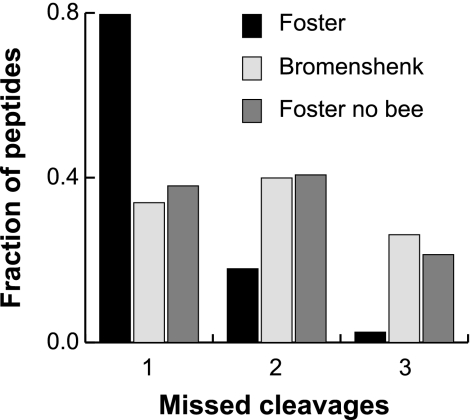
In a recent publication, Bromenshenk et al. claim that an iridovirus, Invertebrate Iridescent Virus-6 (IIV-6)1, is tightly linked to colony collapse disorder (CCD, the cause of many of the bee losses over the past four winters) based on proteomic analyses of bees from CCD-afflicted and unafflicted colonies (1). We believe that there are fundamental flaws in the interpretation of their data based on the following rationale. First, liquid chromatography-tandem MS (LC-MS/MS) tends to identify the most abundant proteins much more frequently and the major capsid protein of IIV-6 constitutes at least 17% of total virion protein (2) yet of the 792 IIV-6 peptides reported by the authors, only four (0.5%) are from protein 274L, the major capsid protein. This is especially troubling because the authors rely on spectral counting to correlate IIV-6 levels with CCD. Second, in the list of identified peptides provided by the authors there is a high frequency of missed cleavage sites. Trypsin is a very reliable protease (3) and, indeed, if we examine some of our own recent large-scale bee proteomic data sets (available at http://www.ebi.ac.uk/pride/), we find that nearly 80% of all peptides are perfect tryptic peptides, with ∼18% containing one missed cleavage and a few percent containing two (Fig. 1, black bars). The peptides from Bromenshenk et al. are skewed dramatically toward greater numbers of missed cleavages (Fig. 1, light grey bars), which could be explained in one of two possible ways: (1) that the tryptic digest was inefficient, or (2) that many of the peptide identities are incorrect (i.e. a high false discovery rate (FDR)). Because there is no independent “gold standard” MS/MS data from IIV-6 proteins to compare against it is difficult to definitively evaluate the efficacy of trypsin from these data. However, other aspects of the described Methods suggest that the second possibility, a high FDR, is the more likely explanation: the authors state that they did not consider bee protein sequences when interpreting their MS/MS spectra, only pathogen protein sequences. Others have shown that when identifying proteins using a search engine such as SEQUEST or Mascot it is important to consider all the protein sequences that might be present in the sample or risk a high FDR (4). If we take the above-mentioned, large-scale LC-MS/MS dataset acquired on an linear trap quadrupole (LTQ)-OrbitrapXL, that should have similar fragmentation characteristics to the LTQ data reported by the authors, and search all 692,336 MS/MS against a database comprised only of proteins from IIV-6 and all other known bee viruses (i.e. no Apis mellifera sequences), we can also “identify” 103 IIV-6 peptides. However, if we include A. mellifera protein sequences in this search, as well as the virus sequences, then only a single IIV-6 peptide is found at an FDR of 1% based on reversed database searching: the other 102 spectra that matched IIV-6 peptides in the absence of bee sequences match considerably better to bee peptides than to IIV-6 peptides. In other words, at least 102 of the 103 matches were false discoveries when bee proteins were not considered. Interestingly, if one then plots the distribution of missed trypsin cleavages in the false IIV-6 peptides that we have “discovered,” the distribution is almost identical to that of the peptides from Bromenshenk et al. (Fig. 1, dark grey bars). We believe that there is currently insufficient evidence to conclude that bees are a natural host for IIV-6, let alone that the virus is linked to CCD.
Fig. 1.
Missed cleavages in peptides. A large-scale honey bee LC-MS/MS dataset was acquired on an LTQ-OrbitrapXL as described (5) and searched using MaxQuant against two different protein libraries: (1) all Apis mellifera protein sequences plus sequences from Israeli Acute Paralysis Virus, Kashmir Bee Virus, Black Queen Cell Virus, Invertebrate Iridescent Virus 6, Deformed Wing Virus, and Acute Bee Paralysis Virus, or (2) just the above mentioned virus sequences. The number of missed trypsin cleavages (defined as the count of internal R or K residue except those followed by a P) was then evaluated in the results from these two searches (black bars for search #1, dark grey bars for search #2), as well as the list of peptides provided by Bromenshenk et al. (light grey bars).
Footnotes
1 The abbreviations used are:
- IIV-6
- invertebrate iridescent virus-6
- CCD
- colony collapse disorder
- FDR
- false discovery rate
- LTQ
- linear trap quadrupole.
REFERENCES
- 1. Bromenshenk J. J., Henderson C. B., Wick C. H., Stanford M. F., Zulich A. W., Jabbour R. E., Deshpande S. V., McCubbin P. E., Seccomb R. A., Welch P. M., Williams T., Firth D. R., Skowronski E., Lehmann M. M., Bilimoria S. L., Gress J., Wanner K. W., Cramer R. A., Jr. (2010) Iridovirus and microsporidian linked to honey bee colony decline. PLoS One 5, e13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ince I. A., Boeren S. A., van Oers M. M., Vervoort J. J., Vlak J. M. (2010) Proteomic analysis of chilo iridescent virus. Virology 405, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Olsen J. V., Ong S. E., Mann M. (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 4. Cargile B. J., Bundy J. L., Stephenson J. L., Jr. (2004) Potential for false positive identifications from large databases through tandem mass spectrometry. J Proteome Res 3, 1082–1085 [DOI] [PubMed] [Google Scholar]
- 5. Parker R., Melathopoulos A. P., White R., Pernal S. F., Guarna M. M., Foster L. J. (2010) Ecological adaptation of diverse honey bee (apis mellifera) populations. PLoS ONE 5, e11096. [DOI] [PMC free article] [PubMed] [Google Scholar]