Abstract
Biomedical research requires protein detection technology that is not only sensitive and quantitative, but that can reproducibly measure any set of proteins in a biological system in a high throughput manner. Here we report the development and application of a targeted proteomics platform termed index-ion triggered MS2 ion quantification (iMSTIQ) that allows reproducible and accurate peptide quantification in complex mixtures. The key feature of iMSTIQ is an approach called index-ion triggered analysis (ITA) that permits the reproducible acquisition of full MS2 spectra of targeted peptides independent of their ion intensities. Accurate quantification is achieved by comparing the relative intensities of multiple pairs of fragment ions derived from isobaric targeted peptides during MS2 analysis. Importantly, the method takes advantage of the favorable performance characteristics of the LTQ-Orbitrap, which include high mass accuracy, resolution, and throughput. As such it provides an attractive targeted proteomics tool to meet the demands of systems biology research and biomarker studies.
The success of proteomics research depends on the ability to reliably identify and quantify any protein or set of proteins in a biological system. Currently “shotgun” mass spectrometry (MS), paired with stable isotope labeling of proteins or peptides, is an attractive and widely applied approach for quantitative proteomics measurements (for reviews, see (1–6)). While such shotgun MS-based quantitative proteomics platforms have been used with great success to quantify significant fractions of proteomes, the sensitivity and reproducibility of the approach does not meet the demands of many proteomics studies. To address these issues, recent efforts have focused on developing MS-based methods to monitor specific sets of proteins. Such targeted proteomics platforms are expected to play important roles in clinical applications (7) as well as in basic science studies where sets of proteins need to be consistently quantified under different conditions. One particularly promising targeted approach involves the use of selected reaction monitoring (SRM1, also termed MRM for multiple reaction monitoring) MS of specific sets of parent and fragment ions (transitions) for each targeted peptide using triple quadrupole instruments (8–15). Other targeted methods involve the use of inclusion lists with high mass accuracy scanning mass spectrometers such as the LTQ-Orbitrap to focus MS analysis on predetermined precursor ions (16–17).
Although successful applications have been reported, a number of features of current targeted proteomics approaches are not optimal. For instance, although inclusion list methods can provide enhanced sensitivity and reproducibility, the gains are limited by the need to detect the precursor ion in question in an MS1 survey scan to trigger the generation of fragment ion spectra (MS2 or MS/MS). This can be problematic when the targeted peptides are of low abundance in biological samples with high complexity and dynamic range. SRM is a sensitive, reproducible, and quantitative targeted approach (18). However, the throughput of SRM is limited by a need to balance duty cycle with sensitivity. To improve throughput, segmented methods are often applied in which transitions for specific groups of peptides are measured in a given time segment of a liquid chromatography (LC)-MS experiment. While this increases throughput to ∼1,000 transitions corresponding to ∼100 targeted peptides (assuming 3 transitions X 2 isotopic forms per peptide) in a typical LC-SRM experiment, it imposes a requirement for highly reproducible chromatography. In comparison, during a standard LC-MS/MS run on an LTQ-Orbitrap, 2000 features corresponding to ∼1000 peptides can be targeted for MS2 analysis without rigorous time segmentation (16). Furthermore, because detection is typically based on a limited number of transitions per peptide made on triple quadrupole instruments with modest mass accuracy and resolution, the ability to confidently identify and accurately quantify individual peptides in a complex mixture may be compromised. Finally, until recently, application of SRM has been limited by a prerequisite assay optimization process that typically involves selecting the most suitable transitions for each targeted peptide (18). Given these issues with the current targeted proteomics approaches, the development of a targeted quantitative proteomics platform that permits reproducible and accurate measurements in a high throughput manner, without the need for a priori selection of optimal transitions and rigorous retention time requirements, is highly desirable.
In this study, we report a novel targeted quantitative proteomics platform termed Index-ion triggered MS2 ion quantification (iMSTIQ) that alleviates many of the issues associated with current targeted methods. iMSTIQ involves the use of isotopically heavy “index” peptides to reproducibly trigger the acquisition of full MS2 spectra for the isotopically light target peptides, independently of the target peptides' ion intensities. This results in enhanced sensitivity and reproducibility compared with inclusion list-based approaches. In addition, it does not require the highly reproducible chromatography required for segmented SRM-based studies nor the a priori selection of optimal transitions. The acquisition of full MS2 spectra for targeted peptides permits confident peptide identification as well as accurate quantification based on the relative intensities of multiple pairs of fragment ions derived from isobaric peptides. Importantly, we demonstrate iMSTIQ's ability to accurately quantify peptides in the low fmol range on the LTQ-Orbitrap, an instrument capable of targeting hundreds of peptides with high mass accuracy and resolving power per analysis. Finally, we demonstrate the utility of the method by applying it to measure the temporal release of targeted inflammatory mediators from macrophages in response to lipopolysaccharide (LPS) stimulation.
EXPERIMENTAL PROCEDURES
Synthetic Peptides
All heavy synthetic peptides used in this study were purchased from Sigma custom AQUA peptides carrying C-term Lys (13C615N2) with > 95% purity. The labeled amino acid contains 98% 13C and 98% 15N isotopic content. The light peptides used in the ITA and iMSTIQ analyses were custom synthesized at crude purity level by Peptide 2.0 Inc. (Chantilly, VA) or Genscript Co. (Scotch Plains, NJ).
iMSTIQ Labeling Reagents
mTRAQ® reagents (Δ0, Δ4, and Δ8, Applied Biosystems, Foster City, CA) were used as MSTIQ labeling reagents (see Fig. 1 for schematic detail). In addition, we generated heavy and light NHS ester variants of BOC-Ala and used them for MSTIQ labeling for the experiment presented in Fig. 4 (structures shown in supplemental Fig. S2): one equivalent each of l-Alanine-N-T-BOC (13C315N1) (Cambridge Isotope Laboratories, Andover, MA), N-Hydroxysuccinimide (NHS, from Sigma, St. Louis, MO), and N,N′-diisopropylcarbodiimide (Sigma) in DMF (Sigma) were mixed and incubated at room temperature over night. Following overnight incubation, the diisopropylcarbodiimide adduct (O-acylisourea) precipitated and the clear supernatant containing the BOC-Ala-O-NHS was used. The isotopically light labeling reagent was directly purchased for Sigma Aldrich (t-BOC-Ala-OSu).
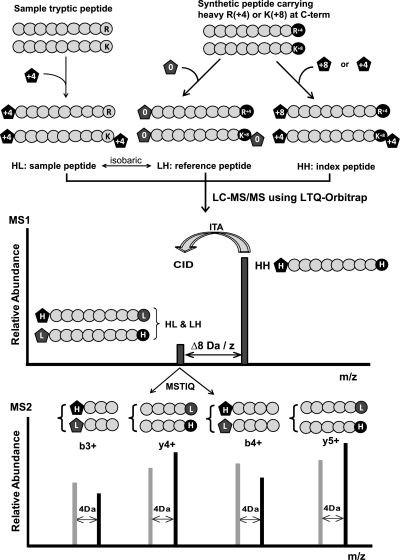
Fig. 1.
Schematic overview of the iMSTIQ strategy. See text for details.
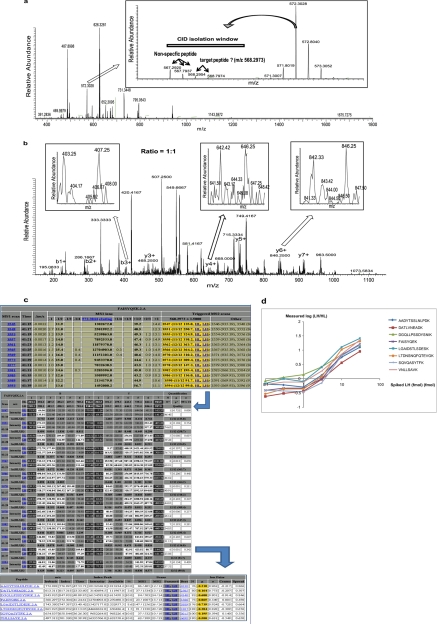
Fig. 4.
iMSTIQ peptide quantification in a complex sample. Eight synthetic peptides were labeled by MSTIQ and combined in six titers containing HH peptides (100 fmol), HL peptides (crude, ∼3 fmol), LH peptides (0.1, 0.3, 1, 3, 10, or 30 fmol, respectively), and a human N-glyco plasma peptide mixture (∼1 μg). A, Full MS1 spectrum (350–1800 m/z) at 41.41 min. Inset: expanded view (566–575 m/z) showing the index ion for a representative peptide “FAISYQEK” ([M+2H]2+, 572.3028 m/z). Targeted peptides ([M+2H]2+, 568.2973 m/z) and a co-eluting off-target peptide ([M+2H]2+, 567.2920 m/z) are within the CID isolation window (567.2–570.2 m/z, bar). B, Full LTQ MS2 spectrum (145–1150 m/z) triggered by the index peptide in (A). Positively identified peptide-specific fragment ion pairs are marked. The targeted MSTIQ fragment ion pairs have lower intensities than the fragment ions corresponding to the off-target peptide (singlet fragment ion pattern). Insets: expanded views of MS2 spectra for representative fragment ion pairs (4 Da apart) each with a relative abundance close to 1:1. C, Screenshots of ISBquant output for quantification of the peptide “FAISYQEK” in the 3 fmol (LH) sample. MS1 scans during elution of the index ion (upper panel) that triggered MS2 scans and met ISBquant scan selection criteria (see Experimental procedures) were selected for quantification (gold). The measured intensity of the index ion for each scan is indicated in the column labeled, “572.3044 eluting.” Individual fragment ion intensities (middle panel) at all selected scans meeting strict quality criteria (white; see Experimental procedures) are averaged to compute the peptide abundance ratio. Quantification of all eight peptides is summarized in the lower panel including the final log-ratio (gold). Links to individual MS2 spectra (active in the web interface) are shown in blue. D, Plots of log (LH/HL) versus the known amount of each LH peptide (log scale). A consistent linear regression is observed for 1 ≤ LH ≤ 30 fmol.
Sample Preparation
ITA Assay
Yeast strain BY4741 grown in YPD media to log phase was harvested and bead-beaten in lysis buffer (50 mm HEPES, pH8.0, 1% SDS, 150 mm NaCl) with protease inhibitor mixture (Roche, Mannheim, Germany). The protein extract was then acetone precipitated and dissolved in 10 mm Tris-HCl, pH 8.0 containing 1M Urea. The protein mixture was then treated with 5 mm dithiothreitol and alkylated by 15 mm iodoacetamide and trypsin digested overnight. The trypsin digested peptides were purified by C18 spin column (The Nest Group, Southboro, MA) prior to use for mass spectrometry analyses.
65 lysine-terminal AQUA peptides corresponding to mouse proteins were alkylated with 6 mm methyl methanethiosulfonate and then labeled with heavy (Δ4) or light (Δ0) mTRAQ reagents (Applied Biosystems) following the manufacturer's protocol to generate the heavy and light peptide pairs with 8 Da mass differences. Another 21 lysine-terminal AQUA peptides (used as heavy forms) were also included along with their synthetic light peptide variants. The total 86 pairs of the heavy peptide forms (at 500 fmol each) and light peptide forms (at 0.75, 3.75, 7.5, and 15 fmols, respectively) were spiked into 1 μg yeast peptide mixture.
MSTIQ Assay
RAW264.7 cells (ATCC, Manassas, VA) were grown at 37 °C with 5% CO2 on stable isotope labeling with amino acids in cell culture (SILAC)-specific RPMI 1640 media (Caisson Laboratories, North Logan, UT) supplemented with 10% dialyzed fetal bovine serum (Invitrogen, Carlsbad, CA), 1% Pen/Strep (Invitrogen), 2 mm l-glutamine (Invitrogen), and with either light Lys (12C614N2) and Arg (14N4) amino acids or heavy Lys (13C615N2) and Arg (15N4) amino acids. Following 5 passages, cells were activated by 100 ng/ml LPS (Sigma) for 4h prior to harvest. The collected cells were lysed in 25 mm HEPES, 150 mm NaCl, 5 mm MgCl2, 1% TritonX-100, 0.05% SDS, 1 mm EDTA, and protease inhibitor mixture (Roche) on ice for 30 min. Following centrifugation at 18,200 × g for 10 min at 4 °C, the supernatant (cell lysate) was collected and stored at −80 °C until further processed. Proteins in the cell lysate were denatured with 1 mg/ml (w/v) RapiGest (Waters, Milford, MA) and 0.1% SDS (Mediatech, Herndon, VA) at 95 °C for 10 min, reduced with 5 mm Tris(2-carboxyethyl) phosphine (Thermo scientific, Waltham, MA) at 60 °C for 1 h, and alkylated with 12.5 mm iodoacetamide (Sigma) at 37 °C in the dark for 30 min. The sample was diluted 10-fold with 100 mm triethylammonium bicarbonate (Thermo Scientific) prior to digestion with sequencing grade modified trypsin (Promega) at 1:50 (w/w) enzyme to substrate ratio overnight at 37 °C. The digested heavy or light peptides (labeled via SILAC at C termini) were then labeled with light (Δ0) or the heavy (Δ4) mTRAQ reagents (Applied Biosystems) respectively to generate isobaric peptides. The resulting two peptide samples were mixed at ratios of 1:30, 1:10, 1:3, 1:1, 3:1, 10:1, and 30:1. The peptide mixtures were purified on MCX μElution plates (Waters, Denver, CO) prior to LC-MS/MS analysis.
iMSTIQ Assay
Ten milligrams (137 μl) human serum (Bioclamation, Hicksville, NY) was used for the hydrazide-based solid-phase capture of the N-glycosylated peptides as previously described (19). The resulting N-glycosylated peptide mixtures were dissolved in 137 μl 0.1% formic acid (∼1 μg/μl). Eight tryptic peptides carrying either the heavy (13C615N2) or light (12C614N2) C-terminal Lys were chemically synthesized. The heavy peptides were then labeled with either the heavy version (13C315N1) of the t-BOC-Ala-NHS reagent to generate the index peptides (HH), or the light version (12C314N1) of the t-BOC-Ala-NHS reagent to generate the LH peptides. Similarly the light synthetic peptides were labeled with the heavy t-BOC-Ala-NHS reagent to generate the HL peptides. Specifically, 30 μl 0.15 m t-BOC-Ala-NHS reagents in dimethylformamide were added to 30 μg peptides dissolved in 20 μl 0.5 m HEPES, pH 8.0. Following 30 min, the t-BOC was removed by adding 45 μl 37% HCl for 30 min. The labeled peptides were purified on MCX μElution plates before being combined. The final peptide mixture used in a single LC-MS/MS run consisted of the following: 100 fmol of the HH peptides (index), 3 fmol of the LH peptides, 1 μg of the N-glycopeptides, and the HL peptides at 0.1, 0.3, 1, 3, 10, or 30 fmols.
Macrophage Protein Release Assay
RAW264.7 cells were grown on regular RPMI 1640 media (Invitrogen) containing 10% fetal bovine serum (Invitrogen), 1% Pen/Strep (Invitrogen), and 2 mm l-Glutamine (Invitrogen) at 37 °C with 5% CO2. To activate the cells, the culture media were replaced by OptiMEM (Invitrogen) supplemented with 100 ng/ml LPS (Sigma). Conditioned media were collected at 1, 2, 4, 8, and 18 h. As a control, cells were exposed to OptiMEM supplemented with phosphate buffered saline (PBS) and media were collected after 18 h. The media were concentrated by centrifugation (3500 rpm for 30 min) using Appllo-20 ml concentrators (Orbital Biosciences) and the buffer was exchanged by repeated resuspension in PBS and centrifugation. Equal amounts of protein (15 μg) from each sample were denatured by addition of SDS to 0.2% followed by incubation at 95 °C for 5 min. 2,2,2-trifluoroethanol (Fluka) was added to 50% followed by incubation at 60 °C for 30 min. The denatured proteins were then reduced with 5 mm Tris 2-carboxyethyl phosphine at 60 °C for 30 min followed by alkylation with 12.5 mm iodoacetamide at 37 °C in the dark for 30 min. The samples were diluted 1:10 with 50 mm triethylammonium bicarbonate and then digested with trypsin at 1:50 (w/w) enzyme to substrate ratio overnight at 37 °C. The digested peptides were desalted on a C18 cartridge (Waters), dried by SpeedVac, and de-glycosylated by addition of 1 μl N-glycanase (Prozyme, San Leandro, CA) following the manufacturer's protocol. The resulting peptide mixtures were then labeled with the heavy (Δ4) mTRAQ reagents (Applied Biosystems) to generate the HL peptides according to the mnaufacturer's protocol. To generate the LH and HH peptides, the 24 selected peptides (14 peptides corresponding to targeted proteins known to be released during inflammation for validation, and 10 peptides corresponding to nonspecific proteins that were used in unrelated studies in the lab) were synthesized via the AQUA platform (Sigma) carrying heavy Lys (13C615N2) at their C termini and labeled with light (Δ0) and heavy (Δ4) mTRAQ reagents (Applied Biosystems) respectively. The labeled peptides were combined containing 500 fmol of the HH peptides, 33.3 fmol of the LH peptides, and the HL peptides from ∼3 μg sample. The final peptide mixtures were purified on MCX μElution plates, dried, and resuspended in 0.1% formic acid prior to LC-MS/MS analysis.
Reverse Phased LC-MS Analysis
Peptide samples were analyzed by reversed phase high performance liquid chromatography (Agilent 1100 series) electro-spray ionization LC-MS using LTQ-Orbitrap (Thermo scientific). The high performance liquid chromatography column (75 μm × 15 cm) was packed in house with C18 resin (Magic C18 AQ 5 μm, Michrom BioResources, Auburn, CA). Peptides were resolved by running a gradient of Buffer A (0.1% formic acid) to Buffer B (0.1% formic acid, 99.9% acetonitrile) as follows: 8–25% Buffer B over 34 min (for ITA analysis) or 53 min (for other experiments); 25–35% Buffer B over 6 min (for ITA) or 10 min (for others); 35–80% Buffer B over 8 min (for ITA) or 10 min (for others). A fixed flow rate of 350 nl/min was applied.
In general, MS1 scans were acquired by the Orbitrap with a resolution of 30,000 at 400 m/z. MS2 scans were acquired by the LTQ using normal scan mode except in the MSTIQ assay where MS2 scans were acquired by the Orbitrap with a resolution of 7500 at 400 m/z. For each Orbitrap MS1 scan, 5 × 105 ions were accumulated over a maximum time of 500 ms. For each LTQ MS2 scan, 5 × 103 ions were accumulated over a maximum time of 250 ms. For each Orbitrap MS2 scan, 2 × 105 ions were accumulated over a maximum time of 1000 ms. The normalized collision energy for collision-induced dissociation (CID) was set at 35%. In the ITA assay (see Fig. 2), up to 5 MS2 scans were acquired following each MS1 scan with dynamic exclusion set for 10 s in an attempt to maximize MS2 data acquisition for target peptides. The threshold for MS2 analysis was set at 2000 counts which was significantly below the background level. A CID isolation window of 1.1 m/z was used. Charge-state screening was applied with only +2 or +3 charged ions being analyzed per MS run. For the inclusion list method, the m/z values of the light peptides were put in the “parent mass list” to trigger CID on themselves upon detection during MS1; For the ITA method, the m/z values of the heavy peptides were put in the “parent mass list” to trigger CID on potential co-eluting isotopically light peptides by enabling the “Add/Subtract” feature. For the +2 charged ions, a mass of “-4.0000” was set in the “Add/Subtract” feature; for the +3 charged ions, a mass of “-2.6667” was applied.
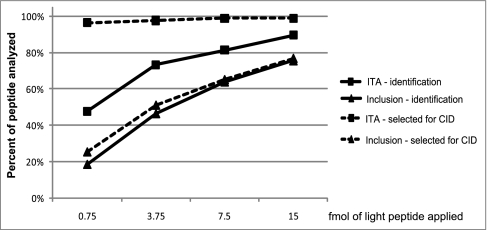
Fig. 2.
ITA (index-ion triggered analysis) improves peptide selection for CID (collision induced dissociation) and identification compared with an inclusion list method. Eighty-six peptides were spiked into a tryptic digest derived from 1 μg of a whole cell yeast protein extract at the indicated amounts and targeted for identification using either ITA (squares) or an inclusion list method (triangles). The percentage of peptides selected for CID (dashed lines) or identified (solid lines) by each method are plotted as a function of the quantity of peptides that were spiked into the sample.
In the MSTIQ assay (see Fig. 3), commonly used parameters for data dependent acquisition were used: up to 4 MS2 scans were acquired on the most abundant precursor ions detected in each MS1 scan; the selected precursor ions were dynamically excluded for 60 s; MS1 signals exceeding 500 counts were chosen for CID; and an isolation window of 1.0 m/z was used. In the iMSTIQ assay (see Fig. 4), because only eight peptides were targeted, up to three MS2 analyses were carried out following each MS1 scan without dynamic exclusion, which allowed acquisition of multiple MS2 scans for each target peptide. Precursor ion signals exceeding 100 counts triggered CID, during which an isolation window of 3.0 m/z was used. Charge state screening was used to limit ion selection to +2 charge states only. A “parent mass list” containing the m/z values of the index peptides was used to trigger CID on ions with m/z values equal to m/z (index peptide) −3.6000 Da using the “Add/Subtract” feature. In the macrophage protein release assay (see Fig. 5), up to five MS2 spectra were acquired following each MS1 scan. Dynamic exclusion was disabled. An intensity threshold of 1000 counts was required to trigger CID. A CID isolation window of 1.0 m/z was used. Charge state screening was used to limit MS2 analysis to +2 ions. The “Add/Subtract” feature was set to trigger CID on ions with m/z values equal to m/z (index peptide) −3.7500 Da. The parameters used for this experiment are recommended for application of iMSTIQ to biological samples, although adjustment of dynamic exclusion settings may improve performance as the target list increases.
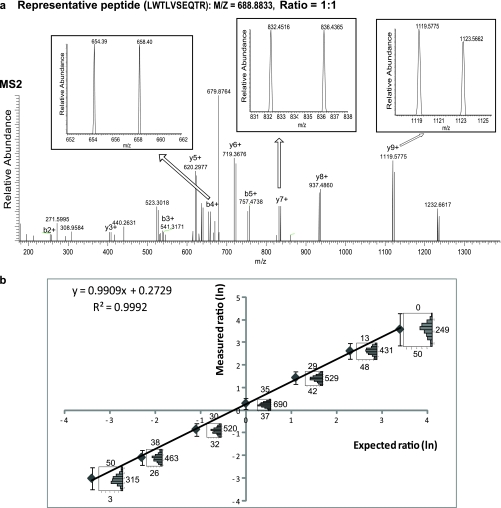
Fig. 3.
Peptide quantification by MSTIQ. Tryptic peptides derived from mouse macrophage lysates were labeled by MSTIQ, combined in seven titers (1:30, 1:10, 1:3, 1:1, 3:1, 10:1, and 30:1), and analyzed by LC-MS/MS. A, Full scan Orbitrap MS2 spectrum (175–1390 m/z) of the precursor peptide “LWTLVSEQTR” ([M+2H]2+, m/z 688.88). Multiple b- and y-ion pairs are shown. Insets: expanded view of representative pairs of 1+ fragment ions, 4 Da apart; as expected relative peak intensities are ∼1:1. B, Expected versus measured abundance ratio (ln(HL/LH)) for all seven titers. Peptides were quantified with ISBquant software. Medians (M, dots) and range (M ± 2s, see Experimental procedures; bars) are shown for each titer. Histograms show values in the range; counts above, below, or beside each histogram represent peptides with ratios higher, lower, or within the range, respectively.
Fig. 5.
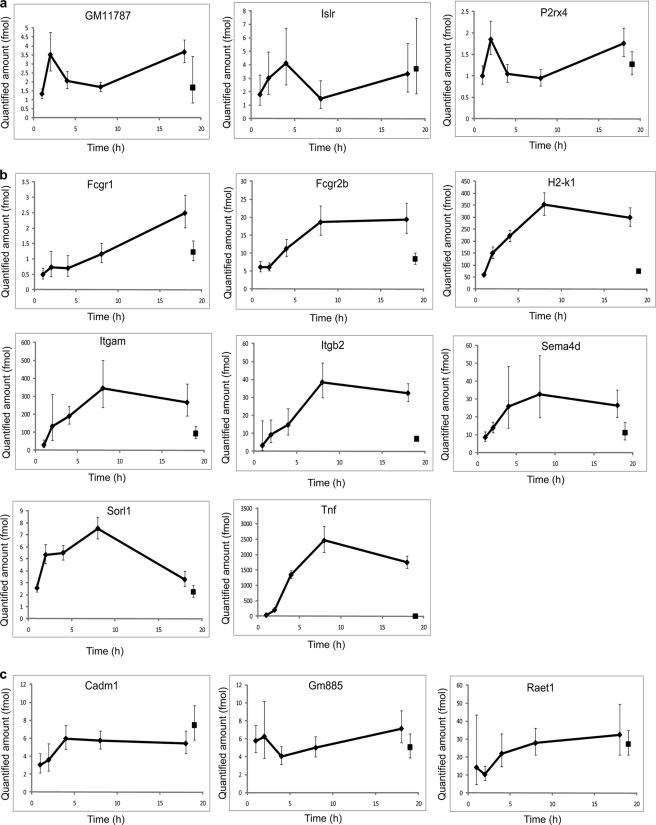
Quantification of inflammatory proteins by iMSTIQ. Peptides derived from conditioned media harvested at 1, 2, 4, 8, and 18 h post-treatment of mouse macrophages with LPS were analyzed using iMSTIQ. As a control, peptides were prepared from media harvested from macrophages 18 h following treatment with PBS. Absolute abundance (y axis) of 14 detected peptides in all six samples is plotted as a function of time points (x axis). Data from the control sample (collected at 18-h time point) are shown at the 19-h time point for clarity. A, Plots for the three nonspecific peptides (group 2 in Table I). B, Plots for the eight targeted peptides that display LPS-dependent release. C, Plots for three targeted peptides that do not display LPS-dependent release.
Peptide Identification and Quantification
RAW files from the LTQ-Orbitrap were converted to mzXML by ReAdW (version 4.3.1) with default parameters except whenever ITA was used the “precursorFromFilterLine” option was applied.
For the ITA experiment, the acquired MS2 spectra were searched against a yeast database (yeast.nci.20080206; 11,319 entries searched) supplemented with the sequences of the targeted peptides using X!Tandem 2 (2007.07.01.3). The following search parameters were applied: full tryptic cleavage specificity; mass tolerance of ± 20 ppm for precursor ions and ±0.4 Da for fragment ions; no missed cleavage allowed; fixed modification on Cys (+45.9877 Da) and variable modifications on Met (+15.9949), Lys (+148.1092), and N termini (+140.095). Peptide identification was achieved by processing the search results with the Trans-Proteomic Pipeline (TPP, http://tools.proteomecenter.org/wiki/index.php?title = Software:TPP).
For the MSTIQ assay, the seven datasets were each searched twice against the mouse International Protein Index database (v.3.56;56,294 entries searched) using X!Tandem2 as described above with the following parameter changes: a fixed modification of +57.021464 on Cys was used; one search was conducted with fixed modifications on N termini (+144.1021) and Lys (+144.1021) to account for HL peptide modifications; the other search was conducted with fixed modifications on N termini (+140.095), Lys (+148.1092), and Arg (+3.98814) to account for LH peptide modifications. Each search was repeated against the reversed database to estimate the false positive rate. Peptide identification was achieved by processing the search results with the TPP. Confidently identified peptides (fully-tryptic, 2+ and 3+ ions with no missed cleavages, PeptideProphet probability ≥ 0.9) in at least one of the seven titers were considered target peptides.
Each target peptide was quantified in all seven datasets using ISBquant (available on request). ISBquant considered a scan for quantification of a target peptide if 1) the theoretical and observed parent m/z's were within 20 ppm, and 2) at least 7 HL or 7 LH fragment ions in the MS2 scan were observed above background level (BG, defined as the 25th percentile of all non-zero intensities in the scan) within 0.5 units of their predicted m/z's. This enabled some peptides to be quantified in titers in which they did not pass the PeptideProphet threshold of 0.9. Scan selection was further restricted by identifying the highest quality scan, and only scans with a retention time ≤ ±1 min from the highest quality scan were used for quantification. For this purpose, scan quality was judged by the number of observed HL or LH MSTIQ ions (whichever was greater); ties were broken by the median observed MSTIQ ion intensity in each scan. In addition to automatic scan selection (applied in Fig. 3), ISBquant allows for manual scan selection by the user.
To compute peptide abundance ratios, the intensities of each fragment ion pair i were quantified as and outliers were detected with the “MAD-Median Rule” (see reference (20), p.101): xi is an outlier if |xi − M| > 3.321*m; the choice of the constant 3.321 corresponds to a 5% chance of rejecting a nonoutlier from a normally distributed sample), where M = median{xi} and m = MAD(xi) = 1.4826*median{|xi − M|} denotes a robust estimate of the standard deviation based on the sample's Median Absolute Deviation (R mad() function). The final quantification for each identified peptide is mean {xi} following rejection of outliers. This “trimmed” mean estimate has known standard error , where sW is the sample Winsorized variance (see reference (20), p.63). Finally, peptides were only considered “quantified” if at least two MSTIQ-labeled fragment ion pairs contributed to the trimmed mean. A MSTIQ ion was considered “observed” if its intensity ≥ SNR*BG; unless otherwise noted, SNR = 2. Some potential MSTIQ fragment pairs were excluded from quantification for one of four reasons: (1) all b1+ and y1+ fragment ion pairs were excluded, (2) any fragment ion pair containing a MSTIQ ion with a predicted m/z within ±1.5 Da of another predicted MSTIQ ion, or a neutral loss from a predicted ion (b-ions, loss of NH3; y-ions, loss of H2O), was excluded, (3) any fragment ion pair for which the intensity of at least one MSTIQ ion was not “observed” was excluded. A fragment ion pair was deemed quantifiable if the intensity of one ion (HL- or LH-derived) was above background and the intensity of the sibling ion was nonzero.
For the iMSTIQ assay automated scan selection for quantification is achieved using the same constraints described for MSTIQ. In addition, the index ion elution profiles were used to assist in scan selection. Ion intensities for the predicted monoisotopic index ion peak (±20 ppm), as well as intensities for peaks at +1/z and −1/z were extracted. The peak at −1/z is because of the slight impurity of the heavy isotopes used in labeling (see Discussion). Elution of an index ion was distinguished from other ions based on two criteria. First, a scan was considered to contain an index ion if intensities of 0.05–0.35× (-1/z peak) and 0.5–1.5× (+1/z peak) the predicted index ion's monoisotopic peak intensity were present. Second, only MS2 scans triggered from MS1 scans with an index ion intensity of ≥10% of the peak index ion elution intensity were considered. MS2 scans without this evidence of an intense, co-eluting index ion were rejected. Other than these differences, scan selection and quantification were the same as for MSTIQ. Scans were selected automatically for the iMSTIQ study in Fig. 4 and entirely manually for the macrophage study in Fig. 5.
RESULTS
Overview of the iMSTIQ Platform
For iMSTIQ analysis, sample peptides are labeled with an isotopically heavy amine labeling reagent such as the +4 Da form of the mTRAQ reagent (ABI) (Fig. 1). For each target sample peptide, a quantitative reference peptide is synthesized with isotopically heavy arginine (+4 Da) or lysine (+8 Da) at its C-terminus and labeled with the isotopically light form of the amine labeling reagent. Relative to the reference peptide, the sample peptide is isotopically heavy at its N-terminus but isotopically light at its C-terminus (HL). This labeling scheme generates isobaric pairs of reference and target sample peptides that in turn produce pairs of fragment ions separated by 4 Da/z units during MS2 analysis; b ions derived from the sample HL peptide appear in the spectrum at 4 Da/z units larger than the corresponding b ions from the reference LH peptide, and y ions derived from the reference LH peptide appear in the spectrum at 4 Da/z units larger than the corresponding y ions from the sample HL peptide. Furthermore, a corresponding index peptide is also prepared by labeling the synthesized reference peptide with the isotopically heavy form of the amine labeling reagent, (isotopically heavy at both termini, HH). Importantly, the sample, reference, and index forms of the target peptide are virtually chemically identical and co-elute during LC. Prior to LC-MS/MS analysis, the index and reference peptide are spiked into the sample at appropriate levels (see below). During LC-MS/MS analysis on the LTQ-Orbitrap, detection of the index peptide triggers MS2 analysis on the m/z of the targeted isobaric sample and reference peptides regardless of whether these peptide ions are detectable in the survey scan (ITA).
Importantly, the index peptide is spiked in at a level sufficient to ensure its detection during MS1, but only the sample and reference forms of the target peptide are selected for CID. The reference peptide is spiked in at a level expected to be comparable to the target sample peptide to minimize the dynamic range required during MS2 quantification. Quantification by the MSTIQ method is achieved by measuring the relative intensities of pairs of fragment ions derived from the targeted sample and reference peptides, both of which are present in the MS2 spectrum.
ITA Enhances Detection of Peptides to the Sub-fmol Level in Complex Mixtures
ITA was specifically designed to improve the reliability of MS2 data acquisition for targeted peptides and thus enhance the sensitivity of their detection. We compared the capability of ITA to select predetermined peptides for MS2 analysis and to identify them to an inclusion list method. We spiked both heavy (+8 Da, index) and light (target) isotopic variants of 86 synthetic peptides corresponding to mouse or human proteins into a tryptic digest of 1 μg of a yeast whole cell extract. In four titration experiments, the index peptides were each spiked in at 500 fmol, and the target peptides were spiked in at a low fmol level (0.75, 3.75, 7.5, or 15 fmol, respectively). These samples were analyzed separately by either ITA or the inclusion list method on the LTQ-Orbitrap. For the inclusion list method, the parent mass list contained the m/z of each target peptide ion, and detection of an ion at a listed m/z triggered fragmentation at the listed m/z. For ITA, the parent mass list contained the m/z of each index peptide ion, and detection of an ion at a listed m/z triggered fragmentation at the corresponding target peptide m/z (e.g. for a +2 ion, Δ m/z = −4 m/z). Data from both triggering methods were searched with X!Tandem (21, 22) against a yeast database augmented with the target peptide sequences. Correctly identified peptides from each analysis are shown in supplemental Table S1 (false discovery rate <5%).
ITA outperformed the inclusion list method in terms of correct selection of targeted peptides for MS2 analysis, as well as their positive identification in all four titration experiments (Fig. 2). ITA showed the most improvement at the lowest amounts of target peptide: at 0.75 fmol of target peptide, ITA correctly triggered MS2 analysis on nearly all targets (97%), whereas the inclusion list method typically failed to trigger (26%). Improved triggering led to improved rates of peptide identification: at 0.75 fmol of targeted peptides, 48% of the peptides were identified via ITA but only 19% of the peptides were identified via inclusion list. Importantly, ITA showed consistently high rates of correct MS2 triggering in all four experiments (97%, 98%, 99%, and 99% respectively). These results demonstrate that ITA ensures reliable triggering of MS2 events on targeted peptides, and improves rates of peptide identification. The performance enhancement is particularly striking for MS2 analysis and identification of low abundance peptides in complex mixtures.
The MS2 Ion Quantification (MSTIQ) Strategy for Peptide Quantification
Quantification constitutes a critical component of targeted proteomics platforms. While it is possible to quantify ITA-targeted peptides using isotope dilution strategies at the MS1 level (23), we sought to develop a strategy for quantification of ITA targeted peptides that takes advantage of the favorable signal to noise characteristics of MS2 spectra compared with MS1 spectra, as well as the enhanced selectivity of quantifying peptide-specific fragment ions (24–28). Reporter ion-based isobaric tagging reagents (iTRAQ, TMT) are used for MS2-based quantification (29, 30), but their usefulness is limited by (1) the need to detect reporter ions in the low mass range, which limits the range of suitable instruments; and, more importantly, (2) the potential for co-eluting, tagged peptide ions to fall within the CID isolation window. Since the same reporter ions are generated from any tagged peptide present during CID, co-eluting peptides can alter the quantification result.
The MSTIQ strategy uses peptide-specific fragment ions instead of nonspecific reporter ions for quantification, allowing the targeted peptide to be distinguished from co-eluting peptides (Fig. 1). As described above, a synthetic LH reference peptide and the HL sample peptide are isotopically distinct at both their N terminus and C terminus; b- and y-ion fragments appear in the MS2 spectrum as pairs (separated by 4 Da/z), one from the reference peptide and one from the sample peptide. Alternatively, the LH and HL peptides can be prepared from tryptic digestion of proteins that are differentially labeled by metabolic incorporation of isotopically heavy or light lysine and arginine (SILAC) (23); this allows two samples to be directly compared. Quantification is achieved by comparing the relative intensities of corresponding fragment ion pairs; each ratio of intensities is an independent measurement of the relative abundance of the HL and LH peptides. Ratios from all observed fragment ion pairs (across multiple scans, when available) are analyzed together to reject outliers, then averaged to improve accuracy (see Experimental procedures).
Evaluation of MSTIQ as a Tool for Quantitative Proteomics
We examined MSTIQ's effectiveness by preparing two differentially labeled protein mixtures from mouse macrophage lysates, mixing the samples at known relative amounts, and measuring the relative levels of all peptides in each mixture. To avoid possible bias due to evaluating a preselected set of targeted peptides, SILAC-labeled mouse macrophages were used to prepare a large number of MSTIQ peptides for LC-MS/MS analysis. Isotopically heavy or light arginine ([15N4] or [14N4]) and lysine ([13C615N2] or [12C614N2]) were metabolically incorporated into the proteome during cell culture. After digesting the SILAC-labeled proteins with trypsin, the resulting peptides were labeled with either light or heavy mTRAQ reagents to create isobaric peptides. The labeled peptides were combined in seven different ratios from 1:30 to 30:1 (HL:LH), and analyzed by LC-MS/MS using intensity-based data-dependent acquisition (DDA) followed by database searching to identify a set of target peptides for quantification. Next, our in house software ISBquant (Isobaric quantification), identified MS2 scans corresponding to these target peptides in each titer based on accurate mass measurements and the presence of at least 7 HL or 7 LH fragment ions above background level, and computed an abundance ratio (HL/LH) for each peptide using the intensities of the quantifiable peptide-specific fragment ion pairs (see Experimental Procedures). We note that both in this experiment and in the iMSTIQ approach identification of scans corresponding to target peptides is not based on sequence database search algorithms. This is important because we noticed that some peptide spectra received lower X!Tandem scores in the 1:1 titer than the corresponding spectra in the 1:30 or 30:1 titers, indicating that the presence of doublet fragment ions can negatively impact X!Tandem scores.
Fig. 3 shows the MS2 spectra from the 1:1 mixture for one representative peptide, “LWTLVSEQTR”. A series of b- and y- ions appearing as isotopic pairs is seen in the spectrum (Fig. 3A). The spectra for three representative fragment ion pairs (b4+, y7+, and y9+) are shown in expanded views (insets). Two peaks separated by 4 m/z units are observed for each fragment ion pair and their relative peak intensities are ∼1:1, as expected.
We then evaluated relative abundances of the HL and the LH peptides in each titration experiment by examining a total of 1080 peptides that were positively identified (PeptideProphet p ≥ 0.9, false discovery rate ≤ 1.5%) in at least one titration. The ratios for all peptides quantified in each titer by ISBquant are summarized in Fig. 3B. Excellent linearity between the MSTIQ-measured and expected ratios is observed over relative abundances of 1:30 to 30:1 (R2 = 0.9992). The accuracy of the method is indicated by the histograms of individual peptide quantifications within 2 standard deviations of the calculated median; 50% of the quantifications have a relative error ≤13% (Suppl. Tables S2, S3). These results demonstrate that MSTIQ is an effective method for accurate quantification of peptides in complex mixtures over a wide dynamic range.
The iMSTIQ Platform Permits Quantification of Low Abundance Peptides in Complex Mixtures
A combination of the enhanced sensitivity and reproducibility of ITA and the accurate quantification of MSTIQ generates a powerful new proteomics platform for quantification of targeted proteins (Fig. 1). In this platform, index peptides containing isotopically heavy moieties at both their N and C termini (“HH” peptides) are used to trigger MS2 analysis of the co-eluting pair of MSTIQ-labeled target peptides (HL sample and LH reference peptides). Multiple MS2 scans are acquired in the ion trap for each targeted m/z (every 1–2 s across the entire elution peak). Due to a decrease in sensitivity when the Orbitrap is used for MS2 analysis, MS2 scans were acquired in the ion trap. It is possible that the use of higher-energy C-trap dissociation (HCD) (42) or future instrument improvements will allow acquisition of high resolution MS2 spectra in the Orbitrap with sensitivity similar to that of the ion trap. MS2 scans containing the target HL and reference LH peptides are selected for MSTIQ quantification based on co-elution of the index ion, accurate index ion m/z, and the presence of peptide-specific fragment ions in MS2 spectra (see Experimental Procedures). The iMSTIQ platform is particularly useful for targeted proteomics analyses in which reproducible and accurate quantification of a specific set of proteins is desired in complex samples, such as in time course or biomarker studies in which a set of proteins is repeatedly measured in multiple samples.
To evaluate iMSTIQ's ability to quantify low abundance peptides in complex samples, we generated a pair of MSTIQ peptides [“FAISYQEK”, HL (95% purity) and LH (crude)] and spiked ∼3 fmol of each into 1 μl N-glycopeptide mixture (∼1 μg) isolated from 1 μl human plasma (∼70 μg/μl) (19). This N-glycopeptide mixture is similar to that used in studies which investigate the sensitivity of peptide detection in complex mixtures by other MS approaches (9, 31). One hundred fmol of the HH peptide was also spiked into the same sample to serve as the index peptide. Fig. 4A shows a representative full MS1 scan at the time when the HL, LH, and HH peptides eluted; an expanded view of 566 to 575 m/z is shown in the inset. Detection of the peak at 572.3028 m/z (corresponding to the index peptide ion) triggered CID on ions within a window of 567.2–570.2 m/z according to the preset ITA program. As shown in the inset, the targeted peptide ion (568.2973 m/z) is either not detectable or overlaps with an isotopic peak (568.2954 m/z) of a co-eluting, off-target ion (567.2920 m/z) and could be missed for CID if the inclusion list method was applied. Nonetheless, ITA correctly triggered CID as shown by the presence of a series of fragment ion pairs specific to the targeted peptide in the MS2 spectrum (Fig. 4B). As a result, the targeted peptide was positively identified and quantified. Consistent with the observed complexity of the MS1 spectrum, in the MS2 spectrum the intensities of the target peptide fragment ion pairs (identified doublets; insets) are much lower than the intensities of the fragment ions derived from the co-eluting ion (observed as singlets). Strikingly, iMSTIQ permits identification and quantification of target peptides in complex mixtures even if they co-elute with other ions. If a reporter ion strategy such as iTRAQ or TMT was used, it would not be possible to distinguish the reporter ions derived from the target versus the co-eluting, off-target peptides, and this could compromise the accuracy of quantification. The abundance ratio of this peptide was determined using the ISBquant software, which was specifically developed for the iMSTIQ platform. ISBquant used multiple peptide-specific pairs of fragment ions observed in multiple MS2 scans across the elution peak of the index peptide to quantify the peptide (Fig. 4C).
To gain insight into the reliability of MS2 data acquisition and the range of quantification that iMSTIQ can provide in a complex mixture, we further examined quantification of the “FAISYQEK” peptide at various amounts of the spiked-in LH peptide, as well as seven other synthetic peptides. In these titration experiments, the LH peptides (synthesized via the AQUA platform (32) with 95% purity) were spiked in to ∼1 μg of an N-glycopeptide mixture at quantities ranging from 0.1 to 30 fmol, whereas the HH (AQUA) and HL (crude) peptides were spiked in at fixed amounts (100 fmol and ∼3 fmol, respectively), to generate expected relative abundances (LH:HL) of 1:30 to 10:1. The ratios measured via ISBquant for all eight peptides are plotted against the spiked amount of the LH peptides (Fig. 4D, data for each peptide is presented separately in supplemental Fig. S1 and supplemental Table S4). Consistent with the results presented in Fig. 2, ITA triggered MS2 data acquisition for all eight target peptides in every titer. In samples containing 1 to 30 fmol of the LH peptides, there is good agreement between measured and expected ratios for all eight peptides. For LH peptides spiked in at less than 1 fmol the measured ratio is independent of LH, suggesting that the limit of quantification has been reached. The average coefficient of variation for these 8 peptides is 26%; this is likely an upper estimate of iMSTIQ's coefficient of variation in typical use since the experiments were performed close to the limit of quantification of the technique. We then quantified the actual amount of each HL peptide (crude) in the sample based on the linear relationships observed for LH peptide (accurately quantified AQUA peptide) amounts of ≥3 fmol (see supplemental Fig. S1). The estimated amount of each HL peptide was between 1 and 3 fmol, which is consistent with the expected amounts of these crude peptides based on the information provided by the supplier. These experiments show that the iMSTIQ platform reliably triggers MS2 data acquisition and accurately quantifies low fmol amounts of target peptides in complex samples over a >30-fold range of abundances. In addition, they demonstrate that iMSTIQ can be used to determine absolute quantities of target peptides in a complex mixture.
iMSTIQ Abundance Profiling of Targeted Inflammatory Proteins During LPS Activation of Macrophages
Next we applied iMSTIQ to analyze temporal patterns of protein release from mouse macrophages treated with the inflammatory stimulus, LPS. Release of inflammatory mediators such as cytokines and chemokines by macrophages is a key component of the inflammatory response (33). Previously, using shotgun proteomics combined with iTRAQ (29) and N-glycopeptide enrichment (19), we detected a set of proteins that were inducibly released (secreted or proteolytically shed) from macrophages upon treatment with LPS for 4 h (unpublished data). We investigated whether iMSTIQ could be used to directly profile the abundance of these inflammatory proteins in conditioned media during a comprehensive time course experiment. Samples of conditioned media were collected at various times following treatment of macrophages with LPS. As a control, macrophages were treated with PBS and conditioned media were harvested at 18 h. Proteins from the concentrated conditioned media were digested with trypsin, deglycosylated with N-Glycanase, and then labeled with the heavy mTRAQ reagents to generate HL peptides. We specifically targeted 14 peptides corresponding to proteins that were found to be inducibly released from macrophages upon LPS treatment in our previous screen (Table I). We also targeted 10 peptides corresponding to proteins that are not known as macrophage-released proteins (referred to as nonspecific peptides). These 24 peptides were synthesized with isotopically heavy lysine at their C terminus, and then modified with either light or heavy mTRAQ to generate quantification reference (LH) or index peptides (HH), respectively. The sample peptide mixtures containing the HL peptides were then spiked with the LH peptides (33.3 fmol each) and the HH peptides (500 fmol each) and analyzed on the LTQ-Orbitrap using ITA.
Table I.
| Gene symbol | Protein | Peptide | Proteins of interest | Detection of HL | Group |
|---|---|---|---|---|---|
| Cadm1 | Cell adhesion molecule 1 | FQLLN*FSSSELK | targeted | Yes | 1 |
| Fcgr1 | Fc receptor, IgG, high affinity I | EVVN*ATK | targeted | Yes | |
| Fcgr2b | Fc receptor, IgG, low affinity IIb | SQVQASYTFK | targeted | Yes | |
| Gm885 | Predicted gene 885 | VN*VSNLMK | targeted | Yes | |
| H2-k1 | histocompatibility 2, K1, K region | WASVVVPLGK | targeted | Yes | |
| Itgam | Integrin alpha M | YLN*FTASEMTSK | targeted | Yes | |
| Itgb2 | Integrin beta 2 | LTDNSNQFQTEVGK | targeted | Yes | |
| Raet1 | Retinoic acid early Transcript 1, A-E | CN*LTIK | targeted | Yes | |
| Sema4d | Semaphorin 4D | AAN*YTSSLNLPDK | targeted | Yes | |
| Sorl1 | Sortilin-related receptor, LDLR class A repeats-containing | GIGN*WSDSK | targeted | Yes | |
| Tnf | Tumor necrosis factor | VNLLSAVK | targeted | Yes | |
| Gm11787 | Predicted gene 11787 | GSLLDFLK | nonspecific | Yes | 2 |
| Islr | Immunoglobulin superfamily containing Leucine-rich repeat | FQAFAN*GSLLIPDFGK | nonspecific | Yes | |
| P2rx4 | Purinergic receptor P2X, ligand-gated ion channel 4 | AAEN*FTLLVK | nonspecific | Yes | |
| Alcam | Activated leukocyte cell adhesion molecule | N*ATGDYK | targeted | No | 3 |
| Anpep | Alanyl (membrane) aminopeptidase | N*ATLVNEADK | targeted | No | |
| Havcr2 | Hepatitis A virus cellular receptor 2 | N*VTYQK | targeted | No | |
| Bst1 | Bone marrow stromal cell antigen 1 | N*CTAIWEAFK | nonspecific | No | 4 |
| Chmp2a | Chromatin modifying protein 2A | SN*NSMAQAMK | nonspecific | No | |
| Dclk1 | Doublecortin-like kinase 1 | NVNPN*WSVNVK | nonspecific | No | |
| Serpinf1 | Serine (or cysteine) peptidase inhibitor, clade F, member 1 | SSFVAPLEK | nonspecific | No | |
| Smc3 | Structural maintenace of chromosomes 3 | ALDQFVN*FSEQK | nonspecific | No | |
| Vcam1 | Vascular cell adhesion molecule 1 | SLEVTFTPVIEDIGK | nonspecific | No | |
| Ythdf2 | YTH domain family 2 | VQN*GSVHQK | nonspecific | No | |
N*: N-linked glycosylation site replaced by D in synthetic peptides.
Consistent with previous results, MS2 data acquisition was reproducibly triggered by ITA for all 24 target peptides at each time point (Fig. 5 and data not shown) which permitted identification of all LH peptides used as quantitative reference peptides by ISBquant. 10 sample peptides (HL peptides), including three from the targeted group and seven from the nonspecific group (Table I, group 3 and 4 respectively), were not consistently detected in all six samples and thus were not further analyzed. The other 14 peptides, including 11 targeted peptides (Table I, group 1) and three nonspecific peptides (Table I, group 2), were successfully detected in all of the samples and the abundance ratio between the peptides in the sample and the spiked reference peptides (HL/LH) was determined for each peptide at all monitored time points (supplemental Table S5).
Fig. 5 shows absolute quantification (fmol) of each peptide (y axis) at each monitored time point (x axis). Evaluation of the 3 nonspecific peptides (group 2) indicates that they are present in similar amounts at all measured time points following LPS or PBS treatment (Fig. 5A). In contrast, 8 of the 11 targeted peptides (group 1) showed the expected overall pattern of inducible release by macrophages over the time course of LPS treatment (Fig. 5B). Proteins corresponding to these peptides include TNFα, Itgb2 (CD18) and Itgam (Mac-1, CD11b). TNFα is a type 1 pro-inflammatory cytokine known to be released by macrophages in response to inflammatory stimuli (34, 35). Itgb2 and Itgam are components of a heterodimeric complex found on the surface of leukocytes that plays important roles in inflammation (36), and was previously reported as being shed during an inflammatory response (37, 38). Unexpectedly, the amounts of the 3 other targeted peptides, corresponding to Gm885, Raet1, and Cadm1, showed no significant increase after LPS treatment (Fig. 5C), suggesting that they are not specifically released in response to LPS. These studies highlight the importance of systematically validating potential proteins of interest identified in snapshot discovery screens. Overall, these results demonstrate the utility of iMSTIQ as a powerful approach to reproducibly quantify specific sets of proteins in complex mixtures, a capability that is an essential component of systems biology research and biomarker studies.
DISCUSSION
Comprehensive investigation of biological systems requires the ability to measure any protein or set of proteins in an unbiased, sensitive, reproducible, and quantitative manner. To address these requirements, we have developed iMSTIQ, a novel proteomics platform that permits reproducible detection (via ITA) and accurate quantification (via MSTIQ) of targeted peptides in complex mixtures.
Application of ITA significantly increased the frequency at which targeted peptides were selected for CID compared to an inclusion list approach (16–17) and thus resulted in improved sensitivity and reproducibility of peptide identification (Fig. 2). The ability of ITA to reliably trigger MS2 data acquisition was further demonstrated in our time course experiment in which we measured the release of targeted proteins from macrophages after LPS stimulation (Table I and Fig. 5) and the N-glycoplasma spike-in experiment (Fig. 4C).
Another benefit of the ITA strategy is that the use of index peptides enhances the confidence of identification of the targeted peptides. The index peptides themselves are readily identified by accurate mass measurement, retention time, and the presence of a minor peak −1/z units from the index peptide's monoisotopic m/z. This minor peak results from the use of isotopically heavy labeling reagents that contain a small fraction of 12C and 14N atoms in them to produce the index peptides. Considering the isotopic impurity of the reagents (∼1–2%) and the fact that index peptides (the HH form) contain at least two modifications per peptide, the intensity of this “-1” peak is estimated to be ∼10–20% of the monoisotopic peptide peak, depending on the precise purity of the reagents, natural isotopic variation, and composition of the peptide. We do indeed observe such unique index peptide features in our data (Fig. 4A, see peak at 571.8019 m/z), and we have incorporated this special feature into the ISBquant software to assist in identifying index peptides. Because the index and target peptides co-elute during LC, positive identification of the index peptides provides a parameter, in addition to full MS2 spectra, that can be used to confidently identify target peptides. This is particularly useful when analyzing peptides of very low abundance in samples of high complexity and dynamic range such as human plasma. Further improvements in target peptide identification are expected upon modification of the LTQ-Orbitrap operating software to permit acquisition of MS2 spectra for both the index peptide and the targeted peptides during iMSTIQ analysis.
MSTIQ is a quantitative method with improved selectivity compared with many other quantitative proteomics platforms. Similar to other isobaric, MS2-based quantitative approaches such as iTRAQ (29) and TMT (30), MSTIQ takes advantage of the mass filtering effect of MS1. An important and distinguishing feature of MSTIQ is its use of peptide-specific fragment ions for quantification. Compared with isobaric approaches that use common reporter ions for quantification, the use of peptide-specific fragment ions permits quantification even in the presence of a more intense contaminating peptide present during CID (Fig. 4B). Quantification using multiple peptide-specific fragments ions, together with statistical analysis, should also improve the accuracy of quantification. These improvements are particularly important when quantifying peptides in samples with high complexity and large dynamic range. We note however that should the user prefer to use a reporter ion approach for quantification, ITA analysis is also compatible with these approaches.
Other MS2-based quantitative proteomics approach have been described that use peptide-specific fragment ions for quantification (27, 28, 39). However, in these approaches an enlarged precursor isolation window (e.g. 10 m/z units) is used to include both light and heavy isotopically labeled precursor peptides for simultaneous CID and subsequent quantification based on the intensities of a subset of fragment ions (y-ions). While the accuracy of this strategy likely benefits from the use of multiple fragment ions for quantification, the wide isolation window that is required for this approach is a concern due to the potential for inclusion of unrelated peptide ions and chemical noise in the collision cell that may interfere with quantification. MSTIQ alleviates these limitations by using isobaric precursors, compatible with a standard CID isolation window (2–3 m/z units). In addition, MSTIQ uses both b- and y- ions for quantification instead of the y-ions alone. The potential for more measurements per peptide should improve quantitative accuracy. Another benefit specific to MSTIQ is the ability to detect potential errors introduced during the isotopic labeling step: systematic inconsistency between quantifications of b-ions versus y-ions may point to a problem that was introduced during the labeling.
During the preparation of our manuscript, an independent study was reported by Koehler et al. using a similar approach for quantifying isobaric peptides during MS2 (40). In that study, isobaric peptides were generated by chemical labeling of LysC digested peptides with both succinic anhydride (d0 or d4) at the N termini and 2-methoxy-4,5-dihydro-1H-imidazole (d4 or d0) at the C-terminal lysine residues, respectively. Consistent with our results, their study provides independent evidence for the utility of isobaric peptides labeled at both ends for quantification. However, the approach described by Koehler et al. is restricted to peptides ending with lysine, and requires two amine-based labeling steps. In addition, in their method protein abundance is only semiquantitatively estimated from Mascot scores (41) rather than from direct measurement of fragment ion intensities. Most importantly, this approach lacks a mechanism to ensure reliable MS2 data acquisition of target peptides. We note that although both MSTIQ and the approach described by Koehler et al. are not useful for quantification of peptides containing internal lysine or arginine residues (∼11% of tryptic peptides derived from the murine proteome that can be reliably identified by MS2 analysis are predicted to contain internal lysine or arginine due to the presence of KP/RP), this is not a significant limitation for targeted analyses as most proteins will produce many peptides upon enzymatic digestion that can be measured.
Integration of ITA and MSTIQ generates the iMSTIQ targeted proteomics methodology, which takes advantage of the sensitivity, reproducibility, and accurate quantification provided by the two approaches. A powerful example of the effectiveness of iMSTIQ is presented in Fig. 4, in which accurate peptide quantification was achieved despite the inability to clearly detect the target peptide in the MS1 spectrum. If an inclusion list method had been used under these circumstances, it is likely that CID would not have been triggered for the target ion due to low abundance and/or inability of the instrument software to distinguish the targeted ion from the co-eluting off-target ion. Another example of the utility of iMSTIQ is presented in Fig. 5 in which we were able to identify and quantify several peptides (Sorl1, Sema4d, Fcgr1, Fcgr2b, Itgam, Itgb2, Raet1, Cadm1, and Gm885) in unfractionated samples that previously required either glycopeptide enrichment or fractionation by isoelectric focusing to be detected using shotgun proteomics. In addition, all of these targeted peptides were successfully quantified at all six time points. Therefore, iMSTIQ provides a powerful targeted quantitative proteomics platform that allows for systematic analysis of protein levels or validation of proteins of interest discovered from shotgun analyses.
Several features of the iMSTIQ targeted proteomics platform are noteworthy. First, iMSTIQ has been developed and primarily implemented on LTQ-Orbitrap instruments. As such, it takes advantage of the high mass accuracy, resolution and throughput offered by this instrument, allowing at least two hundred peptides to be accurately targeted per MS run without stringent retention time requirements (16). The upper limit for targeting is primarily determined by column capacity which is typically ∼1 microgram. Assuming the use of ∼250 fmol of each index peptide to target peptides in 1 microgram of sample, 200 index peptides would only account for ∼10% of the total sample quantity. This compares favorably to stable isotope dilution (SID) SRM studies on a triple quadrupole instrument in which segmented methods that demand highly reproducible chromatography are required to target ∼100 peptides (assuming 3 transitions × 2 isotopic forms per peptide) with high sensitivity. Second, with iMSTIQ confident peptide identification is achieved by acquisition of full MS2 spectra and observation of a co-eluting index peptide ion. In contrast, high throughput SID-SRM studies typically rely on observation of a limited number of co-eluting transitions from isotopic peptide pairs, along with retention time coordinates, for peptide identification. Third, selection of peptides for targeted analysis benefits from the fact that iMSTIQ can be performed on ion trap instruments, the same instruments that have been used to acquire most of the profiling data by the proteomics community. Fourth, unlike high throughput SRM, iMSTIQ does not require predetermination of transitions for optimal measurements, relying instead on consistency of quantification to identify all reliable transitions available under the specific experimental conditions. Finally, iMSTIQ provides opportunities to target proteins that have never been detected by mass spectrometry, independent of any existing MS data. Currently, we are developing the second stage of the iMSTIQ platform in which isotopically heavy synthetic proteins of interest are directly spiked into samples as internal standards for quantification. All possible tryptic peptides of each targeted protein within the scanning range will be analyzed in a single MS run. In this way we will be able to directly analyze the targeted proteins without the constraints of preselecting a limited number of peptides and/or transitions.
Similar to SID SRM-MS, implementation of iMSTIQ requires peptides that contain isotopically heavy lysine or arginine at their C termini. While these peptides are currently costly, they can be readily obtained by chemical synthesis that is expected to become much more affordable in the near future. In addition, iMSTIQ currently requires a chemical labeling step in which both sample peptides and synthetic heavy peptides are modified with labeling reagents (Fig. 1). Labeling with mTRAQ reagents to generate both the index (HH) and reference (LH) peptide (100 μg each) costs ∼$50 USD, but once they are prepared, they serve as a resource for hundreds of analyses. Thus, once the index and reference peptides are prepared, the main sample preparation expense for an iMSTIQ analysis is the cost of mTRAQ reagents for labeling the sample peptides (∼$4 USD per 10 μg).
We note that it is possible to perform inclusion list-based targeted quantitative proteomics analysis with either MSTIQ or isobaric reporter ion-based approaches such as iTRAQ by using an appropriately labeled synthetic peptide that serves as both the MS2 triggering index peptide and the quantitative reference peptide. While this approach is expected to be effective for targeted analysis of abundant peptides, it is not optimal for less abundant peptides since, in many cases, the high levels of synthetic peptide necessary to ensure efficient MS2 triggering will not be ideal for quantification due to dynamic range limitations of the mass spectrometer. In addition, as discussed above, potential quantification errors due to contributions from co-eluting, tagged peptides that fall within the CID isolation window are possible with reporter ion-based approaches. With iMSTIQ, both of these issues are alleviated.
In conclusion, we have developed a novel targeted quantitative proteomics technology that has enhanced reproducibility and sensitivity compared with “shotgun” MS and inclusion list-based MS methods. The method alleviates potential quantification errors caused by co-eluting peptides and chemical noise to which other methods are susceptible. The technology has been developed for use with a high mass accuracy mass spectrometer capable of analyzing hundreds of peptides per run without the stringent retention time requirements of high throughput SID SRM-MS studies, but can be adapted to any MS platform that permits targeted, full MS2 analysis. We have demonstrated here that iMSTIQ accurately quantifies peptides in complex biological samples such as samples derived from cell extracts, cell culture media and plasma. By targeting all of the potentially observable peptides of a protein of interest, iMSTIQ provides a way to measure any protein with high sensitivity and reproducibility without pre-existing proteomics data. As such, iMSTIQ holds great promise as a robust method for facilitating efforts to systematically profile specific sets of proteins and even complete proteomes.
Acknowledgments
We thank Robert Moritz and Mark Gillespie for helpful comments on the manuscript, Min Yuan at the ISB proteomics facility for assistance with LC-MS analysis, and Vinzenz Lange, Ulrike Kusebauch, Simon Letarte, Yong Zhou, and Julian Watts for providing peptides.
W.Y. and J.L. jointly conceived the study and performed the experiments. W.Y., J.L. and J.R. designed the experiments. M.R., J.L. and J.E. developed data analysis software, and W.Y., J.L. and M.R. analyzed the data. W.Y., J.R. and M.R. wrote the paper. J.R. and R.A. supervised the project.
Footnotes
* This work was supported by a grant from the National Institute of General Medical Sciences grant (PM50 GM076547/Center for Systems Biology), and contracts from the National Heart, Lung, and Blood Institute, US National Institutes of Health (N01-HV-28179) and the University of Luxembourg.
 This article contains supplemental Figs. S1 and S2 and supplemental Tables S1 to S5.
This article contains supplemental Figs. S1 and S2 and supplemental Tables S1 to S5.
1 The abbreviations used are:
- iMSTIQ
- index-ion triggered MS2 ion quantification
- ITA
- index-ion triggered analysis
- MSTIQ
- MS2 ion quantification
- SRM
- selected reaction monitoring
- LPS
- lipopolysaccharide
- SILAC
- stable isotope labeling with amino acids in cell culture.
REFERENCES
- 1. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review, Analyt. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 3. Panchaud A., Affolter M., Moreillon P., Kussmann M. (2008) Experimental and computational approaches to quantitative proteomics: status quo and outlook. J. Proteomics 71, 19–33 [DOI] [PubMed] [Google Scholar]
- 4. Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 5. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 6. Gevaert K., Impens F., Ghesquière B., Van Damme P., Lambrechts A., Vandekerckhove J. (2008) Stable isotopic labeling in proteomics. Proteomics 8, 4873–4885 [DOI] [PubMed] [Google Scholar]
- 7. Carr S. A., Anderson L. (2008) Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory?. Clin. Chem. 54, 1749–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 9. Stahl-Zeng J., Lange V., Ossola R., Eckhardt K., Krek W., Aebersold R., Domon B. (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
- 10. Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lange V., Malmström J. A., Didion J., King N. L., Johansson B. P., Schäfer J., Rameseder J., Wong C. H., Deutsch E. W., Brusniak M. Y., Bühlmann P., Björck L., Domon B., Aebersold R. (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol. Cell Proteomics 7, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeSouza L. V., Taylor A. M., Li W., Minkoff M. S., Romaschin A. D., Colgan T. J., Siu K. W. (2008) Multiple reaction monitoring of mTRAQ-labeled peptides enables absolute quantification of endogenous levels of a potential cancer marker in cancerous and normal endometrial tissues. J. Proteome Res. 7, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 13. Keshishian H., Addona T., Burgess M., Mani D. R., Shi X., Kuhn E., Sabatine M. S., Gerszten R. E., Carr S. A. (2009) Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt A., Gehlenborg N., Bodenmiller B., Mueller L. N., Campbell D., Mueller M., Aebersold R., Domon B. (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol. Cell Proteomics 7, 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaffe J. D., Keshishian H., Chang B., Addona T. A., Gillette M. A., Carr S. A. (2008) Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol. Cell Proteomics 7, 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 19. Tian Y., Zhou Y., Elliott S., Aebersold R., Zhang H. (2007) Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2, 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilcox R. R. (2005) Introduction to robust estimation and hypothesis testing, 2nd ed., Elsevier Academic Press [Google Scholar]
- 21. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 22. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 23. Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 24. Arnott D., Kishiyama A., Luis E. A., Ludlum S. G., Marsters J. C., Jr., Stults J. T. (2002) Selective detection of membrane proteins without antibodies: a mass spectrometric version of the Western blot. Mol. Cell Proteomics 1, 148–156 [DOI] [PubMed] [Google Scholar]
- 25. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 26. Berger S. J., Lee S. W., Anderson G. A., Pasa-Tolić L., Tolić N., Shen Y., Zhao R., Smith R. D. (2002) High-throughput global peptide proteomic analysis by combining stable isotope amino acid labeling and data-dependent multiplexed-MS/MS. Anal Chem. 74, 4994–5000 [DOI] [PubMed] [Google Scholar]
- 27. Zhang G., Neubert T. A. (2006) Automated comparative proteomics based on multiplex tandem mass spectrometry and stable isotope labeling. Mol. Cell Proteomics 5, 401–411 [DOI] [PubMed] [Google Scholar]
- 28. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 29. Ross P. L., Huang Y. N., Marchese J., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. (2004) Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics M400129-MCP400200 [DOI] [PubMed] [Google Scholar]
- 30. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 31. Berven F. S., Ahmad R., Clauser K. R., Carr S. A. (2010) Optimizing performance of glycopeptide capture for plasma proteomics. J. Proteome Res. 9, 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stemmann O., Zou H., Gerber S. A., Gygi S. P., Kirschner M. W. (2001) Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726 [DOI] [PubMed] [Google Scholar]
- 33. Pradervand S., Maurya M. R., Subramaniam S. (2006) Identification of signaling components required for the prediction of cytokine release in RAW 264.7 macrophages. Genome Biol 7, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mohler K. M., Sleath P. R., Fitzner J. N., Cerretti D. P., Alderson M., Kerwar S. S., Torrance D. S., Otten-Evans C., Greenstreet T., Weerawarna K., et al. (1994) Protection against a lethal dose of endotoxin by an inhibitor of tumour necrosis factor processing. Nature 370, 218–220 [DOI] [PubMed] [Google Scholar]
- 35. Mukherjee S., Chen L. Y., Papadimos T. J., Huang S., Zuraw B. L., Pan Z. K. (2009) Lipopolysaccharide-driven Th2 cytokine production in macrophages is regulated by both MyD88 and TRAM. J. Biol. Chem. 284, 29391–29398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coxon A., Rieu P., Barkalow F. J., Askari S., Sharpe A. H., von Andrian U. H., Arnaout M. A., Mayadas T. N. (1996) A novel role for the beta 2 integrin CD11b/CD18 in neutrophil apoptosis: a homeostatic mechanism in inflammation. Immunity 5, 653–666 [DOI] [PubMed] [Google Scholar]
- 37. Evans B. J., McDowall A., Taylor P. C., Hogg N., Haskard D. O., Landis R. C. (2006) Shedding of lymphocyte function-associated antigen-1 (LFA-1) in a human inflammatory response. Blood 107, 3593–3599 [DOI] [PubMed] [Google Scholar]
- 38. Vaisar T., Kassim S. Y., Gomez I. G., Green P. S., Hargarten S., Gough P. J., Parks W. C., Wilson C. L., Raines E. W., Heinecke J. W. (2009) MMP-9 sheds the beta2 integrin subunit (CD18) from macrophages. Mol. Cell Proteomics 8, 1044–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson H., Wong S. C., Simpson D. M., Beynon R. J., Gaskell S. J. (2008) Protein quantification by selective isolation and fragmentation of isotopic pairs using FT-ICR MS. J. Am. Soc. Mass Spectrom 19, 973–977 [DOI] [PubMed] [Google Scholar]
- 40. Koehler C. J., Strozynski M., Kozielski F., Treumann A., Thiede B. (2009) Isobaric Peptide Termini Labeling for MS/MS-Based Quantitative Proteomics. J. Proteome Res. 8, 4333–4341 [DOI] [PubMed] [Google Scholar]
- 41. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 42. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis, Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]