Abstract
Generation 5 poly(amidoamine) (PAMAM) dendrimers were modified by the addition of cyclic RGD targeting peptides and were evaluated for their ability to associate with siRNA and mediate siRNA delivery to U87 malignant glioma cells. PAMAM-RGD conjugates were able to complex with siRNA to form complexes of approximately 200 nm in size. Modest siRNA delivery was observed in U87 cells using either PAMAM or PAMAM-RGD conjugates. PAMAM-RGD conjugates prevented the adhesion of U87 cells to fibrinogen coated plates, in a manner that depends on the number of RGD ligands per dendrimer. The delivery of siRNA through three-dimensional multicellular spheroids of U87 cells was enhanced using PAMAM-RGD conjugates compared to the native PAMAM dendrimers, presumably by interfering with integrin-ECM contacts present in a three-dimensional tumor model.
Keywords: PAMAM dendrimers, siRNA delivery, malignant glioma, tumor spheroids, integrin targeting
INTRODUCTION
Gene silencing using RNA interference (RNAi) is a powerful platform technology being developed clinically for a variety of diseases including cancer, where it is typically considered as part of a multi-pronged approach (1, 2). While RNAi holds promise as an anticancer therapeutic modality, the ability to efficiently and safely deliver siRNA molecules to cells is the main barrier limiting its widespread clinical use (3). The use of synthetic, non-viral delivery vectors such as polymers and liposomes has shown promise in mediating cellular delivery of siRNA molecules (4). Benefits of synthetic delivery vectors include their ability to be manufactured on a large-scale, low immunogenic response compared to their viral counterparts, and the ability to chemically tailor their structure for a particular application.
Poly(amidoamine) (PAMAM) dendrimers are a family of highly branched, synthetic polymers that have garnered interest as potential delivery vectors for nucleic acids, recently including siRNA. Several studies have demonstrated successful delivery of siRNA using PAMAM dendrimers or their derivatives (5–10). The branched architecture of dendrimers makes them particularly attractive for targeted delivery applications as they can present targeting ligands in a manner favorable to promote multivalent binding to target cellular receptors. Multivalent ligand presentation from PAMAM dendrimers has been demonstrated for various targeted systems including folic acid (11), mannose and glucose (12, 13).
Integrin proteins are a family of cell-surface receptors, several of which are known to be over-expressed on the surface of cancer cells. The αvβ3 integrin is particularly known for its role in cancer progression and is overexpressed in melanomas, glioblastoma, ovarian, breast, and prostate cancers (14). The high-affinity interaction between RGD peptides and cancer-related integrins has led to the widespread use of RGD peptide sequences as ligands for integrin-targeted drug and gene delivery applications (15). Several examples of PAMAM dendrimer-RGD conjugates have been reported to enhance the delivery of imaging agents to target carcinoma cells (16, 17). PAMAM-RGD conjugates have also been found to mediate cellular binding and adhesion (18, 19).
As significant promise has been shown for PAMAM-RGD conjugates in drug delivery and imaging applications, the goal of this study was to evaluate this bioconjugate for use as a siRNA delivery vector. Specifically, we sought to investigate the effects of extent of ligand functionalization (multivalency) on cellular delivery. To this end, the ability of dendrimers to deliver siRNA to malignant glioma cells with varying extents of RGD conjugation was evaluated. Furthermore, we hypothesized that the ligand presentation would have a greater impact on delivery in a three-dimensional tumor, where the interactions between the delivery vector, extracellular matrix and cells are more pronounced. To evaluate this hypothesis, the ability of PAMAM-RGD conjugates to interfere with cell-ECM interactions and to mediate siRNA delivery in a three-dimensional cell-culture model of malignant glioma was studied.
EXPERIMENTAL PROCEDURES
Materials
A 22 nt siRNA sequence previously identified as an effective inhibitor of pd1EGFP expression (20) (sense strand: 5′-UUG UGG CCG UUU ACG UCG CCG U-3′, antisense strand: 3′-UGA ACA CCG GCA AAU GCA GCG G-5′) and an irrelevant siRNA sequence (targeted against firefly luciferase; sense strand: 5′-CUU ACG CUG AGU ACU UCG A dTdT-3′, antisense strand: 5′-UCG AAG UAC UCA GCG UAA G dTdT-3′) were purchased from Dharmacon (Chicago, IL). A fluorescently labeled anti-GFP siRNA sequence (5′ Cy3 end modified on the sense strand) was purchased from Integrated DNA Technologies (Coralville, IA, USA). A 20 nt phosphorothioated antisense oligonucleotide targeted against pd1EGFP with a fluorescent label was also purchased from Integrated DNA Technologies (5′-Cy5-TTG TGG CCG TTT ACG TCG CC-3′). The lyophilized powder was resuspended according to the manufacturer’s protocol before use. Generation 5 PAMAM dendrimers with an ethylenediamine core and amine terminal groups were purchased as a 5 wt% solution in methanol from Dendritech (Midland, MI). Unless otherwise stated, all chemicals were purchased from Sigma, and all cell culture products were obtained from Invitrogen (Carlsbad, CA).
Conjugation of PAMAM dendrimers with RGD peptides
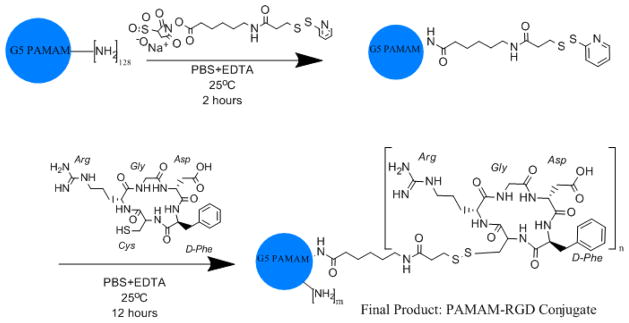
Approximately 6 mg of dry generation 5 PAMAM dendrimers were obtained after the removal of methanol from the storage solution using rotary evaporation. The polymer residue was dissolved in ~2 mL of phosphate buffered saline (PBS) with 1 mM EDTA (reaction buffer). Various molar equivalents (6, 9, 12, or 15) of a Sulfo-LC-SPDP crosslinker (Pierce, Rockford, IL) were added to the polymer solution to yield SPDP-activated PAMAM after a 2 hour reaction at room temperature. Dialysis with a 10,000 MWCO Slide-A-Lyzer dialysis cassette (Pierce, Rockford, IL) was performed against 1 liter of reaction buffer overnight to remove any unreacted SPDP crosslinker. Following dialysis, 1.5 molar equivalents (to SPDP groups added) of cyclic RGDfC peptide (Peptides International, Louisville, KY) were added to the SPDP-activated PAMAM dendrimers to yield PAMAM-RGD conjugates after an overnight reaction (Figure 1). Following conjugation, 10,000 MWCO dialysis was performed against reaction buffer followed by water to remove any unreacted RGD peptide. Purified PAMAM-RGD conjugates were lyophilized overnight to obtain a white powder.
Figure 1.
PAMAM dendrimers are reacted with Sulfo-LC-SPDP to yield SPDP-activated PAMAM intermediates. SPDP-activated PAMAM was reacted with cyclic RGD peptides to obtain final PAMAM-RGD conjugates.
UV spectrophotometry of PAMAM-RGD conjugates
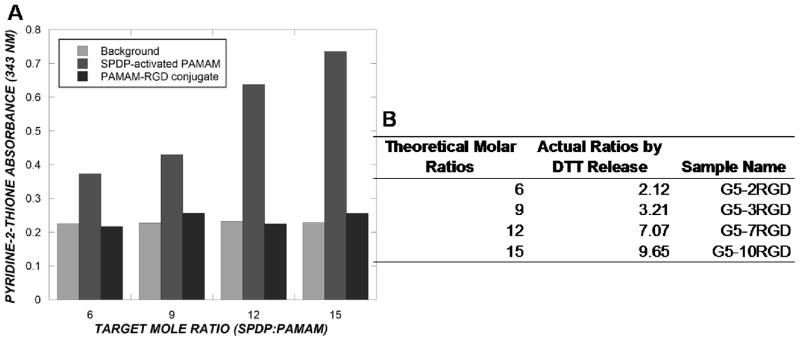
A series of PAMAM-RGD conjugates was characterized using UV spectrophotometry to determine the number of RGD peptides conjugated to each dendrimer. A disulfide reducing agent, DTT, was used to cleave the disulfide bond present in the SPDP crosslinker to release pyridine-2-thione (P2T), which has a UV absorbance at 343 nm. This analysis of pyridine-2-thione release was first performed on the intermediate product, PAMAM-SPDP and used to quantify the average number of SPDP groups added to each dendrimer. After removal of excess SPDP by dialysis, 50 μL of the PAMAM-SPDP solution was added to 450 μL of reaction buffer, and the absorbance of the solution at 343 nm was read using a Unicam UV 300 model UV spectrophotometer (Thermo Spectronics, Rockford, IL) (this reading indicated background absorbance). To the same sample, 5 μL of DTT (Pierce, Rockford, IL) (15 mg/mL) was added, and the absorbance at 343 nm was recorded after a 15 minute incubation at room temperature. The increase in UV absorbance observed after the addition of DTT corresponded to the addition of LC-SPDP groups to dendrimers. The average molar ratio of SPDP:PAMAM was calculated according to the manufacturer’s protocol (Pierce, Rockford, IL). The same analysis was performed on the final PAMAM-RGD conjugate to confirm replacement of all SPDP groups with RGD peptides. After the replacement of P2T groups by RGD peptides, an increase in UV absorbance at 343 nm was not observed.
MALDI-TOF Mass Spectrometry
PAMAM-RGD conjugates were dissolved in water at a concentration of 1 mg/mL prior to MALDI-TOF MS analysis. A matrix solution of 2′-4′-6′-Trihydroxyacetophenone monohydrate (THAP) (10 mg/mL THAP in 50%ACN/(50% H20 with 0.1% trifluoroacetic acid)) was used. The matrix solution and PAMAM dendrimers were mixed 1:1 by volume, and spotted on a 100-well stainless steel sample plate. MALDI-TOF MS analysis was performed in linear positive mode of a Voyager DE Pro instrument (Applied Biosystems) using 25 kV accelerating voltage, 95% grid voltage, 0.3% guide wire voltage, and a delay time of 700 ns. For each spectrum 75 shots were taken, and a minimum of 3 spectra were accumulated for each sample.
PicoGreen assay for PAMAM/siRNA complexation
Complexes were prepared at various charge ratios by mixing equal volumes of PAMAM with siRNA in PBS. Charge ratios (N/P) were calculated as a ratio of the number of primary amines in the polymer to the number of anionic phosphate groups in the siRNA. The samples were then vortexed and incubated at room temperature for 15 minutes to ensure complex formation. The complexes were prepared at a final siRNA concentration of 0.2 μg of siRNA/100 μL of solution and varying polymer concentrations to reach the desired charge ratio. One hundred microliters of each complex were transferred to a 96-well (black-walled, clear-bottom, non-adsorbing) plate (Corning, NY, USA). A total of 100 μL of diluted PicoGreen dye (Molecular Probes, Eugene, OR) (1:200 dilution in Tris-EDTA (TE) buffer) was added to each sample. Fluorescence measurements were made after a 30 minute incubation at room temperature using a DTX800 Multimode Detector (Beckman Coulter, CA), at exitation and emission wavelengths of 485 and 535 nm, respectively. All measurements were corrected for background fluorescence from a solution containing only buffer and PicoGreen dye.
Dynamic light scattering
Dynamic light scattering (DLS) analysis was performed using a Malvern Instruments Zetasizer Nano ZS-90 instrument (Southboro, MA) with reproducibility being verified by collection and comparison of sequential measurements. Polymer/siRNA complexes (siRNA concentration = 100 nM, N/P=15) were prepared using purified water (resistivity=18.5 MΩ-cm). DLS measurements were performed at a 90 ° scattering angle at 37 °C. Z-average sizes of three sequential measurements were collected and analyzed approximately 30 minutes after the polymer/siRNA complexes were formed.
Cell culture
U-87 MG cells (ATCC HTB-14) were maintained in D-MEM medium supplemented with 10% fetal bovine serum (FBS), L-glutamine, and penicillin-streptomycin solution. A U87 cell line containing a stably integrated destabilized EGFP (d1EGFP) transgene (U87-d1EGFP) was generated as described previously (10) and was maintained under constant selective pressure by G418 (500 ug/mL), and the growth medium was supplemented with sodium pyruvate and non-essential amino acids. All cell lines were cultivated in a humidified atmosphere of 5% CO2 at 37°C.
Multicellular spheroid formation
Multicellular tumor spheroids (MCTS) were formed from the U87 or U87-d1EGFP malignant glioma cell lines using the hanging drop method as described previously (21). Cells from a confluent T-25 flask were detached using trypsin-EDTA and resuspended in 5mL of culture medium to a concentration of ~106 cells/mL. Twenty microliter droplets of this concentrated cell suspension were deposited into the lid of a petri dish. The bottom of the petri dish was filled with 2 mL of cell culture medium to facilitate moisture transfer. The lid containing the drops was inverted over the petri dish to form hanging drops. The petri dish was placed in the incubator for a period of 3 days, after which visible cell aggregates were formed in the hanging drops. The cell aggregates were harvested from the hanging drop suspension by adding ~2mL of culture medium to suspend the aggregates. The cell aggregates were individually transferred in 100 μL of culture medium to wells of a 96-well tissue culture plate that was base-coated with 2% agarose. After 24 hours on agarose, cell aggregates formed MCTS of diameters ranging from ~600–800 μm and were subjected to siRNA transfection and confocal imaging analysis.
SiRNA delivery assay
U87-d1EGFP cells were plated at a density of 1.5 × 105 cells/well in 12 well plates ~18 hours prior to transfection. Prior to treatment of cells, PAMAM/siRNA complexes were prepared as described above in 200 μL of PBS (N/P=15). PolyFect (Qiagen, Valencia, CA), a commercially available dendrimer transfection reagent, was used as a positive control. Eight hundred microliters of OptiMEM medium was mixed with each sample to obtain a final siRNA concentration of 100 nM. The serum-containing culture medium was aspirated from the cells, and each well was treated with 1 mL of the PAMAM/siRNA complexes in OptiMEM medium. After a 4 hour incubation period, the transfection mixture was replaced with serum-containing culture medium and maintained under normal growth conditions until the cells were assayed for fluorescence by flow cytometry at various time points after initial treatment. For cells being analyzed for GFP fluorescence, unlabeled siRNA was utilized. To determine intracellular siRNA uptake, non-transformed U87 cells were treated with a Cy-3 labeled siRNA.
To analyze siRNA delivery in multicellular tumor spheroids, a similar transfection protocol was performed as with cells on 12-well tissue culture plates. MCTS transfections were performed in 96-well plates that had been base-coated with 2% agarose. Transfections were performed in a total volume of 150 μL in OptiMEM medium. MCTS were exposed to a Cy3-labeled siRNA sequence to enable confocal imaging of siRNA localization throughout the spheroids at both 4 and 24 hours post-transfection. Spheroids generated from the U87-d1EGFP cell line were treated with a Cy5-labeled (red) antisense oligonucleotide (AON) targeted against d1EGFP so that the label and transgene fluorescence spectra would not overlap. U87-d1EGFP spheroids were imaged at 24, 48, and 72 hours after the initial treatment to evaluate both GFP silencing and localization of the AON with the various dendrimer/ODN formulations conjugates.
Flow cytometry
Cells were washed with PBS, detached with trypsin-EDTA, and collected in growth medium, before they were pelleted by centrifugation for 3.5 min at 200 g and resuspended in 150 μL PBS. Samples were maintained on ice before being subjected to flow cytometry analysis. Ten thousand cells were analyzed on a FACSCalibur two-laser, four-color flow cytometer (BD Biosciences) for GFP fluorescence (FL-1) or Cy3 fluorescence (FL-2). CellQuest software was used to acquire and analyze the results. Viable cells were gated according to their typical forward/side scatter characteristics.
Competitive cell adhesion assay
High-binding 96-well plates (Nunc Maxi SORP) were coated with fibrinogen protein by adding 100 μL/well of fibrinogen solution (100 μg/mL in PBS) overnight at 4°C. The next day, the plate was blocked with 150 μL/well of bovine serum albumin (BSA) solution (10 mg/mL in PBS) for 1 hour at 37°C. The plate was washed in triplicate with cold PBS. Ten microliters of test compounds (RGD-conjugated polymers) at various concentrations were added to wells of the fibrinogen-coated plate on ice in triplicate. Following the addition of test compounds, 90 μL of U87 cells in culture medium (330,000 cells/mL) were added to the plate, and mixed thoroughly with a multi-channel pipette. The plate was incubated at 37°C for 3 hours until the cells attached to the plate. Non-adherent cells were washed from the plate with PBS, and adherent cells were fixed by the addition of 100 μL of 70% ethanol for 30 minutes at room temperature. Following one wash with PBS, 100 μL of PicoGreen dye in TE buffer were added to the plate for 30 minutes to detect adherent cells. PicoGreen fluorescence was measured using a DTX800 Multimode Detector (Beckman Coulter, CA), at excitation and emission wavelengths of 485 and 535 nm, respectively. PicoGreen fluorescence values of wells treated with a RGD-containing competitor was normalized to wells that received no competitor (maximum cells adhering). Data analysis and calculation of IC50 values was performed using GraphPad Prism 4 software (GraphPad Software, La Jolla, CA).
Confocal microscopy of multicellular spheroids
Uptake and distribution of Cy3-labeled siRNA or Cy5-labeled AS ODN in U87 MCTS or U87–d1EGFP MCTS, respectively, were analyzed using confocal microscopy. Imaging was performed at various time points after siRNA transfection of spheroids using an Olympus IX81 model confocal microscope (Olympus, Center Valley, PA). Images of MCTS were performed directly on agarose-coated 96-well plates at 10X magnification. Z-stack imaging was performed to take image slices through spheroids at 20 μm intervals for a total depth of 100 μm. The following excitation and emission wavelengths were used to detect the fluorophores used in this study: GFP fluorescence (excitation= 482 nm, emission= 536 nm), Cy3 siRNA (excitation= 543 nm, emission=593 nm), and Cy5 AS ODN (excitation= 628 nm, emission= 692 nm).
Image analysis of multicellular spheroid fluorescence
The fluorescence intensities of both the Cy5 and GFP channels of the confocal images of the U87-d1EGFP spheroids were quantified using ImageJ software (22).
Statistics
All statistical comparisons among treatment groups were performed using a one way ANOVA test with Fisher’s all-pairs post hoc comparison test. To compare among IC50 fits in the cell adhesion assay, a Monte Carlo procedure was employed. The standard error of measurement was used to randomly perturb individual experimental values around their means. Fits were then applied to the perturbed data and compared across groups. This procedure was repeated 10,000 times. If a particular group had a lower IC50 in at least 95% of Monte Carlo trials, then the fitted IC50 value was considered significant at the p < 0.05 level.
RESULTS
Reaction and characterization of PAMAM-RGD conjugates
Generation 5 PAMAM dendrimers were conjugated with various amounts of a cyclic RGD targeting peptide, RGDfC, using a Sulfo-LC-SPDP crosslinker in a scheme depicted in Figure 1. The PAMAM-RGD conjugates were characterized using UV spectrophotometry at various stages during the crosslinking process. For this purpose, a disulfide reducing agent, DTT, was added to the conjugates to break the disulfide bond present in the SPDP crosslinker, enabling the release of pyridine-2-thione, a molecule that is UV active at 343 nm. PAMAM dendrimers were first reacted with various molar equivalents (6, 9, 12, or 15) of Sulfo-LC-SPDP to attain SPDP-activated PAMAM. After the addition of DTT, increasing amounts of pyridine-2-thione absorbance were detected with increasing SPDP:PAMAM ratios, as expected (Figure 2A). This information was used to calculate the average molar ratios of SPDP:PAMAM yielded in this reaction, ranging from approximately 2 to 10 SPDP:PAMAM. Following reaction with cyclic RGD peptides, the pyridine-2-thione absorbance decreased back to baseline, indicating complete replacement of pyridine-2-thione groups from the crosslinker with RGD peptides. The resulting PAMAM-RGD conjugates are denoted by their experimentally determined extents of RGD conjugation (rounded to whole numbers) as G5-2RGD, G5-3RGD, G5-7RGD, and G5-10RGD.
Figure 2.
UV detection of pyridine-2-thione release from SPDP-activated PAMAM and PAMAM-RGD conjugates after the addition of DTT (A). Experimentally determined extents of RGD conjugation and sample names(B).
PAMAM-RGD conjugates were further characterized using MALDI-TOF mass spectrometry to confirm conjugation of RGD peptides to the dendrimers. Several previous studies have successfully utilized MALDI-TOF MS to characterize PAMAM bioconjugates by observing an increase in molecular weight of dendrimers upon conjugation to other molecules (23, 24). As expected, a shift to the right of MALDI MS curves correlated to increased extents of RGD conjugation, indicating an increase in the molecular weight of the conjugates (Supplementary Data). As MALDI MS has been shown to underestimate the actual molecular weight of high-generation dendrimers (23), it was not used to estimate the molecular weight of PAMAM-RGD conjugates, but as a tool to confirm successful crosslinking of RGD to PAMAM dendrimers. The molecular weight of the PAMAM-RGD conjugates was calculated instead from the RGD crosslinking extent determined using UV spectrophotometry as described above.
Characterization of dendrimer/siRNA complexes
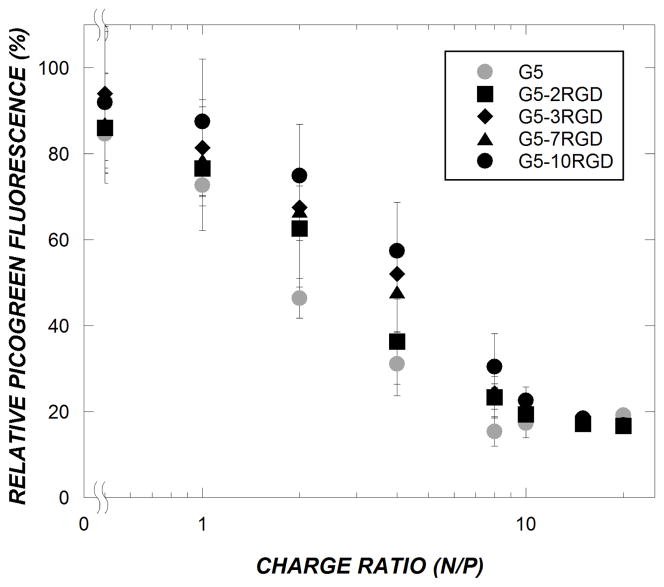
To facilitate siRNA delivery, cationic polymers such as PAMAM dendrimers should form electrostatic complexes with anionic siRNA molecules. The ability of PAMAM-RGD conjugates to complex with siRNA was evaluated using a dye exclusion assay. The amount of unbound siRNA in solutions of PAMAM/siRNA was determined by measuring the fluorescence of a commercially available dye, PicoGreen, that fluoresces upon binding to double stranded DNA or RNA. The fluorescence intensity of PicoGreen decreased following incubation of increasing amounts of polymer with added to a fixed amount of siRNA, indicating association of siRNA with the polymer (Figure 3). For all dendrimer conjugates tested, most of the PicoGreen fluorescence was quenched by N/P=10, indicating that the dendrimers had formed complexes with siRNA. At lower charge ratios, however, it is possible to discern trends in siRNA complexation as a function of RGD crosslinking extent, with higher extents of RGD conjugation corresponding to somewhat less siRNA complexation. To ensure the presence of stable complexes, N/P = 15 was chosen for subsequent experiments.
Figure 3.
PAMAM/siRNA complexes were formed in PBS (final siRNA concentration= 2 μg/mL) and allowed to incubate for 15 minutes before measurement. Relative PicoGreen fluorescence corresponds to unbound siRNA present in solution. The data represent mean ± SEM (n ≥ 3).
Having demonstrated the ability of PAMAM-RGD conjugates to complex with anionic siRNA, the characteristics of the siRNA/PAMAM complexes formed at N/P = 15 were evaluated further by dynamic light scattering (DLS) measurements. Particle size analysis by DLS showed the formation of ~200 nm complexes between dendrimers and siRNA, regardless of extent of RGD conjugation (Table 1).
Table 1.
Particle diameter of dendrimer/siRNA complexes as determined by dynamic light scattering. The data represent mean ± standard deviation (n=3).
| Sample | Particle diameter (nm) |
|---|---|
| G5 | 192 ± 3 |
| G5-2RGD | 166 ± 36 |
| G5-3RGD | 187 ± 14 |
| G5-7RGD | 193 ± 14 |
| G5-10RGD | 182 ± 5 |
siRNA delivery and GFP silencing in U87 cells
The ability of PAMAM-RGD conjugates to deliver siRNA to U87 cells and elicit a gene silencing response was evaluated. A fluorescently labeled siRNA sequence was delivered to U87 cells with PAMAM-RGD conjugates to evaluate siRNA uptake into cells, which was assayed using flow cytometry. Separately, but under the same conditions, a siRNA sequence targeted against the short half-life green fluorescent protein, d1EGFP, was delivered to U87–d1EGFP cells, and relative GFP expression was assayed using flow cytometry.
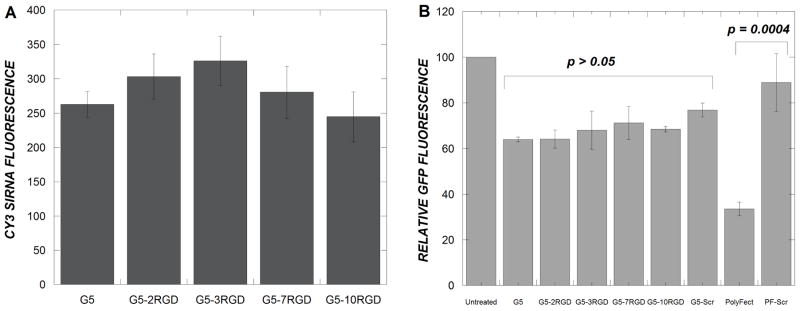
Significant intracellular siRNA levels were observed 24 hours after exposure of U87 cells to PAMAM-RGD/Cy3-siRNA complexes (Figure 4A). If cell binding is a limiting step in dendrimer-mediated siRNA delivery, intracellular siRNA levels would be expected to increase with increasing extent of RGD conjugation onto PAMAM dendrimers, as U87 cells express αvβ3 receptors for which cyclic RGD peptides have high binding affinities (25). In this system, a statistically significant increase in siRNA delivery was not observed upon conjugation of dendrimers with RGD peptides (p > 0.05).
Figure 4.
U87 cells (A) or U87–d1EGFP cells (B) were treated with PAMAM-RGD/siRNA complexes at a final siRNA concentration of 100 nM for 4 hours under serum-free conditions. Cells were analyzed using flow cytometry for Cy3 fluorescence (A) or GFP fluorescence (B) 24 hours after the initial treatment. A 5:1 wt ratio of PolyFect:siRNA was used (B). A scrambled siRNA sequence was delivered with PolyFect and G5 PAMAM dendrimer denoted by PF-Scr and G5-Scr, respectively. Data represent mean ± SEM.
Having observed intracellular siRNA delivery by PAMAM-RGD conjugates, the ability of the system to deliver siRNA to elicit a GFP silencing response was evaluated by flow cytometry. Modest silencing (approximately 40%) of the GFP transgene was observed in U87-d1EGFP cells using unmodified G5 PAMAM dendrimers compared to control cells receiving no siRNA treatment. The extent of GFP silencing achieved was not significantly altered after the conjugation of RGD peptides to PAMAM dendrimers (p > 0.05) (Figure 4B). An irrelevant siRNA sequence not targeted against d1EGFP was used as a control and resulted in ~20% silencing relative to no siRNA treatment. As these values are rather low and not statistically different from each other (p > 0.05), it appears that, even with RGD modification, uptake of G5 PAMAM dendrimers is non-specific and does not result in intracellular trafficking sufficient for robust gene silencing (10). A commercially available dendrimer-based transfection reagent, PolyFect, was used as a positive control in this experiment and elicited ~60% GFP knockdown, which was statistically greater than that induced when the irrelevant sequence was delivered with PolyFect (Figure 4B).
Competitive binding cell adhesion assay
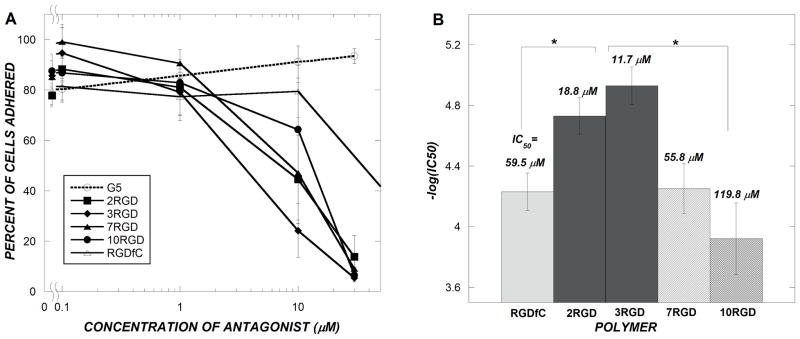
Having observed that the addition of RGD peptides to PAMAM did not significantly alter the ability of PAMAM to deliver siRNA to U87 cells, probably due to significant non-specific cellular uptake of PAMAM/siRNA complexes, we sought to study whether active integrin targeting was occurring to any extent by the use of a cell adhesion assay. Here, the ability of PAMAM-RGD bioconjugates to bind specifically to αvβ3 integrin receptors on cells and prevent their adhesion to a fibrinogen-coated plate was evaluated. Polymers functionalized with various numbers of RGD peptides inhibited adhesion of U87 cells to fibrinogen protein in a concentration dependent manner (Figure 5A). In contrast, unmodified G5 PAMAM dendrimer did not prevent the adhesion of U87 cells to fibrinogen, demonstrating the specificity of this assay to integrin-RGD binding, and the lack of non-specific effects by PAMAM. This result indicated that in all PAMAM-RGD constructs, RGD was presented to the cells in a manner such that it binds to αvβ3 integrin receptor proteins and is sufficient to prevent adhesion of U87 cells to the extracellular matrix protein, fibrinogen.
Figure 5.
Competitive binding curves for U87 cells binding to fibrinogen coated plates in the presence of RGD-containing antagonists (A). Results of IC50 values obtained from binding curves for each antagonist used (B). Different colored bars represent statistically different IC50 values (p<0.05) as determined using a Monte Carlo curve fitting algorithm (B). Data represent mean ± SEM (n ≥ 3) (A) or mean IC50 ± standard deviation (B).
To discern trends in adhesion as a function of RGD conjugation extent, binding curves were fit to a competitive binding model to determine a concentration of RGD at which each dendrimer inhibited 50% of cells from adhering to the plate (IC50) (Figure 5B). Fitted IC50 values were compared statistically using a Monte Carlo procedure described in the Methods. When a low number of RGD peptides was conjugated to the dendrimers (2 or 3), a significantly lower IC50 concentration (or a higher binding avidity) was achieved than with free RGD peptide (p<0.05). However, by increasing the number of peptides per dendrimer (7 or 10), a sharp increase in the IC50 value was observed, corresponding to a decreased net avidity of these PAMAM-RGD conjugates to the integrin protein compared to those conjugates containing just 2 or 3 RGD peptides (p<0.05). Further, a significant difference was observed between the IC50 values of dendrimers containing 7 or 10 RGDs (p<0.05). Using this assay, we found that three RGD peptides per dendrimer were optimal for preventing U87 adhesion to fibrinogen protein on a flat plate.
siRNA delivery through multicellular glioma spheroids
The strong effect of PAMAM-RGD on U87 cell adhesion suggests that these bioconjugates can interfere with integrin-ECM interactions, which would likely influence delivery in a three-dimensional tumor. To investigate this hypothesis, the ability of PAMAM-RGD to deliver siRNA through a three-dimensional multicellular tumor spheroid (MCTS) model of U87 cells was evaluated. As carcinoma cells cultured as 3D spheroids are known to secrete ECM proteins and feature active integrin-ECM interactions (26), we expected that PAMAM-RGD conjugates would be able to facilitate siRNA delivery through MCTS by interfering with integrin-ECM interactions formed by cells in three dimensions. To this end, cellular aggregates of U87 cells were formed using the hanging drop method, and aggregates were then transferred to agarose-coated tissue-culture plates to promote the formation of spheroids. U87 spheroids on agarose were treated with PAMAM-RGD/Cy3-siRNA complexes in a similar manner as for cells cultured on standard 12-well tissue culture plates. PAMAM-RGD polymers containing either 3 or 10 RGDs were used for this study, as G5-3RGD was found to be optimal in the cell adhesion assay and G5-10RGD represents the highest extent of RGD conjugation used in this work. Confocal imaging was performed at four and twenty-four hours after transfection to evaluate the delivery and localization of Cy3-siRNA within U87 spheroids.
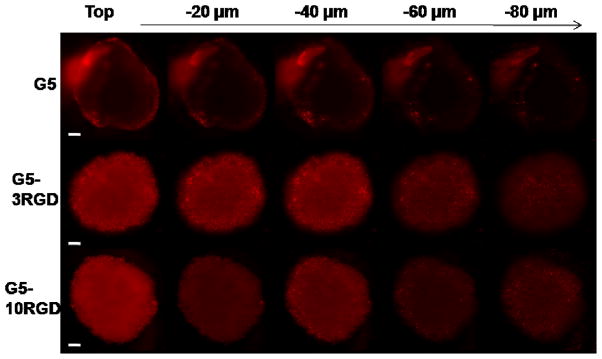
When delivered by G5 PAMAM dendrimers not containing RGD targeting ligands, siRNA fluorescence was observed primarily on the periphery of U87 spheroids (Figure 6). This result is consistent with previous studies on DNA delivery to MCTS using polyethylenimine (PEI), where poor tissue penetration using PEI/DNA complexes was observed (27). However, when PAMAM-RGD conjugates were used, siRNA was delivered successfully throughout the spheroid volume, suggesting that tissue penetration was enhanced by the presence of RGD targeting ligands (Figure 6). Significant siRNA delivery to the center of spheroids was observed when using PAMAM-RGD conjugates containing either 3 or 10 RGD ligands.
Figure 6.
Confocal images of U87 spheroids 24 hours after treatment with dendrimer/siRNA complexes. Cy3 siRNA fluorescence is shown in red. Z-stack images were obtained starting at the top of the spheroid in 20 μm intervals for a total of 100 μm into the spheroids. Images of each treatment were taken from three independent experiments with representative images shown here. The scale bar represents 100 μm.
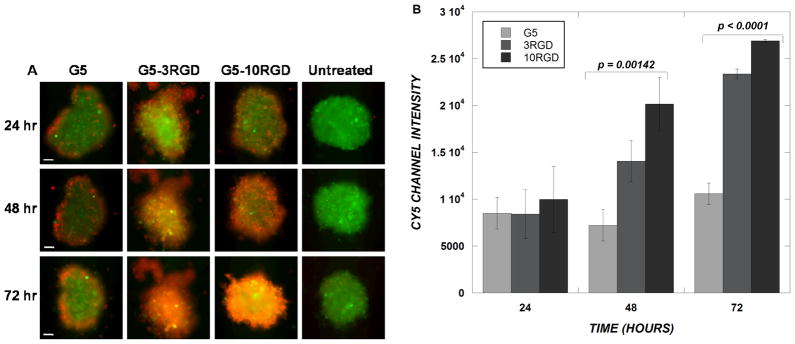
Having observed the ability of PAMAM-RGD conjugates to enhance siRNA delivery through U87 MCTS, spheroids generated from U87–d1EGFP cells were used to study simultaneously the delivery of a fluorescently labeled antisense oligonucleotide (AON) and its target GFP silencing. Consistent with the Cy3-siRNA localization observed in U87 spheroids, AON fluorescence was observed primarily on the spheroid periphery, at a depth of 80 μm into the MCTS, when delivered by unmodified G5 PAMAM (Figure 7A). When delivered by PAMAM-RGD conjugates, however, AON fluorescence was observed throughout the spheroid volume. A statistically significant increase in Cy5 AON fluorescence was observed in the interior of the spheroids when AON was delivered by dendrimers containing RGD peptides compared to the native dendrimers at both 48 (p<0.002) and 72 hours (p<0.0001) after treatment (Figure 7B). Despite the accumulation of AON within the MCTS, quantification of the GFP levels did not reveal significant gene silencing, consistent with the two-dimensional cell culture results (Figure 4B).
Figure 7.
(A) Confocal images of U87 GFP spheroids treated with dendrimer/Cy5-ODN complexes. GFP fluorescence of U87 GFP cells is shown in green, and Cy5 ODN fluorescence is shown in red. All images shown are from a depth of 80 μm into each spheroid. The time points shown indicate the time post-transfection. The scale bar represents 100 μm. (B) Quantified intensities of Cy5 fluorescence. Data represent mean channel intensities of five cross-sectional slices for each spheroid with three spheroids for each condition. Error bars represent mean ± standard deviation.
DISCUSSION
Integrins have proven to be promising targets for macromolecular drug and nucleic acid delivery systems for cancer. Several bioconjugate molecules containing RGD motifs have been successfully targeted to integrins to promote enhanced uptake and bioactivity in carcinoma cells. However, few studies have systematically evaluated how the number of RGD targeting ligands affects the bioactivity of targeted delivery systems. Hence, in this work, we conjugated cationic PAMAM dendrimers with various amounts of cyclic RGD targeting peptides to evaluate the ability of these bioconjugates to deliver siRNA to U87 glioma cells.
PAMAM dendrimers functionalized with varying levels of RGD peptides (2–10 RGDs/PAMAM) were able to deliver siRNA to U87 glioma cells, which express the αvβ3 integrin receptor, to elicit modest GFP silencing. However, contrary to our initial hypothesis, the attachment of RGD to PAMAM dendrimers did not enhance their ability to deliver siRNA to U87 glioma cells. This is likely due to the fact that non-specific uptake of PAMAM dendrimers, mediated by their high cationic charge density, was more considerable than integrin-mediated uptake mediated by the presence of RGD ligands. This result is consistent with other literature reports that have found poor active tumor cell targeting from cationic polymers such as PEI and PAMAM when a short crosslinker was used to conjugate a targeting peptide (5, 28, 29). However, in addition to enhancing uptake of macromolecules into integrin-expressing cells, RGD targeting strategies can serve other purposes, such as interfering with integrin-mediated tumor cell adhesion to ECM proteins (30). This in turn may affect delivery of cargo in a tissue as opposed to a two-dimensional cell culture.
The integrin αvβ3 is intimately involved in tumor cell growth, proliferation, and angiogenesis. Thus, the ability to interfere specifically with the activity of this integrin could help to slow tumorigenesis in malignant glioma, and indeed a cyclic RGD peptide, Cilengitide, is undergoing clinical trials for glioma therapy (31, 32). The PAMAM-RGD conjugates developed in this study inhibited the adhesion of U87 cells to the extracellular matrix protein, fibrinogen. A trend in inhibition of adhesion was observed as a function of RGD conjugation extent, with a maximum inhibition of cell adhesion observed with 3 RGD/PAMAM. Thus, a pronounced effect of RGD conjugation was observed in this cell adhesion model, contrasting the results obtained for siRNA delivery in which a trend was not observed as a function of RGD conjugation. Hence, the results of this assay support the ability of the PAMAM-RGD conjugates to perform active integrin binding.
MCTS models for cancer are garnering significant attention for anticancer drug screening as they mimic the microenvironment of tumors found in vivo more accurately than standard two-dimensional culture models (33, 34), and they have recently been shown to display angiogenic phenotypes akin to those observed in vivo (35). Previous studies using spheroid cell culture models to evaluate nucleic acid delivery by synthetic polymers such as PEI or lipids have found that gene delivery using these carriers was confined to the outer layers of cells in spheroids due to poor penetration through the cell-matrix network (27). However, one study reported that conjugating lactose targeting moieties onto a polymeric micelle delivery system enabled deeper penetration of siRNA into a spheroid model of human hepatocarcinoma (36). Further, the presence of ECM proteins was found to inhibit the transport of nanoparticles through tumor spheroids (37). Since PAMAM-RGD conjugates were found to interfere with cell adhesion mediated by ECM, we hypothesized that using PAMAM-RGD conjugates would facilitate deeper penetration into a spheroid model of malignant glioma than unmodified PAMAM dendrimers via RGD modulation of cell-ECM interactions present in three-dimensional culture. As expected, PAMAM dendrimers conjugated with either 3 or 10 RGD peptides significantly improved the PAMAM-mediated delivery of siRNA to the spheroid interior compared to the native G5 dendrimer, suggesting that the addition of RGD peptides was able to overcome the poor tissue penetration of PAMAM dendrimers through spheroids. Notably, the PAMAM-RGD conjugates, in contrast to the G5 dendrimer, continued to penetrate into the MCTS throughout the 72 hour time course of the experiment, suggesting a dynamic interaction among bioconjugate, ECM and cell. However, despite the significant increases in tumor penetration, silencing levels of GFP remained insignificant. This outcome highlights the fact that cellular accumulation is not sufficient for efficient gene silencing. The siRNA must also be internalized by the cell and trafficked efficiently intracellularly, particularly with regards to endosomal escape. Modification of the dendrimers to enhance these steps is necessary for ultimate application to tumor oncogene silencing.
A major goal of this work was to study how the number of RGD peptides on a PAMAM dendrimer affects its ability to interfere with important cell-ECM interactions. Previous studies have found that controlling RGD ligand presentation from a nanoparticle platform can significantly impact the extent of integrin-targeting that is achieved (38, 39). Here, three RGDs per dendrimer were found to exhibit the strongest integrin-binding response using a competitive binding cell adhesion assay, and a greater number of peptides per dendrimer did not provide any additional benefit. However, in contrast to the trend observed in the competitive binding cell adhesion assay, dendrimers displaying 10 RGD peptides yielded significantly greater delivery of oligonucleotides to U87 spheroids than did dendrimers containing 3 RGD peptides. The three-dimensional architecture of the spheroids likely confers a greater advantage to dendrimers with a greater RGD multivalency, allowing for multiple contacts with ECM molecules within the MCTS. The results of this study highlight the importance of screening novel chemical entities and drug delivery systems in three dimensions.
Taken together, these results suggest that RGD-conjugated dendrimers hold promise for siRNA delivery to solid tumors. Their ability to interfere with integrin-ECM interactions can potentially enable penetration into malignant tumors to improve siRNA delivery, and they may also interfere directly with tumor angiogenesis and integrin-mediated growth signaling by malignant cells. Such multifunctional delivery systems that can modify the biological processes of cancer cells and deliver a therapeutic cargo in concert represent a new venue for cancer therapy. Due to the architecture of dendrimers with many exterior functional groups, the concept can be extended easily to incorporation of conventional drugs and/or imaging agents. Such developments will amplify the impact of siRNA on human disease.
Supplementary Material
Acknowledgments
We are grateful for financial support from the NIH (R01 GM65913), Charles & Johanna Busch Memorial Fund, and an NSF IGERT Fellowship (DGE-0504497) to CW. The authors thank the following people for access to and assistance with various instruments: Dr. Sobin Kim and Daniel Duffield (MALDI-ToF MS), Dr. Rene Schloss and Jeffrey Barminko (confocal microscope), and Dr. Kathryn Uhrich and Sarah Sparks (dynamic light scattering). We also thank Dr. Kathryn Uhrich, Dr. Prabhas Moghe, and Lavanya Peddada for helpful discussions and Paul Gianella for technical assistance.
Footnotes
Supporting Information Available: MALDI-ToF mass spectra of native G5 PAMAM and PAMAM-RGD conjugates are provided. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gartel AL, Kandel ES. RNA interference in cancer. Biomol Eng. 2006;23:17–34. doi: 10.1016/j.bioeng.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–96. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S, Zhao B, Jiang H, Wang B, Ma B. Cationic lipids and polymers mediated vectors for delivery of siRNA. J Control Release. 2007;123:1–10. doi: 10.1016/j.jconrel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 5.Kang H, DeLong R, Fisher MH, Juliano RL. Tat-conjugated PAMAM dendrimers as delivery agents for antisense and siRNA oligonucleotides. Pharm Res. 2005;22:2099–106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- 6.Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjug Chem. 2008;19:1396–403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- 7.Tsutsumi T, Hirayama F, Uekama K, Arima H. Evaluation of polyamidoamine dendrimer/alpha-cyclodextrin conjugate (generation 3, G3) as a novel carrier for small interfering RNA (siRNA) J Control Release. 2007;119:349–59. doi: 10.1016/j.jconrel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem Commun (Camb) 2006:2362–4. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 9.Patil ML, Zhang M, Taratula O, Garbuzenko OB, He H, Minko T. Internally cationic polyamidoamine PAMAM-OH dendrimers for siRNA delivery: effect of the degree of quaternization and cancer targeting. Biomacromolecules. 2009;10:258–66. doi: 10.1021/bm8009973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waite CL, Sparks SM, Uhrich KE, Roth CM. Acetylation of PAMAM dendrimers for cellular delivery of siRNA. BMC Biotechnol. 2009;9:38. doi: 10.1186/1472-6750-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14:107–15. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Wolfenden ML, Cloninger MJ. Mannose/glucose-functionalized dendrimers to investigate the predictable tunability of multivalent interactions. J Am Chem Soc. 2005;127:12168–9. doi: 10.1021/ja053008n. [DOI] [PubMed] [Google Scholar]
- 13.Woller EK, Walter ED, Morgan JR, Singel DJ, Cloninger MJ. Altering the strength of lectin binding interactions and controlling the amount of lectin clustering using mannose/hydroxyl-functionalized dendrimers. J Am Chem Soc. 2003;125:8820–6. doi: 10.1021/ja0352496. [DOI] [PubMed] [Google Scholar]
- 14.Cai W, Chen X. Anti-angiogenic cancer therapy based on integrin alphavbeta3 antagonism. Anticancer Agents Med Chem. 2006;6:407–28. doi: 10.2174/187152006778226530. [DOI] [PubMed] [Google Scholar]
- 15.Dunehoo AL, Anderson M, Majumdar S, Kobayashi N, Berkland C, Siahaan TJ. Cell adhesion molecules for targeted drug delivery. J Pharm Sci. 2006;95:1856–72. doi: 10.1002/jps.20676. [DOI] [PubMed] [Google Scholar]
- 16.Boswell CA, Eck PK, Regino CA, Bernardo M, Wong KJ, Milenic DE, Choyke PL, Brechbiel MW. Synthesis, characterization, and biological evaluation of integrin alphavbeta3-targeted PAMAM dendrimers. Mol Pharm. 2008;5:527–39. doi: 10.1021/mp800022a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, Baker JR., Jr Tumor angiogenic vasculature targeting with PAMAM dendrimer-RGD conjugates. Chem Commun (Camb) 2005:5739–41. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]
- 18.Hill E, Shukla R, Park SS, Baker JR., Jr Synthetic PAMAM-RGD conjugates target and bind to odontoblast-like MDPC 23 cells and the predentin in tooth organ cultures. Bioconjug Chem. 2007;18:1756–62. doi: 10.1021/bc0700234. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Kao WJ. Synthesis and characterization of nanoscale dendritic RGD clusters for potential applications in tissue engineering and drug delivery. Int J Nanomedicine. 2007;2:89–99. doi: 10.2147/nano.2007.2.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee LK, Dunham BM, Li Z, Roth CM. Cellular Dynamics of Antisense Oligonucleotides and Short Interfering RNAs. Annals of the New York Academy of Sciences. 2006;1082:47–51. doi: 10.1196/annals.1348.061. [DOI] [PubMed] [Google Scholar]
- 21.Del Duca D, Werbowetski T, Del Maestro RF. Spheroid preparation from hanging drops: characterization of a model of brain tumor invasion. J Neurooncol. 2004;67:295–303. doi: 10.1023/b:neon.0000024220.07063.70. [DOI] [PubMed] [Google Scholar]
- 22.Rasband WS. National Institues of Health; Bethesda, MD, USA: 1997–2009. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 23.Muller R, Laschober C, Szymanski WW, Allmaier G. Determination of molecular weight, particle size, and density of high number generation PAMAM dendrimers using MALDI-TOF-MS and nES-GEMMA. Macromolecules. 2007;40:5599–5605. [Google Scholar]
- 24.Shi XY, Lesniak W, Islam MT, Muniz MC, Balogh LP, Baker JR. Comprehensive characterization of surface-functionalized poly (amidoamine) dendrimers with acetamide, hydroxyl, and carboxyl groups. Colloids and Surfaces a-Physicochemical and Engineering Aspects. 2006;272:139–150. [Google Scholar]
- 25.Marinelli L, Lavecchia A, Gottschalk KE, Novellino E, Kessler H. Docking studies on alphavbeta3 integrin ligands: pharmacophore refinement and implications for drug design. J Med Chem. 2003;46:4393–404. doi: 10.1021/jm020577m. [DOI] [PubMed] [Google Scholar]
- 26.Sutherland RM. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science. 1988;240:177–84. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 27.Mellor HR, Davies LA, Caspar H, Pringle CR, Hyde SC, Gill DR, Callaghan R. Optimising non-viral gene delivery in a tumour spheroid model. J Gene Med. 2006;8:1160–70. doi: 10.1002/jgm.947. [DOI] [PubMed] [Google Scholar]
- 28.Sun YX, Zeng X, Meng QF, Zhang XZ, Cheng SX, Zhuo RX. The influence of RGD addition on the gene transfer characteristics of disulfide-containing polyethyleneimine/DNA complexes. Biomaterials. 2008;29:4356–65. doi: 10.1016/j.biomaterials.2008.07.045. [DOI] [PubMed] [Google Scholar]
- 29.Clements BA, Bai J, Kucharski C, Farrell LL, Lavasanifar A, Ritchie B, Ghahary A, Uludag H. RGD conjugation to polyethyleneimine does not improve DNA delivery to bone marrow stromal cells. Biomacromolecules. 2006;7:1481–8. doi: 10.1021/bm060073w. [DOI] [PubMed] [Google Scholar]
- 30.Mitra A, Mulholland J, Nan A, McNeill E, Ghandehari H, Line BR. Targeting tumor angiogenic vasculature using polymer-RGD conjugates. J Control Release. 2005;102:191–201. doi: 10.1016/j.jconrel.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Eskens FA, Dumez H, Hoekstra R, Perschl A, Brindley C, Bottcher S, Wynendaele W, Drevs J, Verweij J, van Oosterom AT. Phase I and pharmacokinetic study of continuous twice weekly intravenous administration of Cilengitide (EMD 121974), a novel inhibitor of the integrins alphavbeta3 and alphavbeta5 in patients with advanced solid tumours. Eur J Cancer. 2003;39:917–26. doi: 10.1016/s0959-8049(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 32.Nabors LB, Mikkelsen T, Rosenfeld SS, Hochberg F, Akella NS, Fisher JD, Cloud GA, Zhang Y, Carson K, Wittemer SM, Colevas AD, Grossman SA. Phase I and correlative biology study of cilengitide in patients with recurrent malignant glioma. J Clin Oncol. 2007;25:1651–7. doi: 10.1200/JCO.2006.06.6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4:309–24. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 34.Goodman TT, Ng CP, Pun SH. 3-D tissue culture systems for the evaluation and optimization of nanoparticle-based drug carriers. Bioconjug Chem. 2008;19:1951–9. doi: 10.1021/bc800233a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischbach C, Kong HJ, Hsiong SX, Evangelista MB, Yuen W, Mooney DJ. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc Natl Acad Sci U S A. 2009;106:399–404. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oishi M, Nagasaki Y, Nishiyama N, Itaka K, Takagi M, Shimamoto A, Furuichi Y, Kataoka K. Enhanced growth inhibition of hepatic multicellular tumor spheroids by lactosylated poly(ethylene glycol)-siRNA conjugate formulated in PEGylated polyplexes. Chem Med Chem. 2007;2:1290–7. doi: 10.1002/cmdc.200700076. [DOI] [PubMed] [Google Scholar]
- 37.Goodman TT, Chen J, Matveev K, Pun SH. Spatio-temporal modeling of nanoparticle delivery to multicellular tumor spheroids. Biotechnol Bioeng. 2008;101:388–99. doi: 10.1002/bit.21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. Multivalent effects of RGD peptides obtained by nanoparticle display. J Med Chem. 2006;49:6087–93. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 39.Ng QK, Sutton MK, Soonsawad P, Xing L, Cheng H, Segura T. Engineering clustered ligand binding into nonviral vectors: alphavbeta3 targeting as an example. Mol Ther. 2009;17:828–36. doi: 10.1038/mt.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.