Abstract
Lung cancer continues to be the leading cause of cancer death in the USA and the best example of a cancer with undisputed evidence of environmental risk. However, a genetic contribution to lung cancer has also been demonstrated by studies of familial aggregation, family-based linkage, candidate gene studies and most recently genome-wide association studies (GWAS). The African-American population has been underrepresented in these genetic studies and has patterns of cigarette use and linkage disequilibrium that differ from patterns in other populations. Therefore, studies in African-Americans can provide complementary data to localize lung cancer susceptibility genes and explore smoking dependence-related genes. We used admixture mapping to further characterize genetic risk of lung cancer in a series of 837 African-American lung cancer cases and 975 African-American controls genotyped at 1344 ancestry informative single-nucleotide polymorphisms. Both case-only and case–control analyses were conducted using ADMIXMAP adjusted for age, sex, pack-years of smoking, family history of lung cancer, history of emphysema and study site. In case-only analyses, excess European ancestry was observed over a wide region on chromosome 1 with the largest excess seen at rs6587361 for non-small-cell lung cancer (NSCLC) (Z-score = −4.33; P = 1.5 × 10−5) and for women with NSCLC (Z-score = −4.82; P = 1.4 × 10−6). Excess African ancestry was also observed on chromosome 3q with a peak Z-score of 3.33 (P = 0.0009) at rs181696 among ever smokers with NSCLC. These results add to the findings from the GWAS in Caucasian populations and suggest novel regions of interest.
Introduction
Lung cancer continues to be the leading cause of cancer death in the USA (1) and the best example of a cancer with undisputed evidence of environmental risk. Although 80– 90% of lung cancer incidence is attributed to cigarette smoking, only 15% of smokers develop this disease suggesting that there are individual differences in susceptibility. Differential risk is also seen by race; African-Americans tend to smoke fewer cigarettes (2), yet have higher incidence rates for lung cancer (1), and have higher risk associated with a family history of lung cancer than Caucasians (3). Contributions to the understanding of the genetic susceptibility to lung cancer risk have come from a family-based linkage study (4), candidate gene studies (reviewed in ref. 5) and genome-wide association studies (GWAS) (6–9) conducted in primarily Caucasian populations. Studies of candidate genes have produced mixed results (5), in part reflecting the small size of many of these studies, leaving open the question of the contribution to risk from common low-penetrant genes. The family linkage study and the GWAS have identified several chromosomal regions of interest including 6q23–25 (4), 15q25 (6–8) and 5p15 and 6p21 (10). The 15q25 region contains the nicotinic acetylcholine receptor subunit genes CHRNA3 and CHRNA5 raising the possibility that these genes indirectly affect lung cancer risk through smoking behavior (8,11) versus a direct affect on lung cancer risk (6–9).
Examining genetic risk in African-Americans provides complementary data to localize lung cancer susceptibility genes and explore smoking dependence-related genes because this population has different patterns of cigarette use and linkage disequilibrium (LD) than is seen in other populations. Since admixture between Europeans and Africans occurred relatively recently, large chromosomal regions of European or African origin remain in admixture-induced LD. Admixture mapping represents an alternative strategy for gene discovery that benefits from the LD characteristics of admixed populations (12,13). This chromosomal structure supports the use of ancestry informative single-nucleotide polymorphisms (SNPs) as markers for disease genes that occur differentially between populations of different origin. This approach also requires fewer markers and smaller sample sizes than GWAS (14,15). This study is the first to use admixture mapping to further characterize genetic risk of lung cancer, taking advantage of the higher risks for lung cancer in African-Americans compared with Caucasians.
Materials and methods
Case and control populations
Institutional Review Board approval was obtained at all study sites and informed consent was obtained from all participants. Cases and controls were enrolled through three sites: Wayne State University (WSU), M. D. Anderson Cancer Center (MDA) and the National Cancer Institute (NCI). WSU identified population-based cases through the Metropolitan Detroit Cancer Surveillance System, a participant in the NCI’s Surveillance, Epidemiology and End Results program. Three WSU studies were used in this analysis, all conducted by the same study staff using identical procedures; studies differed only in the eligibility of cases as detailed in other publications. The Family History Study focused on never-smokers and cases diagnosed before age 50 years regardless of smoking habits (16,17), the Women’s Epidemiology of Lung Diseases study focused on non-small-cell lung cancer (NSCLC) in women (18,19) and the Exploring Health, Ancestry and Lung Epidemiology (EXHALE) study is ongoing and includes only African-Americans. With the exception of the EXHALE study, population-based controls were chosen using random digit dialing methods. The EXHALE study relies on volunteer controls identified as friends of the cases and through advertising. All study controls were frequency matched to cases by 5-year-age group, sex and self-reported race. In total, 444 African-American cases and 559 African-American controls from WSU studies were included.
The MDA cases and controls were selected from a large hospital-based case–control study. This population has been described in detail elsewhere (20). Histopathologically confirmed lung cancer cases were recruited from the University of Texas M. D. Anderson Cancer Center and the Michael E. DeBakey VA Medical Center in Houston, TX. Controls were recruited from the Kelsey–Seybold Foundation, a multispeciality physician group practice in the Houston metropolitan area, and were matched to the cases on race, age (±5 years) and sex. In total, 192 African-American cases and 188 African-American controls from MDA were included.
The NCI cases and controls were selected from an ongoing case–control study in Baltimore, MD. The study population and enrollment criteria have been described previously (21). Histopathologically confirmed lung cancer cases were recruited from seven hospitals in the Baltimore, Maryland metropolitan area. Cancer-free population controls were collected through a list derived from the Maryland Department of Transportation. Two hundred and one African-American cases and 228 African-American controls were included from the NCI study.
Genotyping
The WSU and NCI studies used commercially available kits to isolate DNA. The Gentra AutoPure Kit (Qiagen, Valencia, CA) was used to extract DNA from 932 whole blood samples and the Gentra Puregene Kit (Qiagen) was used for 71 mouthwash and saliva samples from WSU. DNA was isolated from whole blood for the NCI samples using the Flexi Gene DNA kit (Qiagen).
For the MDA samples, a standard protocol was used to isolate DNA from the lymphocyte cell pellet collected from the buffy coat by centrifugation of whole blood. Pellets were digested with proteinase K and RNase, and the DNA was extracted with phenol and chloroform/isoamyl alcohol and then precipitated with ethanol.
Concentrated DNA samples for 30 Center d’Etude du Polymorphisme Humain (CEPH) individuals were purchased from the Coriell Institute for Medical Research in Camden, NJ (http://ccr.coriell.org) and used for quality control.
Genotyping was conducted for all samples at the WSU Applied Genomics Technology Center. Isolated DNA samples were diluted to the optimal concentration of ∼75 ng/μl and were run on the commercially available African-American Admixture Panel from Illumina (http://www.Illumina.com). This panel contains 1509 genome-wide SNPs informative for West African versus European ancestry. The Illumina BeadStudio platform was used to cluster the genotypes and provide the completion rates for individual samples and a measure of the predicted accuracy of each SNP (gentrain score).
Quality control
A total of 1864 eligible samples were genotyped. Forty-one samples were excluded after failure to meet sample quality checks (call rates <95% or low DNA concentration). An additional 11 samples had high European ancestry estimates (>85% European ancestry) and were excluded due to potential sample misclassification. Thus, a total of 1812 samples were eligible for analysis.
In addition to excluding entire samples of poor quality, SNPs with poor genotyping accuracy were excluded from all analyses using the following criteria. First, SNPs were excluded from all analyses if they were in the lowest 5% of gentrain scores; the X chromosome SNPs had both poor gentrain scores and high heterozygosity rates in men, so were excluded. Second, SNPs that were out of Hardy–Weinberg equilibrium, as indicated by a P-value <1 × 10−5, were excluded from analysis. Finally, SNPs with >15% of the genotypes varying from the HapMap reported genotypes in 30 CEPH samples were excluded. In the 1257 SNPs with HapMap genotypes available for comparison, there was a concordance rate of 99.9% among the previously genotyped CEPH samples compared with genotyping conducted in this study. After all exclusions, 1344 SNPs were eligible for inclusion in the analyses.
Statistical analysis
Data from 1812 participants and 1344 SNPs were analyzed using ADMIXMAP software. This program fits a Bayesian probability model using a Markov chain Monte Carlo algorithm to compare observed versus expected ancestry across the genome conditional on a priori ancestral genotype frequencies. Regions of excess European or African ancestry suggest that risk variants more common in one of the ancestral populations might be present. Both case–control and case-only analyses were conducted adjusting for age at diagnosis for cases or age at interview for controls, sex, smoking pack-years, family history of lung cancer, history of emphysema and study site. The case-only test compares ancestry at each locus to overall ancestry. The case–control test compares ancestry at each locus between cases and controls while adjusting for individual admixture and covariates included in the model (http://homepages.ed.ac.uk/pmckeigu/admixmap/manual_desc.html). West African Yoruban (YRI) and CEPH Europeans from Utah (CEU) HapMap genotype results were used to estimate prior allele frequencies for the ancestral populations. Analyses were repeated stratified by sex, smoking status and histology type.
Results from the ADMIXMAP analyses are presented in terms of standard normal Z statistics, ancestry risk ratios (aRRs) from case-only analyses, ancestry odds ratios from case–control analyses and P-values. A negative Z-score indicates excess European ancestry at a locus and a positive Z-score indicates excess African ancestry at a locus among cases. Similarly, a risk ratio or an odds ratio >1 indicates increased risk of lung cancer associated with carrying two African alleles in comparison with no African alleles; values <1 indicate reduced risk of lung cancer associated with carrying two African alleles in comparison with no African alleles. Regions with an absolute value Z-score of 3.00 or greater at multiple SNPs (P-value ∼0.0025) in the case-only analysis were further evaluated by conducting repeat analyses that dropped the SNP with the highest absolute Z-score to ensure that results were not driven by a single SNP. Findings are presented for any SNP with an absolute value Z-score ≥ 3.00 in the case-only analysis, which also demonstrated a similar increase in ancestry in the case–control analysis. All SNPs included in these analyses were a minimum of 200 kb apart with a median spacing of 1400 kb. In areas with high absolute Z-scores, ADMIXMAP results evaluating residual LD between SNPs were reviewed. As a result of this review, three SNPs that were tightly linked to adjacent SNPs (score test for residual LD, P < 0.001) on chromosome 1 were removed before final analysis. Additionally, ADMIXMAP was run while excluding every other SNP in the chromosome 1 peak area. This analysis continued to show a chromosome 1 peak that was reduced in size but still very strong. Two-sided tests of significance were applied to case-only and case–control analyses to capture both African and European alleles that may be associated with increased risk of lung cancer.
Results
Table I describes the characteristics of cases and controls (overall and by study site). In the entire sample, females were overrepresented among both cases and controls. Collectively, cases were slightly older, more probably to be smokers and smoked more than controls. Cases and controls were well matched by sex for the WSU and NCI populations; however, MDA cases and controls varied significantly. Overall, 17.9% of cases and 3.7% of controls reported an emphysema diagnosis. Rates of emphysema did not differ significantly across the studies. Adenocarcinoma occurred more frequently in the Detroit population due to the overrepresentation of this histology type in one of the WSU studies and squamous cell carcinoma was more prevalent in the MDA and NCI populations. NSCLC comprised >95% of the known histology types across all three studies. Smoking status did not differ significantly across the studies (P = 0.11); however, average ancestry (P = 0.0003), age (P < 0.0001) and sex (P < 0.0001) were significantly different. The mean proportion of African ancestry in cases and controls combined was 80.7% in the WSU study participants, 78.4% in the MDA participants and 78.8% for the NCI participants. Overall, the population demonstrated six generations of admixture per morgan, and the mean African ancestry did not differ between cases and controls. The average percent information extracted by the marker panel was 74.1%.
Table I.
Subject characteristics for 1812 African-American lung cancer cases and controls
| Total |
WSU |
MDA |
NCI |
|||||||||
| Case (N = 837) n (%) | Control (N = 975) n (%) | P-value | Case (N = 444) n (%) | Control (N = 559) n (%) | P-value | Case (N = 192) n (%) | Control (N = 188) n (%) | P-value | Case (N = 201) n (%) | Control (N = 228) n (%) | P-value | |
| Sex | ||||||||||||
| Male | 373 (44.5) | 401 (41.2) | 0.14 | 163 (36.7) | 202 (36.1) | 0.85 | 110 (57.3) | 84 (44.7) | 0.01 | 100 (49.8) | 115 (50.4) | 0.89 |
| Female | 464 (55.5) | 574 (59.8) | 281 (63.3) | 357 (63.9) | 82 (42.7) | 104 (55.3) | 101 (50.2) | 113 (49.6) | ||||
| Mean age (SD) | 61.8 (10.5) | 60.0 (11.5) | 0.0009 | 60.2 (10.8) | 58.7 (11.4) | 0.04 | 63.2 (9.3) | 55.3 (11.3) | <0.0001 | 64.0 (10.3) | 67.3 (8.2) | 0.0003 |
| Mean African ancestry (%) | 80.1 | 79.4 | 0.16 | 80.8 | 80.6 | 0.75 | 78.2 | 78.5 | 0.83 | 80.6 | 77.2 | 0.003 |
| Smoking status | ||||||||||||
| Never | 69 (8.2) | 339 (34.8) | <0.0001 | 28 (6.3) | 198 (35.4) | <0.0001 | 26 (13.5) | 47 (25.0) | <0.0001 | 15 (7.5) | 94 (41.2) | <0.0001 |
| Ever | 768 (91.8) | 636 (65.2) | 416 (93.7) | 361 (64.6) | 166 (86.5) | 141 (75.0) | 186 (92.5) | 134 (58.8) | ||||
| Mean pack-years (SD) (ever smokers) | 38.3 (27.1) | 23.7 (19.6) | <0.0001 | 36.4 (27.4) | 24.0 (19.7) | <0.0001 | 41.6 (27.0) | 24.6 (21.7) | <0.0001 | 39.7 (26.4) | 21.9 (16.9) | <0.0001 |
| History of emphysema | ||||||||||||
| No | 643 (76.8) | 931 (95.5) | <0.0001 | 363 (81.8) | 534 (95.5) | <0.0001 | 113 (58.0) | 174 (92.6) | <0.0001 | 167 (83.1) | 223 (97.8) | <0.0001 |
| Yes | 150 (17.9) | 36 (3.7) | 81 (18.2) | 24 (4.3) | 35 (18.2) | 7 (3.7) | 34 (16.9) | 5 (2.2) | ||||
| Unknown | 44 (5.3) | 8 (0.8) | 0 | 1 (0.2) | 44 (22.9) | 7 (3.7) | 0 | 0 | ||||
| Histology | ||||||||||||
| Adenocarcinoma | 350 (41.8) | 232 (52.2) | 87 (45.3) | 31 (15.4) | ||||||||
| Small cell carcinoma | 35 (4.2) | 24 (5.4) | 11 (5.7) | 0 (0) | ||||||||
| Squamous cell carcinoma | 172 (20.6) | 87 (19.6) | 52 (27.1) | 33 (16.4) | ||||||||
| Other NSCLC (including NSCLC, nos) | 170 (20.3) | 90 (20.3) | 41 (21.4) | 39 (19.4) | ||||||||
| Unknown | 110 (13.1) | 11 (2.5) | 1 (0.5) | 98 (48.8) | ||||||||
| Family history of lung cancer | ||||||||||||
| No | 653 (78.0) | 838 (86.0) | <0.0001 | 343 (77.3) | 487 (87.1) | <0.0001 | 147 (76.6) | 159 (84.6) | 0.05 | 163 (81.1) | 192 (84.2) | 0.39 |
| Yes | 184 (22.0) | 137 (14.0) | 101 (22.7) | 72 (12.9) | 45 (23.4) | 29 (15.4) | 38 (18.9) | 36 (15.8) | ||||
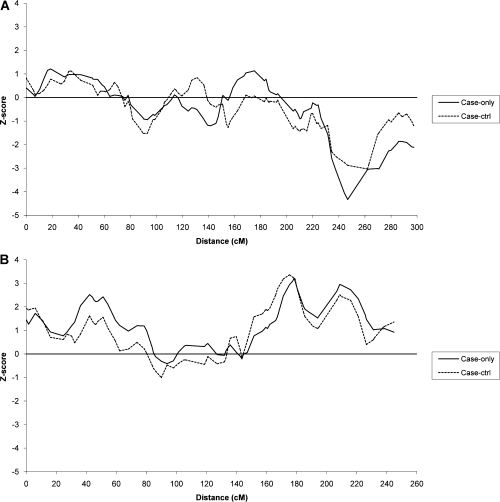
Table II displays the local ancestry results for the peak SNP (i.e. the SNP with the highest absolute Z-score) from the case-only test in two regions of suggestive ancestry association. These regions showed an excess of either European or African ancestry in the case-only analysis that was supported by the corresponding case–control analysis. The strongest evidence for ancestry association was for a region on chromosome 1q42 that displayed excess European ancestry among cases (Figure 1a). The peak occurred at rs6587361 for all subjects, NSCLC only and ever smokers with NSCLC in the case-only analysis (Table II). The maximum Z-score was −4.33 for NSCLC (P = 1.5 × 10−5), with carriers of two African alleles at this locus having a reduction in lung cancer risk of 0.49 (95% confidence interval 0.36–0.68) in comparison with carriers of two European alleles after adjustment for covariates. Excess European ancestry was strongest in women with NSCLC, with a maximum Z-score of −4.82 (P = 1.4 × 10−6) associated with rs6587361 (data not shown). Case–control analysis also indicated excess European ancestry; increased Z-scores for the same SNP demonstrated a large difference in ancestry between the cases and controls (Table II). Again, the association with European ancestry in the case–control analyses was strongest in women with NSCLC in an SNP immediately adjacent to and downstream from rs6587361 (rs2045520, Z-score = −3.93; P = 8.5 × 10−5) (data not shown). Analysis stratified on the most predominant histology type, adenocarcinoma, revealed attenuated Z-scores for both the case-only and case–control analyses. For adenocarcinomas, the peak European ancestry was located at rs493950 (Table II).
Table II.
Admixture mapping peaks associated with risk of lung cancer
| Regiona | Strata | Case-only |
Case–control |
||||||
| Z-scoreb | P-value | aRRb | 95% CI | Z-scoreb | P-value | aORb | 95% CI | ||
| 1q42.13 | rs6587361 | ||||||||
| All cases | −3.38 | 0.0007 | 0.61 | 0.45–0.81 | −2.26 | 0.0240 | 0.78 | 0.63–0.97 | |
| NSCLC | −4.33 | 1.5 × 10−5 | 0.49 | 0.36–0.68 | −2.88 | 0.0040 | 0.72 | 0.57–0.90 | |
| NSCLC ever smokers | −3.96 | 7.7 × 10−5 | 0.51 | 0.36–0.71 | −2.40 | 0.0164 | 0.74 | 0.58–0.95 | |
| rs493950 | |||||||||
| Adenocarcinoma | −3.10 | 0.0019 | 0.49 | 0.31–0.77 | −2.41 | 0.0161 | 0.71 | 0.54–0.94 | |
| 3q25.31 | rs181696 | ||||||||
| All cases | 2.72 | 0.0066 | 1.47 | 1.11–1.94 | 2.63 | 0.0086 | 1.34 | 1.08–1.67 | |
| NSCLC | 3.22 | 0.0013 | 1.64 | 1.21–2.22 | 3.22 | 0.0013 | 1.46 | 1.16–1.84 | |
| NSCLC ever smokers | 3.33 | 0.0009 | 1.72 | 1.25–2.37 | 2.90 | 0.0038 | 1.46 | 1.13–1.89 | |
| Adenocarcinoma | 1.83 | 0.0680 | 1.48 | 0.97–2.25 | 2.51 | 0.0120 | 1.42 | 1.08–1.87 | |
CI, confidence interval.
Results are reported for the peak SNP identified in the case-only analysis.
Z-scores, ancestry risk ratios (aRR) and ancestry odds ratios (aOR) are reported for analyses adjusted for age, sex, pack-years, family history of lung cancer (no/yes), study (WSU, MDA or NCI) and personal history of emphysema (no/yes). Positive Z-scores and aRR/aORs > 1 indicate increased risk of lung cancer associated with carrying two African ancestral alleles in comparison with no African alleles. Conversely, negative Z-scores and aRR/aORs < 1 indicate decreased risk of lung cancer associated with carrying two African ancestral alleles in comparison with none, which is also interpretable as an increased risk of lung cancer associated with carrying two European alleles in comparison with none.
Fig. 1.
(a) Admixture mapping results (Z-scores) for case-only and case–control analyses of chromosome 1 for NSCLC. (b) Admixture mapping results (Z-scores) for case-only and case–control analyses of chromosome 3 for NSCLC.
The second region of suggestive ancestry association was indicated by several SNPs included in a region of excess African ancestry on chromosome 3q25 (Figure 1b). The maximum Z-score for the case-only analysis was associated with rs181696 among ever smokers with NSCLC (Z-score = 3.33; P = 0.0009) (Table II). Carriers of two African alleles at this locus have an increased lung cancer risk of 1.72 (95% confidence interval 1.25–2.37) compared with carriers of two European alleles. Case–control analyses showed an increased risk of lung cancer associated with African ancestry for many SNPs in the region, including rs181696 (Z-score = 3.22; P-value = 0.0013). Analysis of adenocarcinoma cases again revealed a weaker association with ancestry for both the case-only and case–control analyses, with the highest Z-score at rs181696 (Table II).
Supplementary Table I (available at Carcinogenesis Online) highlights additional SNPs that were found in the chromosomes 1 and 3 regions of interest in the NSCLC analysis, as well as two additional regions on 6p24 and 15q12–13.
Discussion
In this admixture analysis of 1812 African-Americans, ADMIXMAP results revealed ancestry associations at 1q42 and 3q25. Both of these areas were identified by increased Z-scores in both the case-only and case–control analyses. This is the first reported admixture analysis of lung cancer.
For the excess European ancestry region on chromosome 1, the most significant Z-scores were found near a gene that has been strongly associated with lung cancer risk, Epoxide Hydrolase 1 (EPHX1), located at 1q42.1 (http://www.ncbi.nlm.nih.gov/gene). This gene metabolizes polycyclic aromatic hydrocarbons, which are found in cigarette smoke as well as other sources. A meta-analysis by Kiyohara showed a significantly decreased risk of lung cancer associated with the low-activity variant of the exon 3 (Tyr113His) polymorphism among whites (22). Only two studies of EPHX1 appear to have included African-Americans (23,24). Wu et al. (24) did not find significant associations between either Tyr113His or His139Arg and lung cancer among African-Americans. When London et al. (23) combined these two polymorphisms, a small decrease in risk was associated with the predicted slow activity genotype. Neither of these two more extensively studied SNPs was included in this admixture panel. It is possible that these or other SNPs in this gene or in this region are responsible for the association we see with lung cancer risk.
Excess African ancestry was displayed on chromosome 3q25. Many studies have reported increased copy number in this region in lung tumors (25). In a 2008 paper, Qian et al. (26) suggest many possible candidate genes in this region, including the oncogene PIK3CA and report that multiple studies have shown amplification of the 3q region to be much more probable in squamous cell carcinoma cases as compared with adenocarcinoma cases. We did not have sufficient numbers of squamous cell carcinoma cases (n = 172) to repeat our analyses in this subpopulation; however, our adenocarcinoma results add support to this finding. When results were restricted to adenocarcinoma cases, Z-scores were significantly lower in this region.
Several recent GWAS for lung cancer have also identified potential susceptibility gene regions at 15q15, 15q25, 5p15 and 6p21 (6–10,27). Within our study, we found excess European ancestry at 6p24 and excess African ancestry at 15q11–13 in the case–control analyses (supplementary Table I is available at Carcinogenesis Online). These areas are a large distance removed from the regions identified in the GWAS (>17 Mb and >14 Mb for the 6p21 and 15q15 regions, respectively). Potential genes of interest in the 15q11–13 region include the genes encoding the inhibitory neurotransmitter GABA receptor gamma 3 (GABRG3, rs8042276), receptor alpha 5 (GABRA5) and receptor beta 3 (GABRB3). Although these 15q11–13 receptor genes have yet to be linked to lung cancer risk or nicotine addiction, GABRA gene clusters on chromosomes 4 and 5 have been associated with nicotine dependence (28,29). Another potential candidate gene is CHRNA7, located further downstream on chromosome 15q14. While CHRNA3 and CHRNA5 were both identified as candidate genes in the lung cancer GWAS, CHRNA7 was not. However, it has been shown to be upregulated in lung cell lines exposed to the tobacco-specific nitrosamine NNK (4-(methylnitrosamino)-1-(3-pyridyl)-1-butonone) and estrogen (30). CHRNA7 is a candidate gene for schizophrenia, where it has been shown to be differentially expressed in smokers compared with nonsmokers (31).
It is possible that we did not see significant associations in the same areas as the GWAS mentioned above because those studies only included white and Asian populations. Furthermore, the regions identified using GWAS methods might be associated with lung cancer risk but might not be as strongly linked to ancestry.
We report both case-only and case–control analysis because these methods complement each other. The case-only analysis is more powerful than the case–control analyses because it compares ancestry estimates at a given locus with its inherent variability to the case’s genome-wide ancestry, which has been estimated over many markers to reduce variability (32). The case–control method compares the admixture at a given locus between the cases and controls (32). Thus, while our Z-scores were sometimes lower for the case–control results, they validate the case-only results in these regions by ruling out the possibility that the findings are due to selection in these regions among African-Americans.
There are several strengths to this study. The admixture mapping approach to gene discovery is a powerful method that takes advantage of the LD patterns among the recently admixed African-American population so that fewer individuals and fewer SNPs are required than for a GWAS. In addition, this study focused on African-Americans, a population that is known to have different smoking patterns and possibly different genetic risks than white and Asian populations. Although racial differences in LD and smoking behaviors warrant the inclusion of African-Americans in genetic studies, few studies have been done in this population.
A proposed cutoff for genome-wide statistical significance has been suggested for admixture mapping studies in African-Americans at P < 10−5 (33). In the case-only NSCLC strata, SNP rs6587361 (P = 1.5 × 10−5) at the 1q42.13 locus approached this significance level, and among women with NSCLC, rs6587361 (P = 1.4 × 10−6) met this threshold. Furthermore, we performed a post-hoc power calculation based on the methods of Hoggart et al. (33) for the case-only analysis. Based on the mean observed marker information (37.98) in our sample across the marker map and a two-tailed genome-wide significance threshold of 10−5, we had 80% power to detect an aRR < 0.40 (or >2.52). This calculation indicates that the sample was reasonably powered to detect the ancestry association signal observed among NSCLC subjects at 1q42.13 (for SNP rs6587361, an aRR of 0.49 among NSCLC subjects overall and an aRR of 0.36 among female NSCLC subjects). Taken together, these results indicate that at 1q42.13, there is a suggestive ancestry association locus for NSCLC.
The study has a large sample size with >1800 cases and controls and used a well-developed set of ancestry informative markers. Women made up 57% of the study population and were disproportionally represented in the WSU group due to the nature of one of the WSU studies. It is possible that the findings in women are driven by study site differences rather than a sex specific effect; however, study stratified analyses did not support a study site-specific finding (data not shown). The only histology categories with large enough numbers for meaningful analyses were NSCLC and adenocarcinoma and therefore, our results might not be generalizable to all histology types. We also had too few never smoking cases for meaningful analyses.
Lung cancer is one of the few cancers for which no substantial progress has been made in early detection and treatment. Given that it is the leading cause of death from cancer, it is exceedingly important to better understand the underlying biology to develop targeted treatments and to identify high-risk populations for targeted behavioral interventions and access to developing screening methods. It is probably that many genes contribute to an individual’s lung cancer risk. Admixture mapping provides an alternative approach to the identification of lung cancer susceptibility genes. New regions were identified in this admixture study. These results add to the findings from the GWAS in Caucasian populations and suggest novel candidate gene areas. These regions need to be confirmed in additional studies with finer mapping.
Supplementary material
Supplementary Table I can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (NIH) (R01-CA87895, R01-CA60691, R01-CA55769, R01-CA127219, R01-CA141716, , R01-CA133996, R01-CA121197, U19-CA148127 and contract numbers N01-PC35145 and P30CA22453); Cancer Prevention Research Institute of Texas (RP10043); Intramural Research Program of the National Cancer Institute/NIH, Center for Cancer Research.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- aRR
ancestry risk ratio
- CEPH
Center d’Etude du Polymorphisme Humain
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- MDA
M. D. Anderson Cancer Center
- NCI
National Cancer Institute
- NSCLC
non-small-cell lung cancer
- SNP
single-nucleotide polymorphism
- WSU
Wayne State University
References
- 1.Ries LAG, et al., editors. SEER Cancer Statistics Review, 1975–2005. Vol. 2008. Bethesda, MD: National Cancer Institute; 2008. [Google Scholar]
- 2.Haiman CA, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N. Engl. J. Med. 2006;354:333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 3.Cote ML, et al. Risk of lung cancer among white and black relatives of individuals with early-onset lung cancer. JAMA. 2005;293:3036–3042. doi: 10.1001/jama.293.24.3036. [DOI] [PubMed] [Google Scholar]
- 4.Bailey-Wilson JE, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am. J. Hum. Genet. 2004;75:460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz AG, et al. The molecular epidemiology of lung cancer. Carcinogenesis. 2006;28:507–518. doi: 10.1093/carcin/bgl253. [DOI] [PubMed] [Google Scholar]
- 6.Amos CI. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 8.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanock SJ, et al. Genomics: when the smoke clears. Nature. 2008;452:537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 12.Patterson N, et al. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty R, et al. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl Acad. Sci. USA. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens JC, et al. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am. J. Hum. Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 15.McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am. J. Hum. Genet. 1998;63:241–251. doi: 10.1086/301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cote ML, et al. Combinations of glutathione S-transferase genotypes and risk of early-onset lung cancer in Caucasians and African Americans: a population-based study. Carcinogenesis. 2005;26:811–819. doi: 10.1093/carcin/bgi023. [DOI] [PubMed] [Google Scholar]
- 17.Wenzlaff AS, et al. GSTM1, GSTT1 and GSTP1 polymorphisms, environmental tobacco smoke exposure and risk of lung cancer among never smokers: a population-based study. Carcinogenesis. 2005;26:395–401. doi: 10.1093/carcin/bgh326. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz AG, et al. Chronic obstructive lung diseases and risk of non-small cell lung cancer in women. J. Thorac. Oncol. 2009;4:291–299. doi: 10.1097/JTO.0b013e3181951cd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cote ML, et al. Tobacco and estrogen metabolic polymorphisms and risk of non-small cell lung cancer in women. Carcinogenesis. 2009;30:626–635. doi: 10.1093/carcin/bgp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etzel CJ, et al. Development and validation of a lung cancer risk prediction model for African-Americans. Cancer Prev. Res. (Phila.) 2008;1:255–265. doi: 10.1158/1940-6207.CAPR-08-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng YL, et al. Bleomycin-induced chromosome breaks as a risk marker for lung cancer: a case-control study with population and hospital controls. Carcinogenesis. 2003;24:269–274. doi: 10.1093/carcin/24.2.269. [DOI] [PubMed] [Google Scholar]
- 22.Kiyohara C, et al. EPHX1 polymorphisms and the risk of lung cancer: a HuGE review. Epidemiology. 2006;17:89–99. doi: 10.1097/01.ede.0000187627.70026.23. [DOI] [PubMed] [Google Scholar]
- 23.London SJ, et al. Lung cancer risk in relation to genetic polymorphisms of microsomal epoxide hydrolase among African-Americans and Caucasians in Los Angeles County. Lung Cancer. 2000;28:147–155. doi: 10.1016/s0169-5002(99)00130-0. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, et al. The association of microsomal epoxide hydrolase polymorphisms and lung cancer risk in African-Americans and Mexican-Americans. Carcinogenesis. 2001;22:923–928. doi: 10.1093/carcin/22.6.923. [DOI] [PubMed] [Google Scholar]
- 25.Huang YT, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J. Clin. Oncol. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian J, et al. Role of chromosome 3q amplification in lung cancer. J. Thorac. Oncol. 2008;3:212–215. doi: 10.1097/JTO.0b013e3181663544. [DOI] [PubMed] [Google Scholar]
- 27.Broderick P, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agrawal A, et al. Gamma-aminobutyric acid receptor genes and nicotine dependence: evidence for association from a case-control study. Addiction. 2008;103:1027–1038. doi: 10.1111/j.1360-0443.2008.02236.x. [DOI] [PubMed] [Google Scholar]
- 29.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Wadei HA, et al. Chronic exposure to estrogen and the tobacco carcinogen NNK cooperatively modulates nicotinic receptors in small airway epithelial cells. Lung Cancer. 2010;69:33–39. doi: 10.1016/j.lungcan.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Mexal S, et al. Differential regulation of alpha7 nicotinic receptor gene (CHRNA7) expression in schizophrenic smokers. J. Mol. Neurosci. 2010;40:185–195. doi: 10.1007/s12031-009-9233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montana G, et al. Statistical tests for admixture mapping with case-control and cases-only data. Am. J. Hum. Genet. 2004;75:771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoggart CJ, et al. Design and analysis of admixture mapping studies. Am. J. Hum. Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.