSurfP/) (32).
RESULTS
Detection of SNO-Modified Proteins
The biotin switch assay was used to reveal the presence of NO-induced modifications on proteins from GSNO treated mitochondria lysates (Fig. 1B). Silver stain gel analysis shows a clear increase in the amount of protein labeled following GSNO treatment when compared with GSH or the other negative control samples, indicating the modification of these proteins by NO (n = 3). The absence of any detectable signal in the unbiotinylated or DTT pretreated samples (Fig. 1B lanes 2 and 3) demonstrates effective and specific capture of biotinylated proteins and NEM blocking of unmodified thiols. Faint bands could be observed in the untreated and GSH treated samples (Fig. 1B lanes 4 and 5), and may be possible residual S-nitrosation or other oxidative modifications. Lane 7 represents untreated and unblocked mitochondria allowing for the labeling of the pool of available cysteines. As an additional positive control, DTT treatment was used to expose and label all cysteines, including those previously engaged in disulfide bonds. A marked increase in the extent and number of proteins labeled was seen in these conditions compared with SNO-modified proteins (Fig. 1B lanes 7 and 8). A complete reaction schema for each of the control and experimental biotin switch reactions is presented in Supplemental Fig. S1.
Determination of SNO-Modified Cysteine Residues
The MS analysis revealed a total of 83 sites of SNO on 60 different mitochondrial proteins (see Supplementary Table S1 for site information and detailed MS data; all proteins identified in the GSNO-biotin switch are listed in Supplementary Table S3). Of those, only three cysteine residues on three proteins, glyceraldehyde-3-phosphate dehydrogenase (Cys 154), isocitrate dehydrogenase 2 (Cys 336), and ADP/ATP translocase 1 (Cys 257) have previously been reported (21, 33, 34). A further 30 sites were identified on 21 proteins reported to be S-nitrosated, but which lacked site-specific information (see Supplementary Table S1 for references). The remaining 50 sites were found on 39 proteins not previously known to be S-nitrosated. The majority of sites (76) were identified in two or more of the replicates including several sites identified by independently using different enzymes (detailed MS data for peptide observations in each of the replicates is presented in Supplementary Table S1). For the seven peptides identified from a single spectrum, the annotated spectra are presented in Supplementary Table S2. False discovery rates for the GSH, GSNO, and positive control MS/MS experiments were determined to be between 0.4% and 1.2% across all experiments. Because of the nature of the biotin switch assay, no characteristic mass tag is left on the isolated peptides. Poor specificity in the streptavidin capture could result in identification of nonspecific cysteine containing peptides. To address this, the presence of noncysteine containing peptides was assessed following elution for each treatment condition as an estimate for the identification of nonspecific cysteine containing peptides (Supplementary Table S5). In each of the GSNO and available Cys data sets, a potential contamination rate of less than 1.4% was observed across all replicates. The contamination rate for GSH samples was slightly higher (10%) because of the smaller number of identified peptides, however a similar absolute number of contaminating peptides was observed. All spectra meeting the minimum Xcorr and probability standard were deposited in the PRIDE public database (http://www.ebi.ac.uk/pride/, accession numbers: 13087–13104.
Determination of SNO-Site Specificity
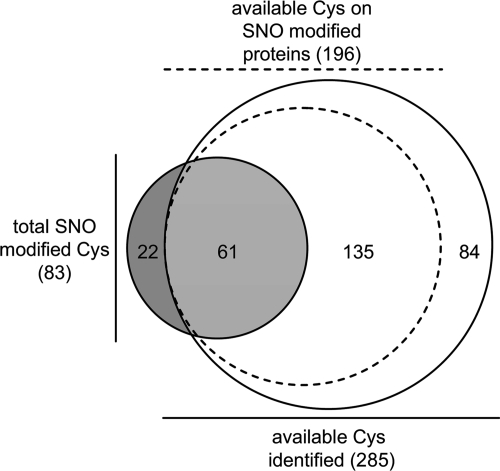
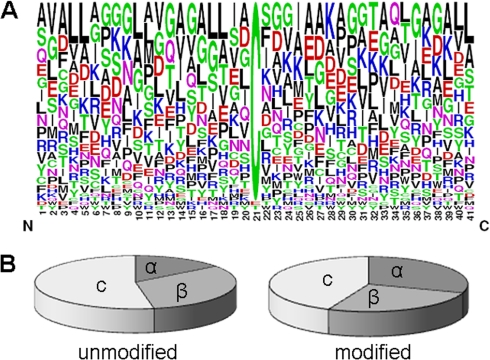
SNO-site mapping revealed considerable cysteine-specificity within a given protein. 80% (48 out of 60) of the SNO-modified proteins possessed a single modified site, despite the presence of multiple cysteine residues in the majority of proteins. To investigate further, an assessment was performed of all the available, nondisulfide bonded, cysteines in the untreated and unblocked samples. A total of 285 sites on 96 mitochondrial proteins were identified; 61 corresponded with SNO-sites identified in this study indicating that only ∼20% of the available mitochondrial cysteines are targets of SNO (Fig. 2; see Supplementary Table S4 for detailed MS data for peptides detected in each replicate). Analysis of the primary amino acid sequence surrounding the SNO-sites showed that these areas did not conform to the previously reported acid/base consensus motif (36) or any other novel consensus motifs (Fig. 3A). Further analysis of the predicted secondary structure immediately surrounding modified and unmodified cysteines indicated that SNO-modified sites might have a weak preference for alpha helical structures (Fig. 3B). It is possible that the receptive nature of these sites is conferred by the local three-dimensional structure although, in a recent study, this type of analysis on a subset of SNO sites failed to find any structural relationship (20).
Fig. 2.
Comparison of the SNO sites to all available cysteines in cardiac mitochondria reveals site-specificity. Identified sites are summarized in a Venn diagram. SNO-modified sites (dark gray) compared with all available cysteines (white). A total of 285 available cysteines were identified, 61 of those corresponded with SNO-modified sites indicating specificity in NO-modification (light gray). Among only the SNO-modified proteins, a total of 196 available sites were found (dashed line), 135 of which were found to be unmodified by NO. Also represented are the 22 SNO-sites not detected in the available cysteine data set. This absence is likely because of the increased complexity of the unblocked samples in the MS analysis.
Fig. 3.
Frequency of amino acids surrounding S-nitrosated cysteines and comparison of secondary structural elements adjacent to SNO-modified and unmodified cysteine residues. A, Analysis of the flanking sequences of 50 S-nitrosated cysteines, randomly chosen from the total of 84 detected modified sites, did not reveal any obvious pattern. However, comparison with a recent analysis on S-nitrosated targets in prostate epithelial cells (52) suggest a preference for aliphatic residues at positions −10 to −7; hydrophobic residues at positions −4 and −1, and two glycines at positions +2 and +3 relative to the modified cysteines. Hydrophobic residues are represented in black; charged residues in red and blue. Other residues are shown in either magenta or green. B, Comparison of the predicted secondary structures flanking nitrosated and unmodified sites might suggest a preference for reduced entropy as indicated by the elevated frequency of α-helix (30% versus 15%) and reduced frequency of coil (44% versus 53%). The modified cysteines appear to have a higher tendency to be buried (96% versus 80%), and show on average an ∼1.9-fold smaller predicted surface accessibility as compared with the set of unmodified cysteines. α: alpha-helix; β: beta sheet; c: coil.
Analysis of NO-Susceptible Functional Processes
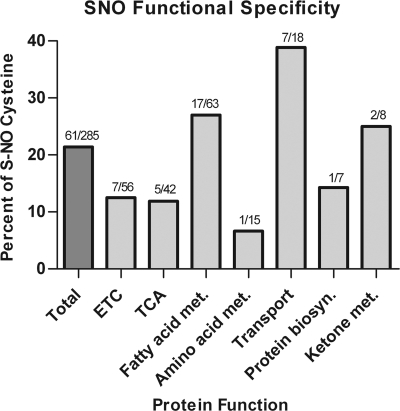
To better understand the implications of these SNO modifications and identify NO-reactive pathways, the results from the site-specificity analysis were combined with functional annotations from the Uniprot database (www.uniprot.org). Functional grouping of the identified proteins revealed a wide range of roles with greater than 60% of the SNO-modifications on proteins associated with energy production, with particular emphasis on proteins involved in fatty acid metabolism (24 sites on 12 proteins). Additionally, several sites of modification were found in proteins of the electron transport chain, the driving force for ATP production in the mitochondria. Of note, seven sites of modification were found on six different protein subunits contained in Complex I. SNO-modifications were also found on proteins involved in transport, signaling, chaperone/assembly, and structural support. Comparison of the SNO-functional groupings to the site-specificity analysis revealed a greater affinity for proteins involved in transport (Fig. 4). Transport protein cysteines were found to be SNO-modified at almost twice the extent of the average (∼40% versus ∼20% of the available cysteines). Conversely, mitochondrial proteins involved in amino acid metabolism were found to be the least susceptible with only ∼6% of the available cysteines were found to be SNO-modified.
Fig. 4.
Summary of cysteine SNO-susceptibility by function. Summary of the percent of SNO modifications compared with the available cysteines for the major functional pathways in cardiac mitochondria. The specific values for each functional group are listed above each bar (number of SNO-cysteines/number of available cysteines).
DISCUSSION
This study presents the first comprehensive analysis of SNO-modifiable proteins in the cardiac mitochondria. Our findings highlight the regulatory potential and suggest a broader role for this modification. We report 83 sites on 60 proteins, 39 of which were not previously known to be affected by SNO-modifications. By combining the SNO-mapping with an assessment of all the available cysteine residues a measure of SNO-selectivity was determined. Pairing this analysis with the function of the affected proteins highlighted some pathways that may be more susceptible to SNO-regulation. The identification of these novel SNO-sites provides deeper insight into the role and scope of SNO-modification in the cardiac mitochondria.
The comparison of the SNO-sites to all the available cysteine residues revealed a unique NO-reactive subset of residues within a larger pool. These unique cysteines were found in a variety of functional pathways to varying degrees. Proteins involved in transport have the highest ratio of modified cysteines, possibly indicating a particular susceptibility for redox-regulation by SNO. Among the SNO-modified transport proteins identified were ADP/ATP translocase 1 (also known as ANT1) and the phosphate carrier protein. These proteins are the fundamental transporters involved in mitochondrial ATP transport. Additionally they have been suggested as potential components/regulators of the mitochondrial permeability transition (MPT) pore (37). The opening of this pore, permitting the flow of small molecules, can lead to mitochondrial swelling triggering cell death. The MPT pore remains closed during periods of ischemia but opens following reperfusion, contributing to reperfusion injury (38). Pore opening has long been implicated in heart disease and its inhibition has been proposed as a mechanism for cardioprotection (37). Oxidation of critical cysteines in ANT1, including Cys 257, has been shown to facilitate calcium induced opening of the MPT pore (39). A similar mechanism has been described for phosphate carrier protein. Oxidation of Cys 86 has been found to shift transporter activity from an antiporter to an uncoupled unspecific uniporter (40). NO-donors are known to be cardioprotective in part by increasing the threshold for a calcium-induced MPT (41). Given these effects of NO, it is conceivable that SNO might occupy these cysteines protecting them from subsequent more severe oxidative damage, thus preventing MPT pore opening and protecting the cell.
SNO-modified proteins were also identified in the respiratory chain, tricarboxylic acid cycle, oxidative defense, and in fatty acid and amino acid metabolism. In the mitochondrial respiratory chain, the main source of energy production in cardiac tissue, 11 novel SNO-sites were mapped to the subunits of complexes I-III, including seven modified residues located in six different subunits of complex I. Previous reports have shown that Complex I is S-nitrosated in preconditioning, and that cardioprotection may stem from reduced production of harmful reactive oxygen species (5, 7, 8, 13). The site-specific information provided here will now allow this hypothesis to be tested. Reduction in energy production has been observed in cardioprotective states (42). Complex V (F1Fo) ATPase activity is reduced upon NO-treatment and the alpha subunit has been found to be SNO-modified in cardiac preconditioning (10). The current study builds on this work by identifying Cys 294 residue in the alpha subunit to be SNO-modified.
Other novel SNO-sites were identified in potentially cardioprotective pathways. Within the tricarboxylic acid cycle, eight SNO-modified enzymes were detected. Of those, 2-oxoglutarate dehydrogenase (2-OGDH) has been found in ischemic preconditioning and GSNO treatment increased its activity (10). This is particularly interesting because 2-OGDH activity has been found to be decreased with ischemia/reperfusion injury, leading to speculation that SNO may protect 2-OGDH from oxidative damage by protecting one or more critical cysteine residues (10, 43). Our finding of Cys 395, the sole SNO-modified residue of 2-OGDH, might provide a link between oxidative regulation of this enzyme and cardioprotection. We also detected SNO-modifications on two proteins involved in the mitochondrial defense against oxidative stress, nicotinamide dinucleotide phosphate (NADP)-dependent isocitrate dehydrogenase (Cys 113, 154, 336) and nicotinamide nucleotide transhydrogenase (Cys 708) (44). Recently, nicotinamide nucleotide transhydrogenase activity was found to be decreased in human heart failure (45). A potential cardioprotective role for SNO may be to activate or prevent the inactivation of these enzymes allowing the continued generation of NADPH, a necessary cofactor for many of the cell's antioxidant processes (45, 46). This would provide cardiac tissue a greater capacity to resist an oxidative insult.
Detecting and mapping SNO-sites requires use of the biotin switch assay. This technique can be very powerful but does suffer from some limitations. The labile nature of the SNO-modifications, requiring a replacement strategy facilitated by ascorbate reduction, does present the potential for artifacts. The selective reducing power of ascorbate for nitrosated cysteines has been investigated by several groups to differing results. Some evidence exists that ascorbate can reduce disulfide bonds (47–49). However, Forrester and colleagues make a compelling case to the contrary (17, 50). In particular, they independently induce S-glutathionylation, S-oxidation (using peroxide), and S-nitrosation modifications to demonstrate the specificity of ascorbate as a SNO-reducing agent (50). They suggest many of the reported artifacts may stem from exposure to indirect sunlight a suggestion that highlights the need for samples to be protected from light during the assay (50). An additional review of S-nitrosation chemistry can be found in (22). The results presented here, support the fairly specific nature of ascorbate as an SNO-reducing agent. In the gel analysis, very little signal is observed in the untreated and control treated samples indicating ascorbate is not responsible for a significant reduction of endogenous disulfide bonds. However, cysteine containing peptides were observed in the GSH treated MS samples. Given the sensitivity of the LTQ instrument, it is possible these peptides represent a small artifactual reduction of glutathionylation or other modification that were not visible in the gel analysis. The stability and potential reversibility of these modifications is likely determined by light exposure and the chemistry of their local environment (51). Studies are underway in our laboratory to develop approaches to better address these limitations in the current method.
Although, the cardioprotective effects of NO have been known for some time, relatively few affected proteins had been identified and even fewer have SNO-site specific information avaliable. In this study, we report multiple novel NO-reactive cysteine residues on proteins in several cardioprotective pathways. Our results highlight the untapped regulatory potential of this post translational modification and may imply a wider role for SNO than currently characterized. Among others, the identified sites suggest SNO-based regulation of MPT, oxidative phosphorylation, the tricarboxylic acid cycle, and oxidative defense pathways. Thus, the broad-based, site-specific analysis presented here, provides the next step in determining the mechanisms of NO-action in the mitochondria. Our findings underscore the significant potential of SNO as a regulatory mechanism for cardioprotection and oxidative phosphorylation in ischemic/reperfusion injury.
Acknowledgments
We would like to thank Bob Cole and Bob O'Meally at the Johns Hopkins Mass Spectrometry and Proteomic Facility for their assistance and D. Brian Foster for insightful discussions during the preparation of our manuscript.
Footnotes
* This work was supported in part by grants to JVE: NIH grants P01-HL081427 and PO1-HL077180. CIM: AHA Pre-doctoral Fellowship 0815145E. LAK: AHA Pre-doctoral Fellowship 0715247U.
 This article contains supplemental Fig. 1.
This article contains supplemental Fig. 1.
1 The abbreviations used are:
- SNO
- S-nitrosation
- GSH
- glutathione
- GSNO
- S-nitrosoglutathione
- MPT
- mitochondrial permeability transition
- MS
- mass spectrometry
- NEM
- N-ethylmaleimide
- NO
- nitric oxide
- DTT
- dithiotreitol.
REFERENCES
- 1. Jones S. P., Bolli R. (2006) The ubiquitous role of nitric oxide in cardioprotection. J. Mol. Cell Cardiol. 1, 16–23 [DOI] [PubMed] [Google Scholar]
- 2. Foster D. B., Van Eyk J. E., Marban E., O'Rourke B. (2009) Redox signaling and protein phosphorylation in mitochondria: progress and prospects. J. Bioenerg. Biomembr. 2, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2, 150–166 [DOI] [PubMed] [Google Scholar]
- 4. Sun J., Murphy E. (2010) Protein S-nitrosylation and cardioprotection. Circ. Res. 2, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borutaite V., Brown G. C. (2006) S-nitrosothiol inhibition of mitochondrial complex I causes a reversible increase in mitochondrial hydrogen peroxide production. Biochim. Biophys. Acta. 5–6, 562–566 [DOI] [PubMed] [Google Scholar]
- 6. Chouchani E. T., Hurd T. R., Nadtochiy S. M., Brookes P. S., Fearnley I. M., Lilley K. S., Smith R. A., Murphy M. P. (2010) Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem. J. 1, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dahm C. C., Moore K., Murphy M. P. (2006) Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria. J. Biol. Chem. 15, 10056–10065 [DOI] [PubMed] [Google Scholar]
- 8. Burwell L. S., Nadtochiy S. M., Tompkins A. J., Young S., Brookes P. S. (2006) Direct evidence for S-nitrosation of mitochondrial complex I. Biochem. J. Pt 3, 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Foster M. W., Stamler J. S. (2004) New insights into protein S-nitrosylation. Mitochondria as a model system. J. Biol. Chem. 24, 25891–25897 [DOI] [PubMed] [Google Scholar]
- 10. Sun J., Morgan M., Shen R. F., Steenbergen C., Murphy E. (2007) Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ. Res. 11, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 11. Burwell L. S., Brookes P. S. (2008) Mitochondria as a target for the cardioprotective effects of nitric oxide in ischemia-reperfusion injury. Antioxid. Redox. Signal. 3, 579–599 [DOI] [PubMed] [Google Scholar]
- 12. Prime T. A., Blaikie F. H., Evans C., Nadtochiy S. M., James A. M., Dahm C. C., Vitturi D. A., Patel R. P., Hiley C. R., Abakumova I., Requejo R., Chouchani E. T., Hurd T. R., Garvey J. F., Taylor C. T., Brookes P. S., Smith R. A., Murphy M. P. (2009) A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 26, 10764–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nadtochiy S. M., Burwell L. S., Ingraham C. A., Spencer C. M., Friedman A. E., Pinkert C. A., Brookes P. S. (2009) In vivo cardioprotection by S-nitroso-2-mercaptopropionyl glycine. J. Mol. Cell. Cardiol. 6, 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yellon D. M., Downey J. M. (2003) Preconditioning the myocardium: from cellular physiology to clinical cardiology. Physiol. Rev. 4, 1113–1151 [DOI] [PubMed] [Google Scholar]
- 15. Halestrap A. P., Clarke S. J., Khaliulin I. (2007) The role of mitochondria in protection of the heart by preconditioning. Biochim. Biophys. Acta. 8, 1007–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murphy E., Steenbergen C. (2007) Preconditioning: the mitochondrial connection. Annu. Rev. Physiol. 51–67 [DOI] [PubMed] [Google Scholar]
- 17. Forrester M. T., Foster M. W., Benhar M., Stamler J. S. (2009) Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic. Biol. Med. 2, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2, 193–197 [DOI] [PubMed] [Google Scholar]
- 19. Jaffrey S. R., Snyder S. H. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 86, l1. [DOI] [PubMed] [Google Scholar]
- 20. Lam Y. W., Yuan Y., Isaac J., Babu C. V., Meller J., Ho S. M. (2010) Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. PLoS One. 2, e9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hao G., Derakhshan B., Shi L., Campagne F., Gross S. S. (2006) SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. U.S.A. 4, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hogg N. (2002) The biochemistry and physiology of S-nitrosothiols. Annu. Rev. Pharmacol. Toxicol. 585–600 [DOI] [PubMed] [Google Scholar]
- 23. Pedersen P. L., Greenawalt J. W., Reynafarje B., Hullihen J., Decker G. L., Soper J. W., Bustamente E. (1978) Preparation and characterization of mitochondria and submitochondrial particles of rat liver and liver-derived tissues. Methods Cell Biol. 411–481 [DOI] [PubMed] [Google Scholar]
- 24. McDonald T., Sheng S., Stanley B., Chen D., Ko Y., Cole R. N., Pedersen P., Van Eyk J. E. (2006) Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol. Cell Proteomics. 12, 2392–2411 [DOI] [PubMed] [Google Scholar]
- 25. Agnetti G., Kaludercic N., Kane L. A., Elliott S. T., Guo Y., Chakir K., Samantapudi D., Paolocci N., Tomaselli G. F., Kass D. A., Van Eyk J. E. (2010) Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ. Cardiovasc. Genet. 1, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Derakhshan B., Wille P. C., Gross S. S. (2007) Unbiased identification of cysteine S-nitrosylation sites on proteins. Nat. Protoc. 7, 1685–1691 [DOI] [PubMed] [Google Scholar]
- 27. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 5, 850–858 [DOI] [PubMed] [Google Scholar]
- 28. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics. 7, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 29. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 20, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 30. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 17, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 31. Rigoutsos I., Floratos A. (1998) Combinatorial pattern discovery in biological sequences: The TEIRESIAS algorithm. Bioinformatics. 1, 55–67 [DOI] [PubMed] [Google Scholar]
- 32. Petersen B., Petersen T. N., Andersen P., Nielsen M., Lundegaard C. (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. B.M.C. Struct. Biol. 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 34. Lee J. H., Yang E. S., Park J. W. (2003) Inactivation of NADP+-dependent isocitrate dehydrogenase by peroxynitrite. Implications for cytotoxicity and alcohol-induced liver injury. J. Biol. Chem. 51, 51360–51371 [DOI] [PubMed] [Google Scholar]
- 35. Vizcaino J. A., Cote R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) A guide to the Proteomics Identifications Database proteomics data repository. Proteomics. 18, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stamler J. S., Toone E. J., Lipton S. A., Sucher N. J. (1997) (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 5, 691–696 [DOI] [PubMed] [Google Scholar]
- 37. Halestrap A. P., Pasdois P. (2009) The role of the mitochondrial permeability transition pore in heart disease. Biochim. Biophys. Acta. 11, 1402–1415 [DOI] [PubMed] [Google Scholar]
- 38. Halestrap A. P., Clarke S. J., Javadov S. A. (2004) Mitochondrial permeability transition pore opening during myocardial reperfusion–a target for cardioprotection. Cardiovasc. Res. 3, 372–385 [DOI] [PubMed] [Google Scholar]
- 39. McStay G. P., Clarke S. J., Halestrap A. P. (2002) Role of critical thiol groups on the matrix surface of the adenine nucleotide translocase in the mechanism of the mitochondrial permeability transition pore. Biochem. J. Pt 2, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stappen R., Kramer R. (1993) Functional properties of the reconstituted phosphate carrier from bovine heart mitochondria: evidence for asymmetric orientation and characterization of three different transport modes. Biochim. Biophys. Acta. 1, 40–48 [DOI] [PubMed] [Google Scholar]
- 41. Wang G., Liem D. A., Vondriska T. M., Honda H. M., Korge P., Pantaleon D. M., Qiao X., Wang Y., Weiss J. N., Ping P. (2005) Nitric oxide donors protect murine myocardium against infarction via modulation of mitochondrial permeability transition. Am. J. Physiol. Heart Circ. Physiol. 3, H1290-H1295 [DOI] [PubMed] [Google Scholar]
- 42. Burwell L. S., Nadtochiy S. M., Brookes P. S. (2009) Cardioprotection by metabolic shut-down and gradual wake-up. J. Mol. Cell Cardiol. 6, 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lundberg K. C., Szweda L. I. (2006) Preconditioning prevents loss in mitochondrial function and release of cytochrome c during prolonged cardiac ischemia/reperfusion. Arch. Biochem. Biophys. 1, 130–134 [DOI] [PubMed] [Google Scholar]
- 44. Murphy M. P. (2009) How mitochondria produce reactive oxygen species. Biochem. J. 1, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sheeran F. L., Rydstrom J., Shakhparonov M. I., Pestov N. B., Pepe S. (2010) Diminished NADPH transhydrogenase activity and mitochondrial redox regulation in human failing myocardium. Biochim. Biophys. Acta. 6–7, 1138–1148 [DOI] [PubMed] [Google Scholar]
- 46. Agledal L., Niere M., Ziegler M. The phosphate makes a difference: cellular functions of NADP. Redox. Rep. 1, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Landino L. M., Koumas M. T., Mason C. E., Alston J. A. (2006) Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem. Biophys. Res. Commun. 2, 347–352 [DOI] [PubMed] [Google Scholar]
- 48. Huang B., Chen C. (2006) An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic. Biol. Med. 4, 562–567 [DOI] [PubMed] [Google Scholar]
- 49. Kallakunta V. M., Staruch A., Mutus B. Sinapinic acid can replace ascorbate in the biotin switch assay. Biochim. Biophys. Acta. 1, 23–30 [DOI] [PubMed] [Google Scholar]
- 50. Forrester M. T., Foster M. W., Stamler J. S. (2007) Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J. Biol. Chem. 19, 13977–13983 [DOI] [PubMed] [Google Scholar]
- 51. Paige J. S., Xu G., Stancevic B., Jaffrey S. R. (2008) Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol. 12, 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lam Y. W., Yuan Y., Isaac J., Babu C. V., Meller J., Ho S. M. Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. PLoS. One. 2, e9075. [DOI] [PMC free article] [PubMed] [Google Scholar]